Extraocular, rod-like photoreceptors in a flatworm express xenopsin photopigment
Figures
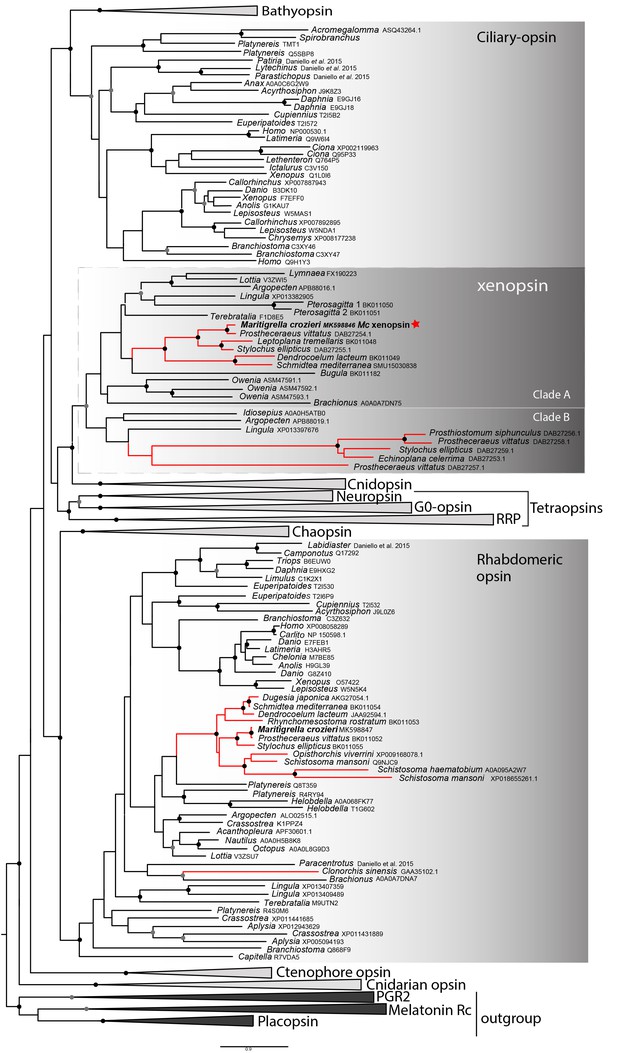
Phylogenetic analysis of metazoan opsins supports flatworm ciliary-like opsins as xenopsins and confirms a clade of flatworm rhabdomeric opsins.
Support for nodes is calculated using 1000 Ultrafast bootstrap replications as well as 1000 SH-aLRT replicates and approximate aBayes single Branch testing. Black dots indicate nodes with support values for three tests ≥ 95% (0.95 for SH-aLRT replicates). Gray dots indicate nodes with support values for three tests ≥ 90% (0.90 for SH-aLRT replicates). Scale bar unit for branch length is the number of substitutions per site. Branches in red correspond to flatworm opsin sequences. See Figure 1—figure supplement 1 for uncollapsed tree and Figure 1—source data 1 for gene accession numbers. The new xenopsin sequences we found in polyclad and triclad flatworms, plus a bryozoan and chaetognath, all fall within clade A of the xenopsins.
-
Figure 1—source data 1
Sequence data for opsins used in phylogenetic analyses for Figure 1.
- https://doi.org/10.7554/eLife.45465.007
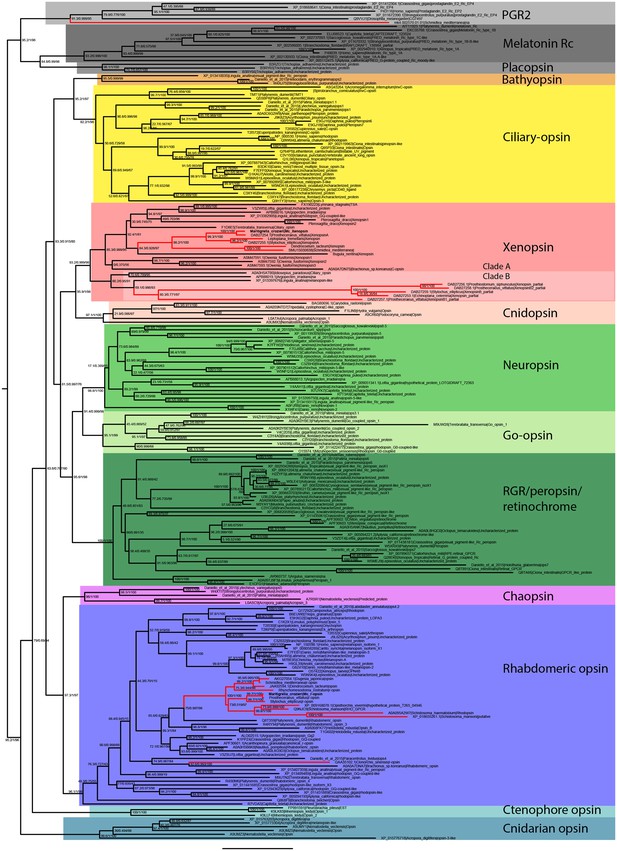
Uncollapsed tree of IQ-TREE phylogenetic reconstruction of opsin relationships.
Node support values correspond to 1000 Ultrafast bootstrap replications, 1000 SH-aLRT replicates and approximate aBayes single Branch testing. Scale bar unit for branch length is the number of substitutions per site. Note low support for all deeper nodes including those uniting xenopsin/cnidopsin with tetraopsins, and the sister relationship between this clade and the c-opsins/bathyopsins. Branches in red correspond to flatworm opsin sequences.
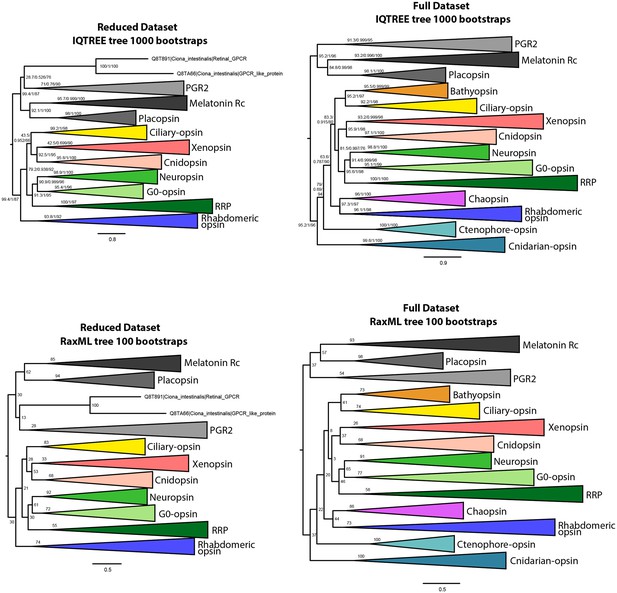
IQtree and RaxML trees showing the influence of the small opsin clades (i.e. chaopsins, bathyopsins, ctenophore and anthozoan opsins) on the position of xenopsins in relation to c-opsins and tetra-opsins (Neuropsin, Go-opsin and RRP); inclusion of these small opsin clades brings xenopsins close to tetraopsins (full dataset), their exclusion brings xenopsins close to c-opsins (reduced dataset).
https://doi.org/10.7554/eLife.45465.005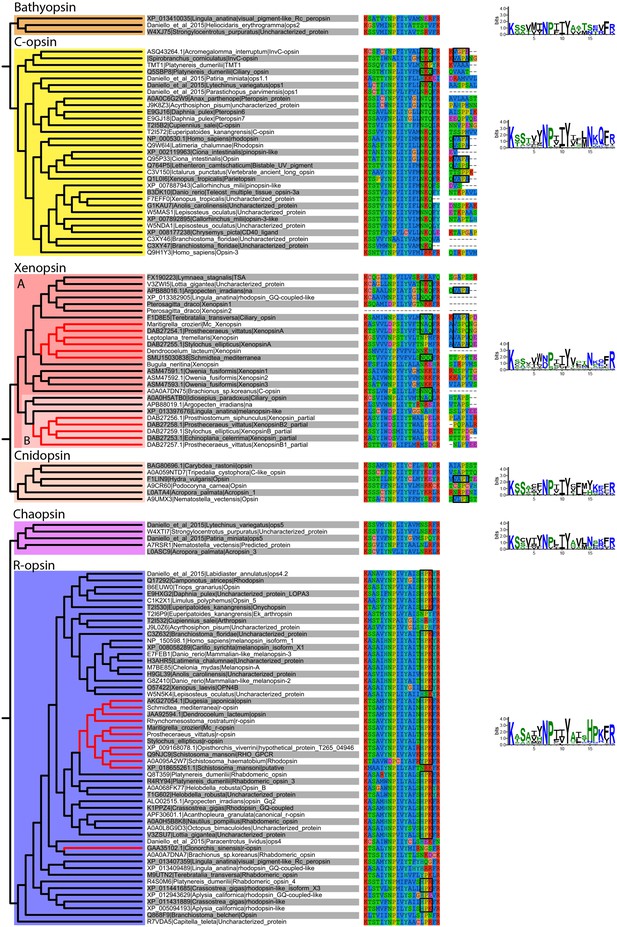
Alignment of major opsin clades showing conserved lysine in transmembrane domain VII, which binds to the retinal chromophore to form a photopigment.
Some xenopsins possess a tripeptide motif, NxQ, which is also found in ciliary opsins and known to be crucial for G-protein activation. A number of flatworm xenopsin sequences in clade A have similar NxQ patterns (including Maritigrella - NAQ), while the motif differs considerably in the polyclad xenopsins of clade B, cnidopsins, bathyopsins and tetraopsins. An alignment of the C-terminal regions of ciliary opsins, xenopsins, cnidopsins, tetraopsins and bathyopsins shows, at a conserved position, similar VxPx motifs in flatworm clade A xenopsins (including Mc xenopsin - VSPQ) as well as a mollusk (A. irradiens) and brachiopod (T. transversa) xenopsin, it is also present in ciliary opsins from non-vertebrate chordates (tunicate and lamprey) and annelids, as well as in cnidopsin sequences. This motif binds the small GTPase Arf4 to direct vertebrate rhodopsin (a ciliary opsin) to the primary cilia. The presence of this motif in some ciliary opsins, xenopsins, cnidopsins may suggest a shared mechanism for the active delivery of these opsins to the cilia in CPRs.
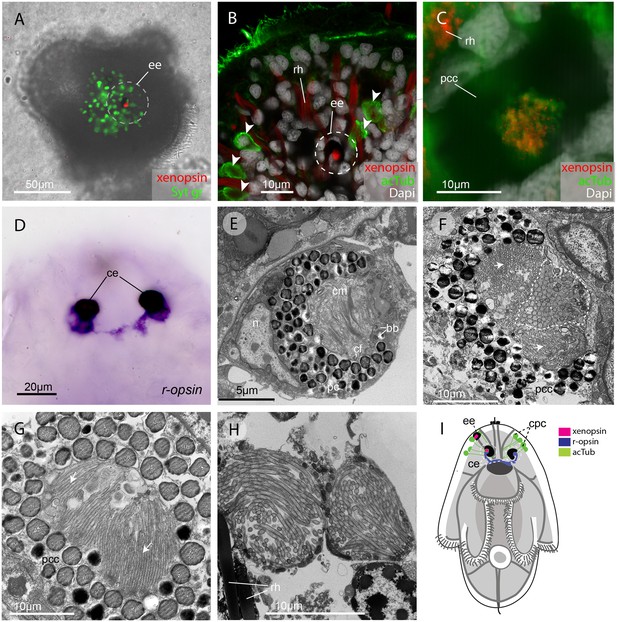
In the larva (1 day post-hatching) xenopsin protein is localized to ciliary cells in the eyes.
(A) Apical view of larva showing xenopsin (red) in the epidermal eye (ee) (100% of individuals examined, n = 50) (OpenSPIM image, Syt gr = Sytox green, staining nuclei and bright-field image also reveal photoreceptor pigments). (B) Confocal optical section showing xenopsin in the epidermal eye (circled) but not in the acetylated tubulin+ (acTub) cells in the epidermis (arrowheads); autofluoresence of rhabdites (rh). (C) Xenopsin is co-localised with acTub in one of the two cerebral eyes providing evidence that xenopsin protein localizes to cilia (this varies between the right and left cerebral eye in different larvae, 50:50, n = 10)(pcc = pigment cup cell). (D) R-opsin is expressed in both cerebral eyes (ce). (100% of individuals examined, n = 30), (E) TEM image showing the epidermal eye which houses elaborated ciliary membranes (cm) inside a pigment cup (pc)(basal bodies, bb; cross section of ciliary flagella, cf, nucleus,n) (100% of individuals examined, n = 3). (F) Ultrastructure of a cerebral eye showing cilia (inside dashed line) and microvilli (arrows) cupped within a pigment cell (pcc). (G) Ultrastructure of another cerebral eye showing microvilli (arrows) cupped inside a pigment cup cell. (H) Multiple cilia projecting into phaosomes (intra-cellular vacuoles) in the epidermis. (I) A schematic of a larva summarizing the expression of xenopsin, r-opsin and acTub. In the larva xenopsin is expressed in two of the three putative ciliary photoreceptor cell types: the epidermal eye, a cerebral eye, but not in the ciliary phaosome cells (cpc).
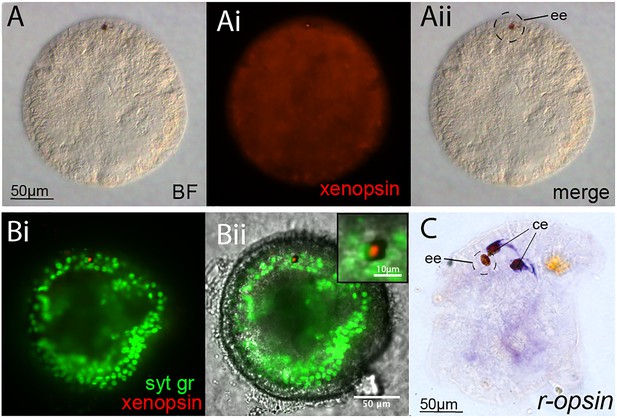
Opsin localization and expression in Maritigrella embryos and larva.
(A and B) Xenopsin expression (red) in the epidermal eye (ee) during Maritigrella crozieri embryogenesis. The epidermal eye develops soon after gastrulation is complete and before development of the cerebral eyes. Syt gr = Sytox Green, staining nuclei. Bright-field (BF) also shows the photoreceptor pigments. Inset is a 3-time magnification. (C) R-opsin expression is expressed in the two cerebral eyes (ce) of the larva but not the epidermal eye.
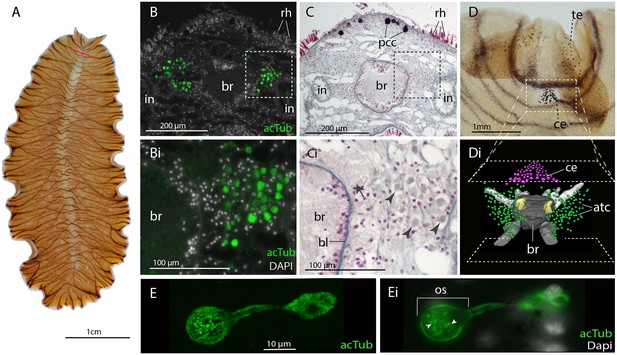
Acetylated tubulin staining identifies two dense clusters of extraocular cells, possible ciliary photoreceptors (CPR), either side of the adult brain.
(A) Live adult, red line shows plane of cross section in B-C. (B and C) Consecutive sections showing; (B) two clusters of acetylated tubulin+ cells and; (C) their distribution between the brain (br) (which is encapsulated in a basal lamina, bl) and intestinal branches (in) (n = 5 individuals). Bi and Ci) Close up showing that these cells (arrowheads) are embedded in extracellular matrix in close proximity to the main nerve tracts (arrow). Pigment cup cells (pcc), rhabdites (rh). (D) Anterior end of adult showing pigmented eyes above the brain (cerebral eyes, ce) and on the tentacles (tentacular eyes, te). (Di) Schematised distribution of acetylated tubulin+ cells (act) and cerebral eyes on a micro-CT reconstruction of the brain and main anterior (white), posterior (gray), and two of the optic (yellow) nerve tracts. (E) Confocal projection of a putative CPR and, Ei) an optical slice of the same cell showing cilia projecting into the intra-cellular vacuole or phaosome (arrowheads) in the outer segment (os).
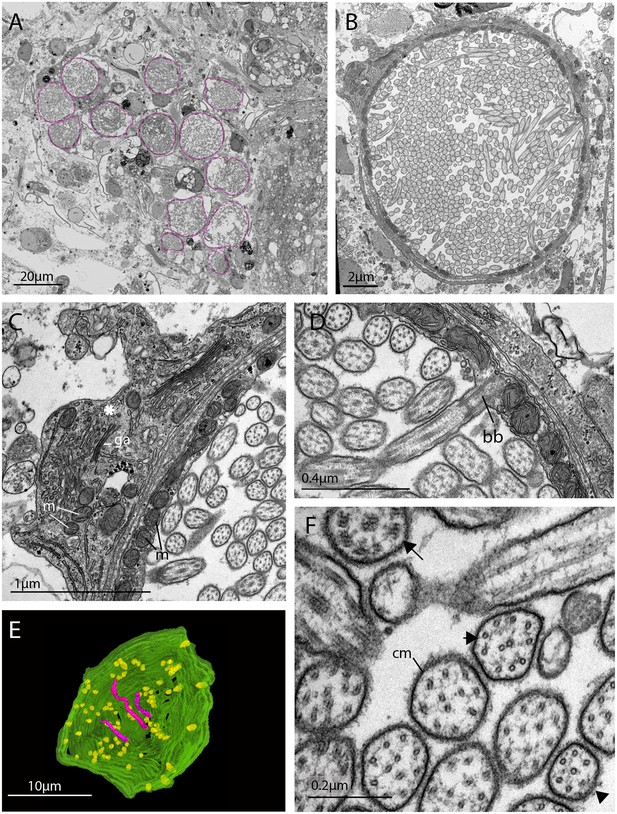
The morphology of the ciliary phaosomal cells in adult Maritigrella crozieri.
(A) A dense cluster of intra-cellular vacuoles (phaosomes – highlighted in magenta) filled with cilia (n = 3 individuals). (B) Multiple cilia in the phaosome. (C) A possible unpigmented supporting cell (asterisk) wrapping around the phaosome cell with mitochondria (m) and Golgi apparatus (ga) in the cytoplasm. (D) Ciliary axonemata (ax) are anchored in the cytoplasmic layer (cl) by basal bodies (bb). (E) 3D reconstruction of the interior of a third of a phaosome, showing that the cilia are unbranched (pink) and the basal bodies (yellow) are distributed all around the phaosome. (F) Cross sections of the ciliary axonemata show various arrangements of microtubules: 9 × 2+ two with dynein arms attached to the A-tubules (arrow), 9 + 2 singlets (double arrowheads), and singlets (triple arrowheads). This variation is related to the distance from the basal body (Figure 4—videos 3), (cm) ciliary membrane.
Serial SEM images (101 × 500 nm sections = 50.5 μm total thickness) showing a cluster of ciliary phaosomes (intra-cellular vacuoles housing multiple cilia) that form the outer segment of the putative extraocular CPR cells in adult Maritigrella.
This cluster is located to the left of the brain.
Serial SEM images (47 × 400 nm sections = 18.8 μm total thickness) showing a complete phaosome.
At least 421 basal bodies were counted projecting cilia into the intra-cellular space that is formed by the cell itself. Note that in a nearby phaosome, the cilia appear to be arranged in a spirally coiled bundle.
Serial SEM images (72 x 100nm sections = 7.2μm total thickness) showing a third of a phaosome.
https://doi.org/10.7554/eLife.45465.014A 3D reconstruction of series in Figure 4—video 3 shows that the cilia (pink) are unbranched and basal bodies (yellow) emerge all around the diameter of the cavity.
https://doi.org/10.7554/eLife.45465.015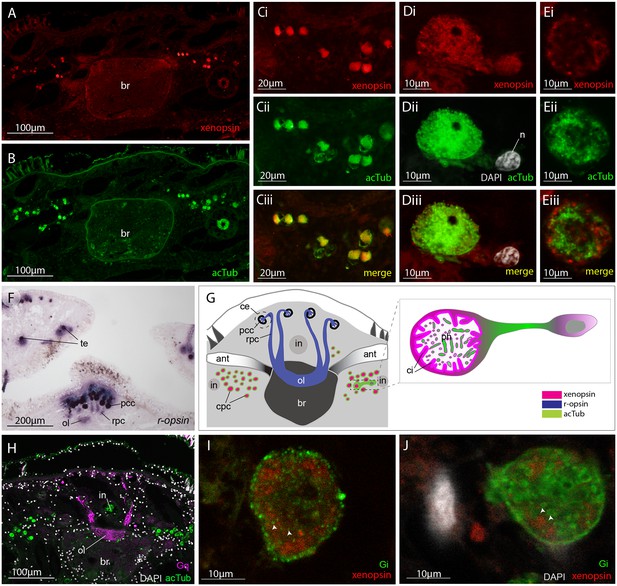
Xenopsin is co-localised with acetylated tubulin and Gαi in extraocular ciliary phaosome cells and r-opsin is expressed with Gαq in eyes.
(A-E) Co-localization of acetylated tubulin and xenopsin is found throughout the cell (n, nucleus) (100% of acTub+ cells express xenopsin (in one cross section, n = 3 individual worms). (E) A cross section of a phaosome shows stronger xenopsin expression near the base of the cilia (n = 8 individuals); (F) r-opsin is expressed in photoreceptor cells (rpc) that extend from the pigment cup cells (pcc) to the optic lobe (ol) of the brain, together forming the cerebral eyes (ce); r-opsin is also expressed in the tentacular eyes (te) (n = 5 individuals); (G) Schematic of xenopsin and r-opsin expression in adult cross-section, with a diagram of a putative ciliary photoreceptor cell (ciliary phaosome cell, cpc) showing co-localization of xenopsin and acetylated tubulin. (H) The position of the putative ciliary photoreceptors (labeled with acetylated tubulin) in relation to the rhabdomeric photoreceptors (labeled with Gαq) (n = 3 individuals). (I and J) Gαi expression in the xenopsin+ cells including on the ciliary membranes (arrowheads).
-
Figure 5—source data 1
Sequence data for G alpha subunits used in phylogenetic analyses for Figure 5.
- https://doi.org/10.7554/eLife.45465.018
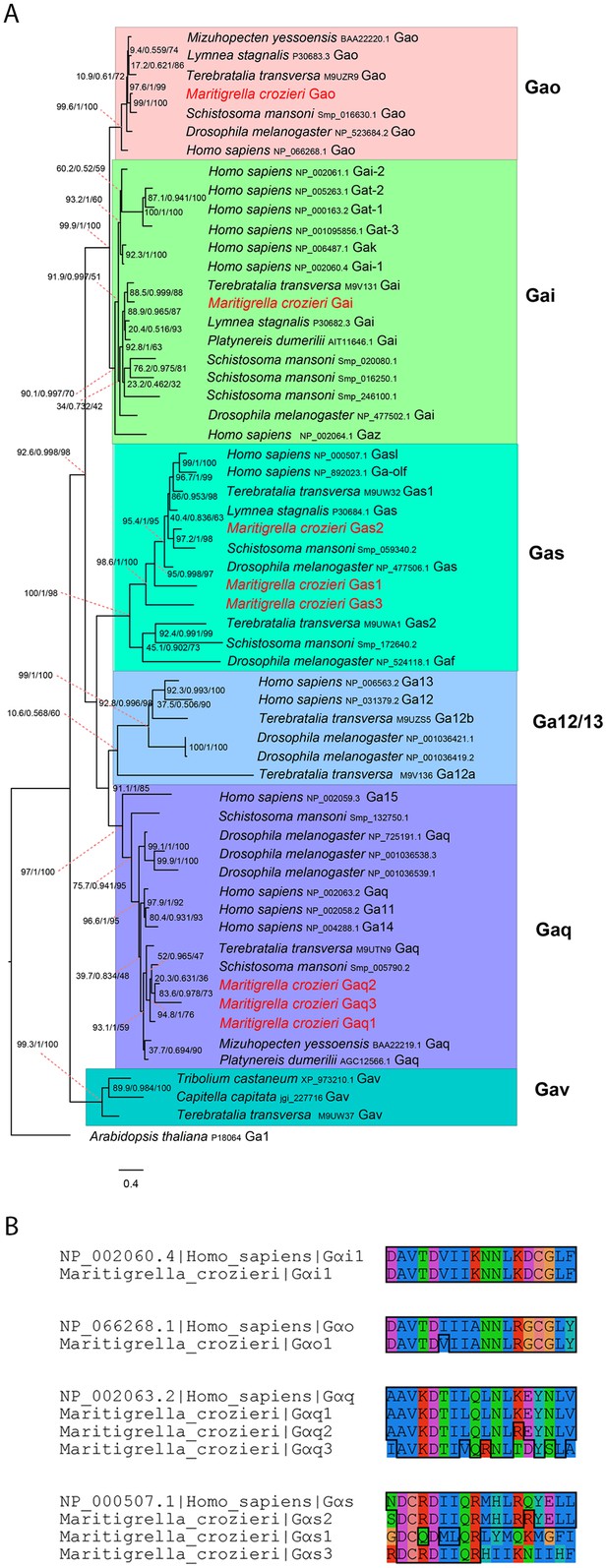
G alpha subunit phylogeny and C terminal end alignment.
(A) IQ-TREE phylogenetic reconstruction of G alpha subunit relationships identifies those expressed in the Maritigrella crozieri larval transcriptome (red). Support for node is calculated using 1000 Ultrafast bootstrap replications as well as 1000 SH-aLRT replicates and approximate aBayes single branch testing. Scale bar unit for branch length is the number of substitutions per site. (B) Alignment of c terminal ends of Human and Maritigrella crozieri G alpha subunits. See Figure 1 additional data file for full amino acid sequences from all taxa.
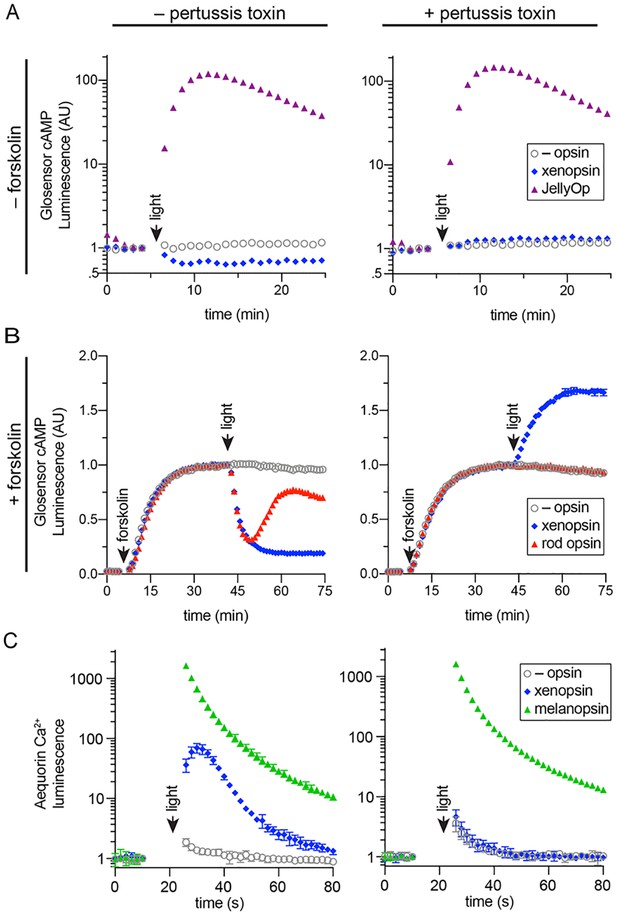
In human cells Maritigrella crozieri xenopsin forms a functional photopigment that predominantly couples to Gαi pathways.
(A,B) HEK293 cells were transfected with Glo22F and indicated opsins, + /- pertussis toxin, and exposed to light. In B, cells were treated with 2 µM forskolin prior to the light flash. (C) HEK293 cells were transfected with mtAequorin and the opsins indicated, + /- pertussis toxin, and exposed to light. Plots show mean luminescence of technical replicates (from one representative of three biological replicates) normalized to the pre-flash timepoint, + /- SEM. Error bars smaller than symbols are not shown. n = 3 technical replicates in A,B; n = 4 technical replicates in C. The other biological replicates are shown in Figure 6—figure supplement 2.
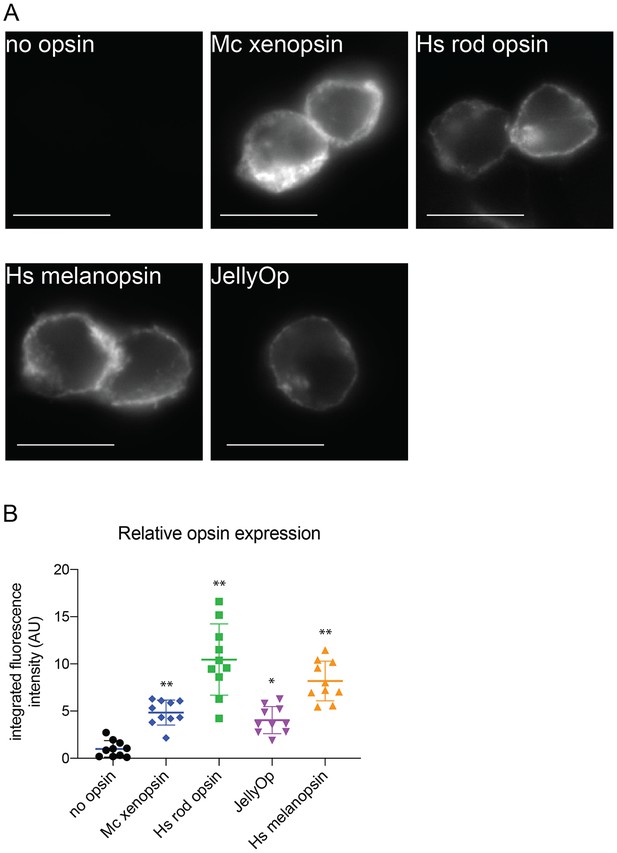
Immunofluorescence to quantify opsin expression in HEK293 cells.
Cells were transfected with opsins, fixed, stained with 1D4 anti-rod opsin antibody and fluorescent secondary antibody, and imaged on a fluorescent microscope. (A) Representative images of cells expressing each opsin and control (scale = 10 µm). (B) Quantification of integrated fluorescence intensity (minus background) for ten randomly selected fields from each sample, normalized to mean of no-opsin control, with mean and SEM. ANOVA and Dunnett’s multiple comparison test show significant differences between no opsin control and positive opsin conditions (*<0.05*, *<0.01). Mc xenopsin (Maritigrella crozieri xenopsin), Hs rod opsin (human rhodopsin), Hs melanopsin (Human melanopsin), JellyOp (Carybdea rastonii jellyfish opsin), SEM (standard error of the mean).
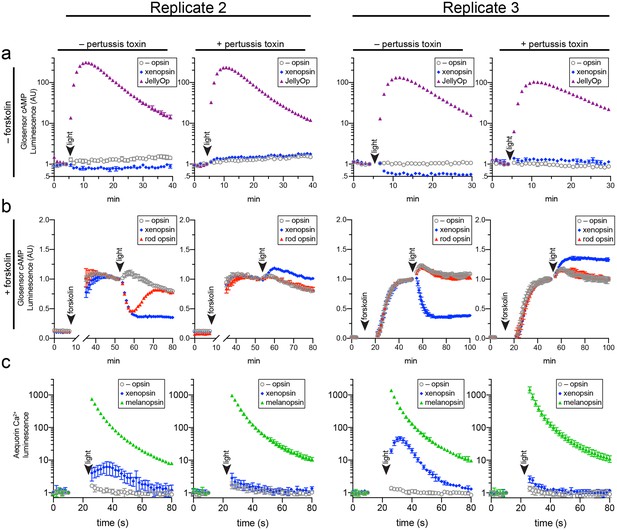
Two further biological replicates of the secondary messenger assays show there was quantitative variation from day to day in the magnitude of responses to light and forskolin, but the qualitative response of each opsin was consistent (excluding one replicate in which rod opsin showed no activity, possibly due to a faulty preparation of plasmid DNA).
https://doi.org/10.7554/eLife.45465.021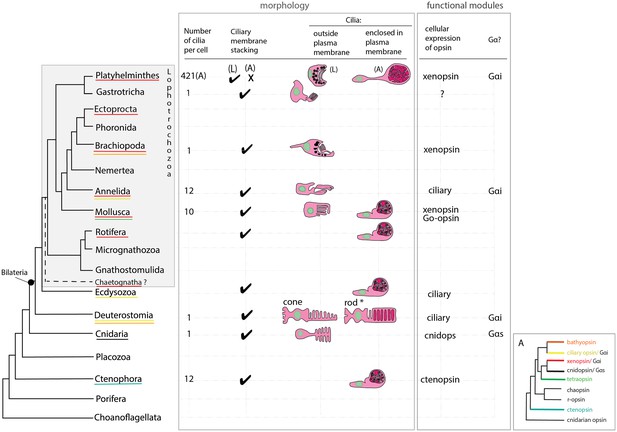
An overview of metazoan ciliary photoreceptor morphology, opsin expression and Gα-protein coupling (determined from cellular assays), highlighting the distinct morphology of the ciliary phaosomes in flatworms and possible convergent evolution of enclosed ciliary membranes in invertebrate phaosomes and jawed vertebrate rods (*).
The colored lines under the phylum names represent the presence of the opsin sub-type in the taxonomic group; note the higher prevalence of xenopsins over ciliary opsins in Lophotrochozoa. (L) = larval photoreceptor, (A) = adult photoreceptor. Box A shows the opsin relationships according to our phylogeny and the known Gα-binding of opsins expressed in ciliary photoreceptors.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | anti-acetylated tubulin (monoclonal, mouse) | Sigma | T7451 RRID:AB_609894 | IF(1:500) |
| Antibody | Gα q/11 (C-19) (polyclonal, rabbit) | Santa Cruz Biotech | sc-392 RRID:AB_631537 | IF(1:300) |
| Antibody | Gαi-1 (R4) (monoclonal, mouse) | Santa Cruz Biotech | sc-13533 RRID:AB_2111358 | IF(1:300) |
| Antibody | Gαi-1/2/3 (35) (monoclonal, mouse) | Santa Cruz Biotech | sc-136478 RRID:AB_2722559 | IF(1:300) |
| Antibody | Gαo (A2) (monoclonal, mouse) | Santa Cruz Biotech | sc-13532 RRID:AB_2111645 | IF(1:300) |
| Antibody | Gαs/olf(A-5) (monoclonal, mouse) | Santa Cruz Biotech | sc-55545 RRID:AB_831819 | IF(1:300) |
| Antibody | Gαs/olf(C-18) (polyclonal, rabbit) | Santa Cruz Biotech | sc-383 RRID:AB_631539 | IF(1:300) |
| Antibody | Gα12 (E-12) (monoclonal, mouse) | Santa Cruz Biotech | sc-515445 | IF(1:300) |
| Cell line (Homo-sapiens) | Embryonic kidney cells | ATCC | CRL3216 RRID:CVCL_0063 | |
| Recombinant DNA reagent | pGlosensor 22 | Promega | E1290 | Live-Cell Biosensors |
| Recombinant DNA reagent | pcDNA3.1 | Invitrogen | V79020 | Mammalian Expression Vector |
| Recombinant DNA reagent | pcDNAFRT⁄TO vector | Thermo | V652020 | Expression vector |
| Chemical compound | 9-cis retinal | Sigma | R5754 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.45465.023





