Dlg1 activates beta-catenin signaling to regulate retinal angiogenesis and the blood-retina and blood-brain barriers
Figures
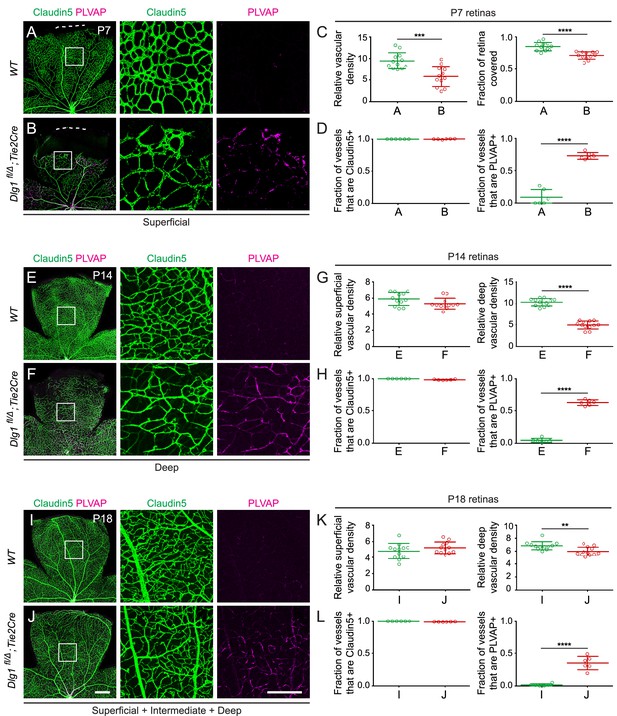
Dlg1 promotes angiogenesis in the mammalian retina.
(A–B) The superficial vascular plexus of P7 retinas from the indicated genotypes (column 1), with boxed regions shown at higher magnification (columns 2 and 3). Dashed white lines indicate the edge of the retina. (C) Quantification of vascular density (left) and vascular coverage (right) in P7 retinas, for the genotypes in (A) and (B). Quantification methodology in this and other figures is described in the Materials and methods section. In this and all other figures, bars represent mean ± SD. Statistical significance, determined by the unpaired t-test, is represented by * (p<0.05), ** (p<0.01), *** (p<0.001), and **** (p<0.0001). (D) Quantification of the fraction of vessel length that is Claudin5+ (left) and PLVAP+ (right) in P7 retinas, for the genotypes in (A) and (B). (E–F) The deep vascular plexus of P14 retinas (column 1) with boxed regions shown at higher magnification (columns 2 and 3). (G) Quantification of vascular density in the superficial plexus (left) and deep plexus (right) in P14 retinas for the genotypes in (E) and (F). (H) As in (D), except with P14 retinas. (I–J) Maximum projection of superficial, intermediate, and deep vascular plexuses of P18 retinas (column 1) with boxed regions shown at higher magnification (columns 2 and 3). (K) As in (G), except with P18 retinas. (L) As in (D), except with P18 retinas. Scale bar for column 1, 400 μm. Scale bar for columns 2 and 3, 200 μm.
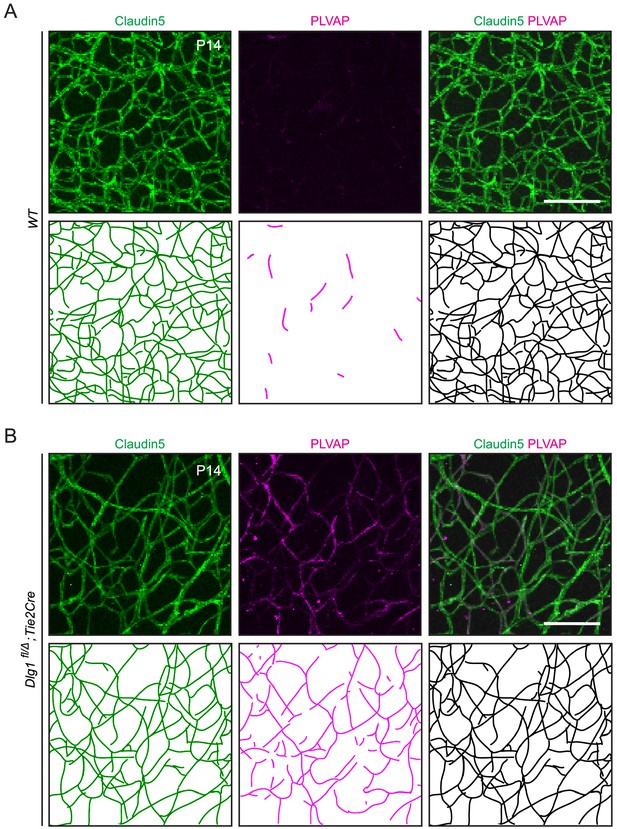
Quantification of Claudin5 and PLVAP accumulation in retinal ECs.
(A,B) Maximum projection of the superficial, intermediate, and deep vascular plexuses of P14 retinas (top) with representative traces of the vasculature for quantification (bottom), as described in Materials and methods. Scale bar, 100 μm.
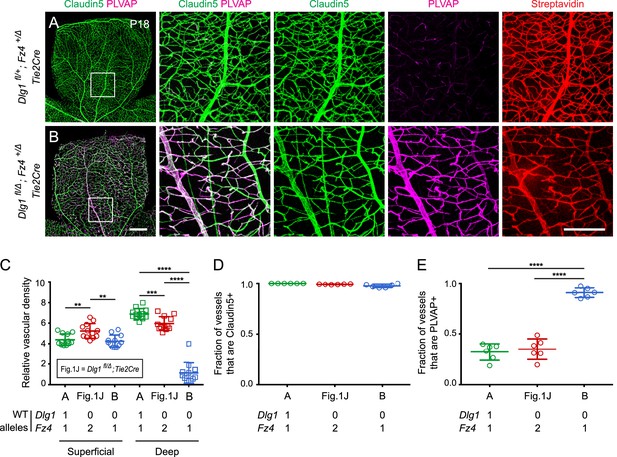
Dlg1 genetically interacts with Fz4 to regulate retinal angiogenesis and the BRB.
(A–B) Maximum projection of the superficial, intermediate, and deep vascular plexuses of P18 retinas from the indicated genotypes (column 1), with boxed regions displayed at higher magnification in columns 2–5. Mice were injected IP with 2–3 mg of sulfo-NHS-biotin 1–2 hr before sacrifice. Scale bar for column 1, 400 μm. Scale bar for columns 2–5, 200 μm. (C–E) Quantification of vascular density in the superficial and deep plexuses (C), the fraction of vessels that immunostain for Claudin5 (D), and the fraction of vessels that immunostain for PLVAP (E), for the genotypes shown in (A), (B), and Figure 1J. In this Figure and Figures 3 and 4, the numbers of WT alleles for the genes of interest are listed below each set of data points.
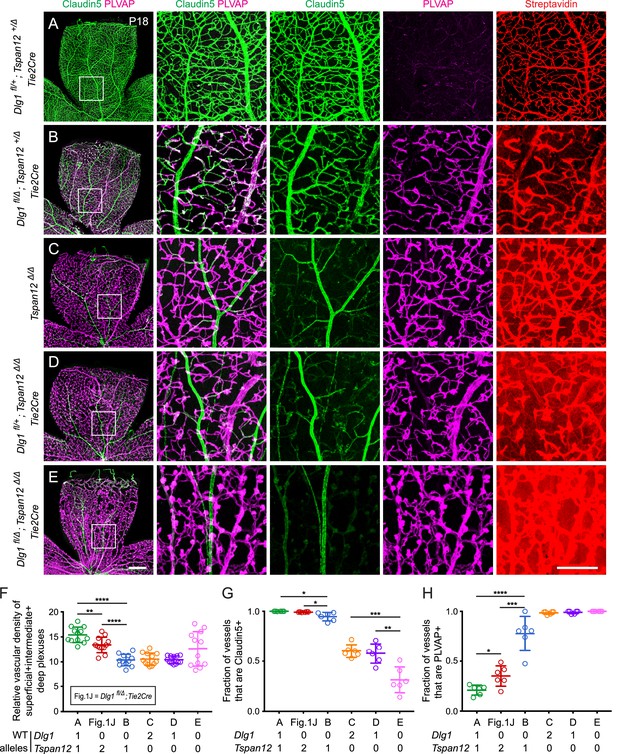
Dlg1 genetically interacts with Tspan12 in the developing retinal vasculature.
(A–E) Maximum projection of the superficial, intermediate, and deep vascular plexuses of P18 retinas from the indicated genotypes (column 1), with boxed regions displayed at higher magnification in columns 2–5. Mice were injected IP with 2–3 mg of sulfo-NHS-biotin 1–2 hr before sacrifice. Scale bar for column 1, 400 μm. Scale bar for columns 2–5, 200 μm. (F–H) Quantification of vascular density (F), the fraction of vessels that immunostain for Claudin5 (G), and the fraction of vessels that immunostain for PLVAP (H), for the genotypes shown in (A–E) and Figure 1J.
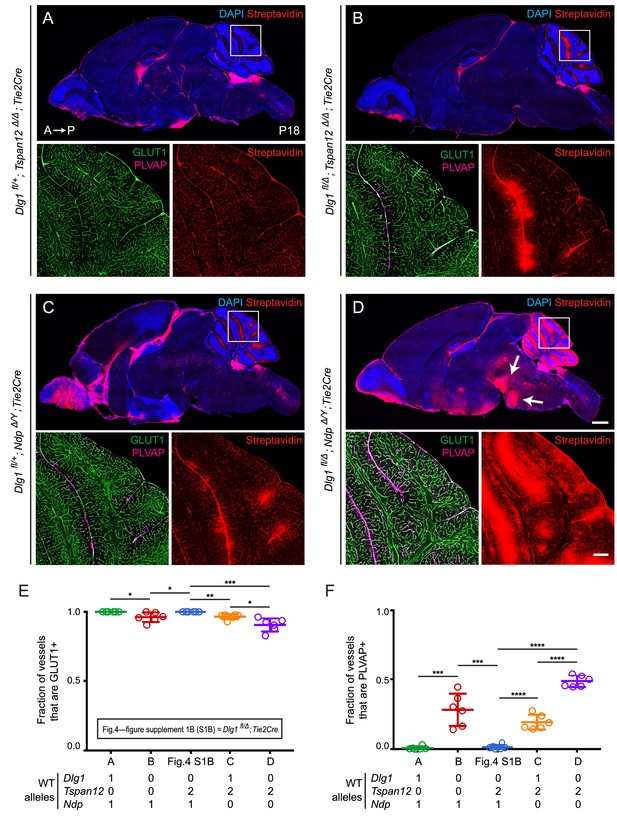
Dlg1 genetically interacts with Tspan12 and Ndp to regulate BBB integrity.
(A–D) Sagittal sections of P18 brains from the indicated genotypes, with color channels for the indicated histochemical stains and/or immunostains indicated in each image. In each set, the boxed region in the cerebellum in the upper image is shown at higher magnification in the lower two images. Mice were injected IP with 2–3 mg of sulfo-NHS-biotin 1–2 hr before sacrifice. (Ndp is an X-linked gene, and Norrin null males are designated NdpΔ/Y.) Arrows in (D) point to biotin leakage in the hindbrain. A, anterior; P, posterior. Scale bar (low magnification images), 1 mm. Scale bar (high magnification images), 200 μm. (E,F) Quantification of the fraction of vessels that immunostain for GLUT1 (E) and PLVAP (F), for each genotype shown in (A–D) and Dlg1 fl/Δ; Tie2-Cre (Figure 4—figure supplement 1B).
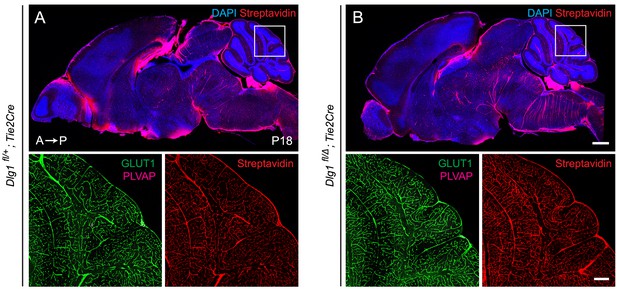
Loss of Dlg1 in endothelial cells has no effect on vascular leakage or vascular markers GLUT1 and PLVAP.
(A,B) Sagittal sections of P18 brains from the indicated genotypes, with color channels for the indicated histochemical stains and/or immunostains listed in the corner of each image. In each image set, the boxed region in the cerebellum in the upper image is shown at higher magnification in the lower two images. Mice were injected IP with 2–3 mg of sulfo-NHS-biotin 1–2 hr before sacrifice. Dlg1 fl/+;Tie2-Cre (A) and Dlg1 fl/Δ;Tie2-Cre (B) brains show no alterations in EC anatomy, EC markers, or BBB integrity. A, anterior; P, posterior. Scale bar for low magnification images, 1 mm. Scale bar for high magnification images, 200 μm.
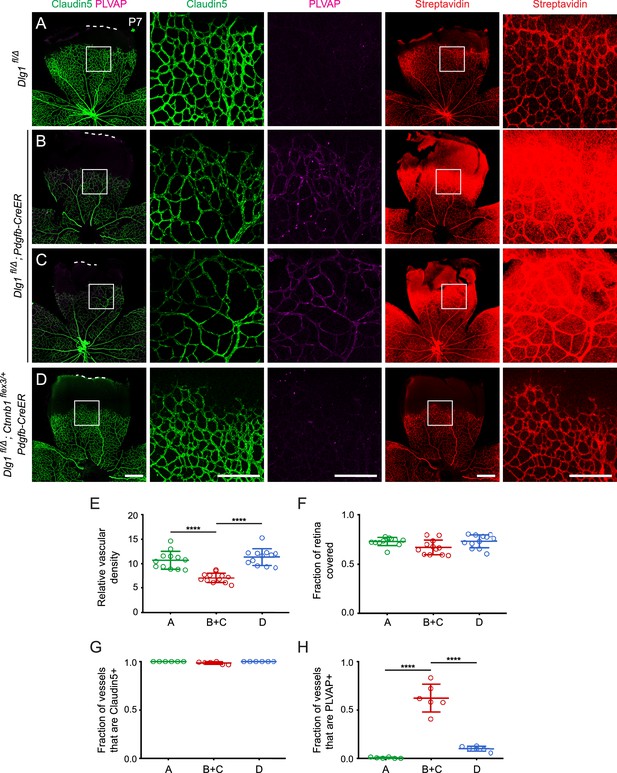
Stabilizing beta-catenin in endothelial cells corrects the retinal vascular defect caused by loss of Dlg1.
(A–D) P7 retinas from the indicated genotypes, with the boxed region in column 1 shown at higher magnification in columns 2 and 3, and the boxed region in column 4 shown at higher magnification in column 5. Mice were injected IP with 2–3 mg of sulfo-NHS-biotin 1–2 hr before sacrifice. Dashed white lines indicate the edge of the retina. (E–H) Quantification of vascular density (E), vascular coverage (F), the fraction of vessels that immunostain for Claudin5 (G), and the fraction of vessels that immunostain for PLVAP (H), for the genotypes shown in (A–D). Scale bars for columns 1 and 4, 400 μm. Scale bars for columns 2, 3, and 5, 200 μm.
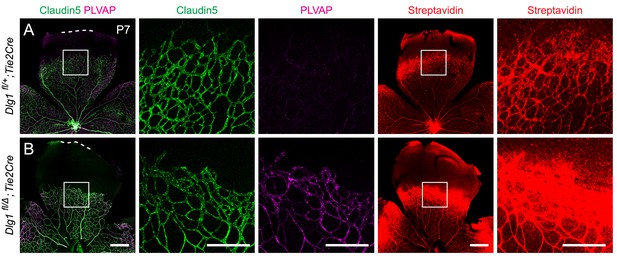
Loss of Dlg1 in endothelial cells in P7 retinas: marker expression and biotin leakage.
(A,B) P7 retinas, with the boxed region in column 1 shown at higher magnification in columns 2 and 3, and the boxed region in column 4 shown at higher magnification in column 5. Mice were injected IP with 2–3 mg of sulfo-NHS-biotin 1–2 hr before sacrifice. Scale bars for columns 1 and 4, 400 μm. Scale bars for columns 2, 3, and 5, 200 μm.
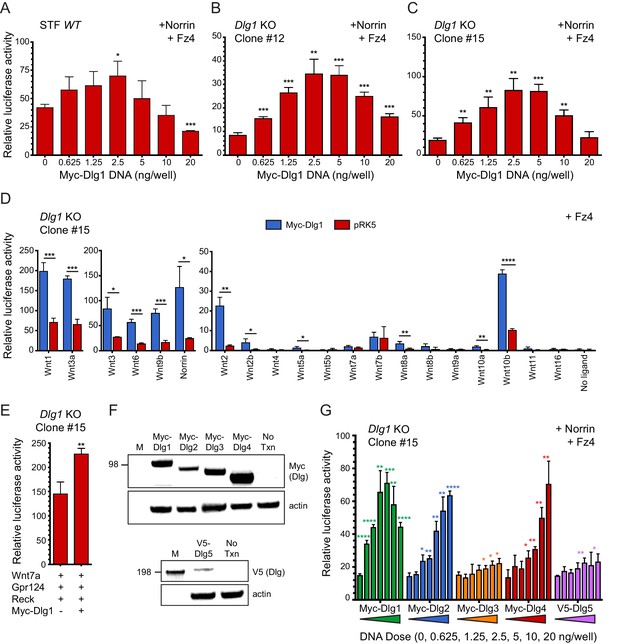
Dlg1 activates Wnt signaling in transfected cells.
(A–C) WT STF (A), Dlg1 KO STF Clone #12 (B), and Dlg1 KO STF Clone #15 (C) were transfected with Norrin and Fz4 plasmids together with the indicated concentrations of Myc-Dlg1 plasmid DNA. The statistical significance of the relative STF firefly luciferase activity (i.e. normalized relative to the transfected Renilla luciferase) was in comparison to the sample with no Myc-Dlg1 transfection, determined from triplicate data using the unpaired t-test. The mean Renilla luciferase values (n = 42 data points per cell line) were 4.54 ± 0.57 and 3.61 ± 0.92 for Clone #12 and Clone #15, respectively, indicating on average an ~25% greater transfection efficiency for Clone #12 compared to Clone #15. (D) Dlg1 KO STF Clone #15 was transfected with Fz4 and each of the 19 mouse Wnts or Norrin, together with Myc-Dlg1 or the empty pRK5 vector at 2.5 ng/well. (E) Dlg1 KO STF Clone #15 was transfected with Fz4, Wnt7a, Gpr124, and Reck, together with Myc-Dlg1 or the empty pRK5 vector at 2.5 ng/well. (F) Immunoblot analysis of Myc-Dlg1, Myc-Dlg2, Myc-Dlg3, Myc-Dlg4, or V5-Dlg5 produced by transient transfection of HEK/293T cells. Actin serves as a loading control. Molecular weight markers are indicated at the left. (G) Dlg1 KO STF Clone #15 was transfected with Norrin and Fz4 plasmids with the indicated concentrations of Myc-Dlg1, Myc-Dlg2, Myc-Dlg3, Myc-Dlg4, or V5-Dlg5 plasmid DNA.
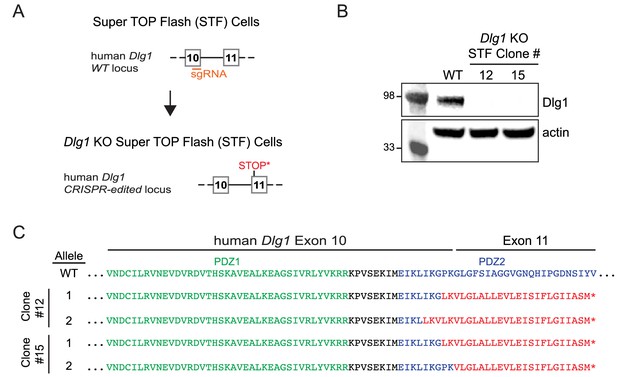
Dlg1 null mutations created by CRISPR/Cas9-mediated gene editing in the STF reporter cell line.
(A) Schematic of the strategy for creating a CRISPR/Cas9-mediated deletion in exon 10 of the Dlg1 gene, leading to a frameshift mutation and termination of the open reading frame in exon 11. (B) By immunoblotting, Dlg1 was undetectable in two CRISPR/Cas9-edited Super TOP Flash (STF) cell lines, Dlg1 KO STF Clones #12 and #15. (C) Predicted amino acid sequences of WT Dlg1 and the CRISPR/Cas9-edited Dlg1 alleles present in Dlg1 KO STF Clones #12 and #15. DNA sequences of the CRISPR/Cas9-edited alleles were obtained from multiple cloned PCR products. Green, PDZ1 sequence; black, inter-PDZ domain sequence; blue, PDZ2 sequence; red, out-of-frame sequence; asterisk, out-of-frame stop codon.
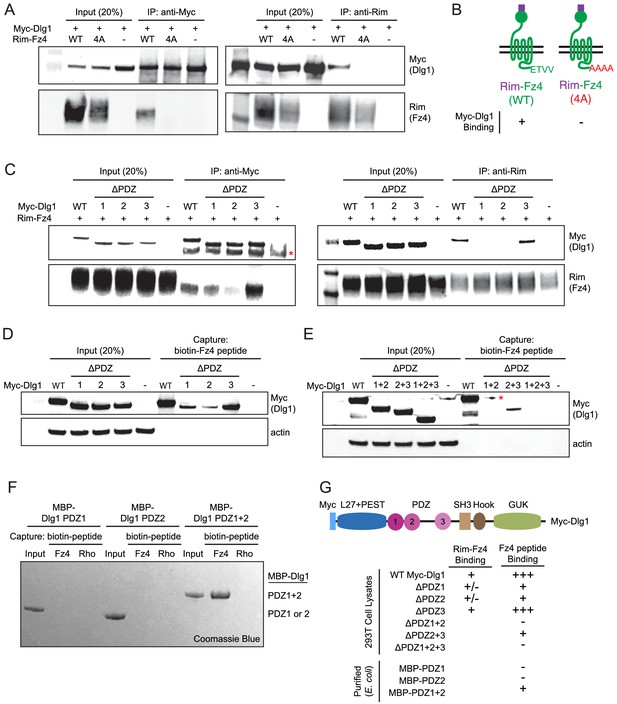
The Fz4 C-terminal PDZ-binding motif binds to PDZ domains 1 and 2 of Dlg1.
(A) Co-immunoprecipitation followed by immunoblotting to assess the interactions between Myc-Dlg1 and Rim-Fz4. HEK/293T cells were co-transfected with Myc-Dlg1 and WT Rim-Fz4, Rim-Fz4(4A), or an empty expression vector. Co-immunoprecipitation was performed with anti-Myc antibody (left) or anti-Rim antibody (right). (B) Schematic summarizing the results in (A). Rim-Fz4 is depicted in the plasma membrane (horizontal black lines) with its Rim-tagged N-terminus in the extracellular space (top) and its C-terminus in the cytoplasm (bottom). (C) HEK/293T cells were co-transfected with Rim-Fz4 and either WT Myc-Dlg1, the indicated PDZ deletion derivatives (ΔPDZ), or an empty expression vector. Co-immunoprecipitation was performed with anti-Myc antibody (left) or anti-Rim antibody (right). In (C) and (E), the red dot indicates a contaminant, which is variably present. (D,E) HEK/293T cells were transfected with WT Myc-Dlg1, the indicated PDZ deletion derivatives (ΔPDZ), or an empty expression vector. Soluble cytosolic proteins from the transfected cells were captured with an N-terminally biotinylated synthetic peptide corresponding to the C-terminal 12 amino acids of Fz4. (F) MBP was fused at its C-terminus to PDZ1, PDZ2, or PDZ1+2 of Dlg1 and the resulting fusion proteins were expressed in E. coli. The MBP fusion proteins were purified and then tested for binding to an N-terminally biotinylated synthetic peptide corresponding to the C-terminal 12 amino acids of Fz4 or the C-terminal 11 amino acids of bovine rhodopsin, which lacks a PDZ-binding motif. (G) Summary of the binding results between Dlg1 and Fz4. For each interaction examined, a score of - (no binding), +/- (little to no binding), + (minimal binding), or +++ (robust binding) was assigned. Blank spaces signify that binding was not assessed.
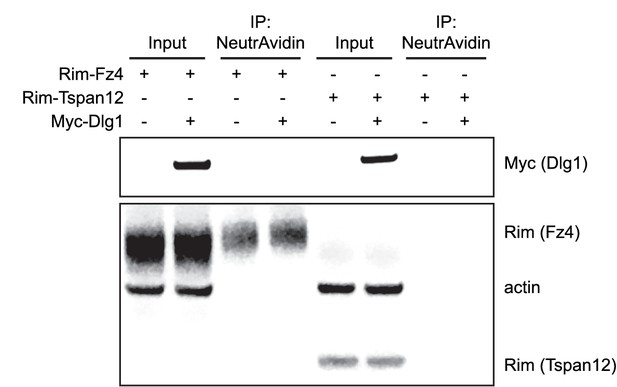
Plasma membrane localization of Fz4 in Dlg1 KO STF Cells.
Surface biotinylation, as described in Materials and methods, of Dlg1 KO STF Clone #15 cells transfected with Rim-Fz4, Rim-Tspan12, and/or Myc-Dlg1.
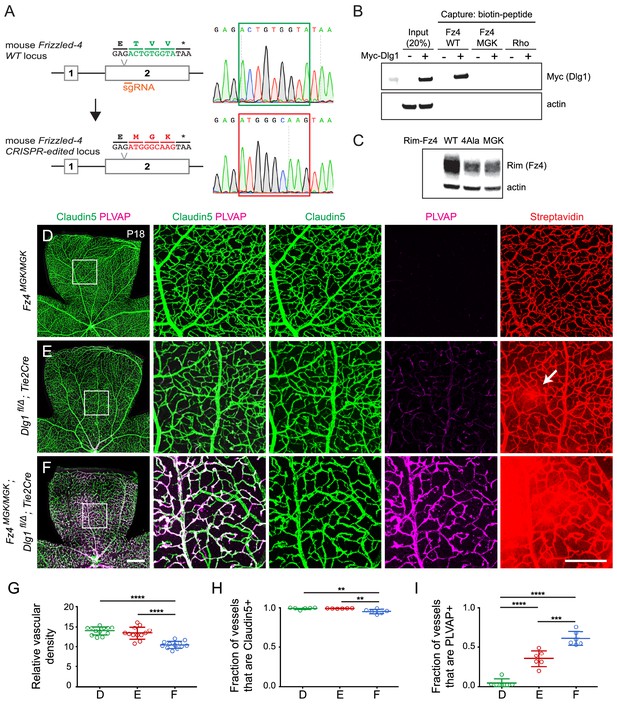
Mutation of the Fz4 C-terminal PDZ-binding domain enhances the severity of retinal vascular defects in a Dlg1 mutant background.
(A) Diagram of the Fz4 allele produced by CRISPR/Cas9 editing (Fz4MGK). Numbers indicate exons. TVV, the C-terminal three amino acids, were substituted with MGK. Right, sequencing chromatograms from mouse tail PCR products. Boxes encompass the three altered codons. Top, WT allele. Bottom, Fz4MGK allele. (B) Protein extracts from HEK/293T cells transfected with WT Myc-Dlg1 (+) or an empty expression vector (–) were subjected to affinity capture using a biotin-Fz4 WT C-terminal peptide, a biotin-Fz4 peptide with the C-terminal three amino acids substituted by MGK, or a biotin-rhodopsin C-terminal peptide. Myc-Dlg1 binds only to the Fz4 WT C-terminal peptide. (C) Immunoblot of protein extracts from HEK/293T cells transfected with Rim-Fz4 WT, Rim-Fz4(4A) or Rim-Fz4(MGK), i.e., Fz4 with the C-terminal three amino acids changed to MGK. (D–F) Maximum projection of the superficial, intermediate, and deep vascular plexuses of P18 retinas from the indicated genotypes (column 1) with boxed regions enlarged in columns 2–5. Mice were injected IP with 2–3 mg of sulfo-NHS-biotin 1–2 hr before sacrifice. Arrow in (E) indicates biotin leakage. Scale bar for column 1, 400 μm. Scale bar for columns 2–5, 200 μm. (G–I) Quantification of summed vascular density (G), the fraction of vessels that immunostain for Claudin5 (H), and the fraction of vessels that immunostain for PLVAP (I), for the genotypes shown in (D–F).
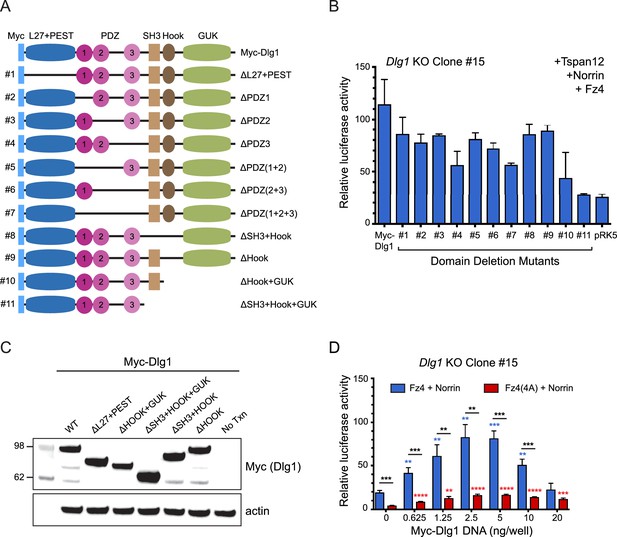
Effect of Dlg1 domain deletions on canonical beta-catenin signaling in STF cells, and effect of WT Dlg1 on signaling by WT Rim-Fz4 vs. Rim-Fz4(4A).
(A) Diagram of WT Myc-Dlg1 and its domain deletion mutants. (B) Dlg1 KO STF Clone #15 cells were transfected with Tspan12, Norrin, and Fz4, together with Myc-Dlg1, the indicated domain deletion mutants of Myc-Dlg1, or the empty expression vector (pRK5). (C) Immunoblot of WT Myc-Dlg1 and various combinations of L27, PEST, SH3, HOOK, and GUK domain deletion mutants shown in the schematic in (A). Immunoblots of the PDZ domain deletions are shown in Figure 7D and E. (D) Dlg1 KO STF Clone #15 cells were transfected with Fz4 WT and Norrin plasmids or Rim-Fz4(4A) and Norrin plasmids, together with the indicated concentrations of Myc-Dlg1 DNA.
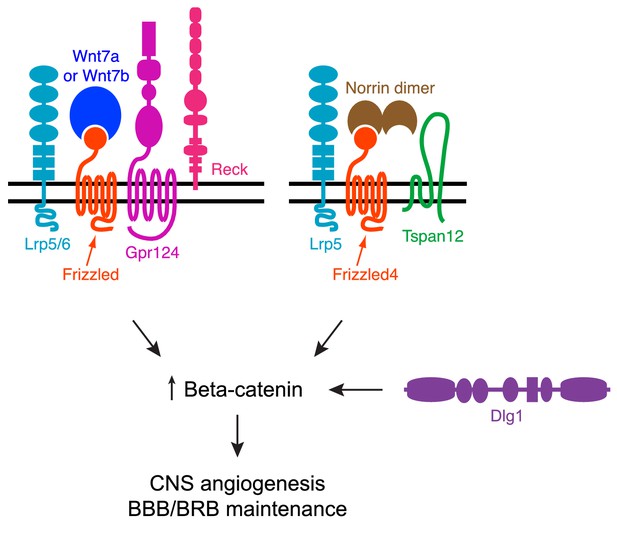
Summary diagram of Dlg1 in beta-catenin signaling for CNS angiogenesis and BBB/BRB maintenance.
https://doi.org/10.7554/eLife.45542.016Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (M. musculus) | Dlg1fl | PMID: 18842882 | Richard Huganir (Johns Hopkins) | |
| Genetic reagent (M. musculus) | Dlg1Δ | this paper | Generated by breeding mice carrying the germline Sox2-Cre transgene with floxed mice | |
| Genetic reagent (M. musculus) | Fz4Δ | PMID: 15035989 | ||
| Genetic reagent (M. musculus) | Fz4MGK | this paper | Please find details under Materials and methods (Gene Targeting) | |
| Genetic reagent (M. musculus) | Tspan12Δ | PMID: 19837033 | Harald Junge (University of Colorado Boulder) | |
| Genetic reagent (M. musculus) | NdpΔ | PMID: 19837032; Jackson Laboratory | Stock #: 012287; RRID:IMSR_JAX:012287 | |
| Genetic reagent (M. musculus) | Ctnnb1flex3 | PMID: 10545105 | Makoto Taketo (Kyoto University) | |
| Genetic reagent (M. musculus) | Tie2-Cre | Jackson Laboratory | Stock #: 008863; RRID:IMSR_JAX:008863 | |
| Genetic reagent (M. musculus) | Pdgfb-CreER | PMID: 18257043 | ||
| Genetic reagent (M. musculus) | Sox2-Cre | PMID: 12617844 | ||
| Cell line (H. sapiens) | HEK/293T | ATCC | Cat. #: CRL-3216 | |
| Cell line (E. coli) | BL21 | New England Biolabs | Cat. #: C2530H | |
| Cell line (H. sapiens) | Super TOP Flash (STF) luciferase reporter cell line | PMID: 15035989 | ||
| Cell line (H. sapiens) | STF Dlg1 KO Clone #12 | this paper | Please find details under Materials and methods (Constructing CRISPR/Cas9 STF cell lines) | |
| Cell line (H. sapiens) | STF Dlg1 KO Clone #15 | this paper | Please find details under Materials and methods (Constructing CRISPR/Cas9 STF cell lines) | |
| Antibody | Rabbit polyclonal anti-Glut1 | Thermo Fisher Scientific | Cat. #: RB-9052-P1; RRID: AB_177895 | 1:400 dilution |
| Antibody | Rat monoclonal anti-PLVAP/MECA-32 | BD Biosciences | Cat. #: 553849; RRID: AB_395086 | 1:400 dilution |
| Antibody | Mouse monoclonal anti-Claudin5, Alexa 488 conjugate | Thermo Fisher Scientific | Cat. #: 352588; RRID: AB_2532189 | 1:400 dilution |
| Antibody | Rabbit polyclonal anti-6xMyc | PMID: 28803732 | 1:10,000 dilution | |
| Antibody | Rabbit monoclonal anti-V5 | Cell Signaling Technology | Cat. #: 13202; RRID: AB_2687461 | 1:1000 dilution |
| Antibody | Mouse monoclonal anti-RIM3F4 | PMID: 9092582 | 1:10,000 dilution | |
| Antibody | Mouse monoclonal anti-actin | Millipore Sigma | Cat. #: MAB1501; RRID: AB_2223041 | 1:10,000 dilution |
| Antibody | Rabbit monoclonal anti-SAP97 | Abcam | Cat. #: ab134156 | 1:1000 dilution |
| Antibody | Goat polyclonal anti-rabbit IgG (H + L) cross-adsorbedsecondary antibody, Alexa 488, 594, and 647 conjugates | Thermo Fisher Scientific | Cat. #s: A-11008, RRID: AB_143165; A-11012, RRID: AB_2534079; A-21244,RRID: AB_2535812 | 1:400 dilution |
| Antibody | IRDye 800CW goat anti-mouse IgG (H + L) secondary antibody | LI-COR | Cat. #: 925–32210 | 1:10,000 dilution |
| Antibody | IRDye 680RD goat anti-rabbit IgG (H + L) secondary antibody | LI-COR | Cat. #: 925–68071 | 1:10,000 dilution |
| Oligonucleotides | Fz4MGK guide RNA: AGGAAAAGGCAACGAGACTG | this paper | Please find details under Materials and methods (Gene Targeting) | |
| Oligonucleotides | Dlg1 guide RNA: AAGCTCATTAAAGGTCCTAA | this paper | Please find details under Materials and methods (Constructing CRISPR/Cas9 STF cell lines) | |
| Recombinant DNA reagents | Rat Myc-Dlg1 cDNA | PMID: 12805297 | Richard Huganir (Johns Hopkins) | |
| Recombinant DNA reagents | pMAL-cR1 expression vector | New England Biolabs | ||
| Recombinant DNA reagents | Mouse Norrin, Wnts, and Frizzleds cDNA | PMID: 23095888 | ||
| Recombinant DNA reagents | Mouse Tspan12 cDNA | PMID: 30478038 | ||
| Recombinant DNA reagents | Mouse Reck cDNA | PMID: 28803732 | ||
| Recombinant DNA reagents | Mouse Gpr124 cDNA | PMID: 28803732 | ||
| Recombinant DNA reagents | Mouse Dlg2 cDNA | Origene | Cat. #: MR222602 | |
| Recombinant DNA reagents | Mouse Dlg3 cDNA | GE Dharmacon | Clone #: 6842105 | |
| Recombinant DNA reagents | Mouse Dlg4 cDNA | GE Dharmacon | Clone #: 4501403 | |
| Recombinant DNA reagents | Mouse V5-Dlg5 cDNA | PMID: 17765678 | Alex Kolodkin (Johns Hopkins) | |
| Recombinant DNA reagents | pSpCasp9(BB)−2A-GFP vector | Addgene | Plasmid #: 48138 | |
| Peptide, recombinant protein | Rhodopsin peptide (SKTETSQVAPA) | this paper | Please find details under Materials and methods (Biotin-peptide binding to proteins expressed in HEK/293T cells) | |
| Peptide, recombinant protein | Fz4 WT peptide (WVKPGKGNETVV) | this paper | Please find details under Materials and methods (Biotin-peptide binding to proteins expressed in HEK/293T cells) | |
| Peptide, recombinant protein | Fz4 MGK peptide (WVKPGKGNEMGK) | this paper | Please find details under Materials and methods (Biotin-peptide binding to proteins expressed in HEK/293T cells) | |
| Commercial assay or kit | MEGAshortscript T7 Kit | Invitrogen | Cat. #: AM1354 | |
| Commercial assay or kit | MEGAclear Transcription Clean-Up Kit | Invitrogen | Cat. #: AM1908 | |
| Commercial assay or kit | mMESSAGE mMACHINE T7 Ultra Transcription Kit | Invitrogen | Cat. #: AM1345 | |
| Commercial assay or kit | Dual-Luciferase Reporter Assay System | Promega | Cat. #: E1910 | |
| Chemical compound, drug | (Z)−4-Hydroxytamoxifen | Sigma-Aldrich | Cat. #: H7904 | |
| Chemical compound, drug | Sunflower seed oil from Helianthus annuus | Sigma-Aldrich | Cat. #: S5007 | |
| Chemical compound, drug | EZ-Link Sulfo-NHS-LC-Biotin | Thermo Fisher Scientific | Cat. #: 21335 | |
| Software, algorithm | ImageJ | https://imagej.nih.gov/ij | ||
| Software, algorithm | Adobe Photoshop CS6 | https://adobe.com/photoshop | ||
| Software, algorithm | Adobe Illustrator CS6 | https://adobe.com/illustrator | ||
| Software, algorithm | GraphPad Prism 7 | http://www.graphpad.com | ||
| Other | FuGENE HD Transfection Reagent | Promega | Cat. #: E2311 | |
| Other | Pierce NeutrAvidin agarose resin | Thermo Fisher Scientific | Cat. #: 29200 | |
| Other | Protein G Dynabeads | Thermo Fisher Scientific | Cat. #: 10004D | |
| Other | Amylose resin | New England Biolabs | Cat. #: E8021S | |
| Other | Streptavidin Dynabeads | Thermo Fisher Scientific | Cat. #: 11047 | |
| Other | Fluoromount G | EM Sciences | Cat. #: 17984–25 | |
| Other | Protease Inhibitor | Roche | Cat. #: 11836170001 | |
| Other | 1x BugBuster Protein Extraction Reagent | Millipore Sigma | Cat. #: 70584 | |
| Other | Texas Red Streptavidin | Vector Laboratories | Cat. #: SA-5006 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.45542.017





