Tumor suppressor SMARCB1 suppresses super-enhancers to govern hESC lineage determination
Figures
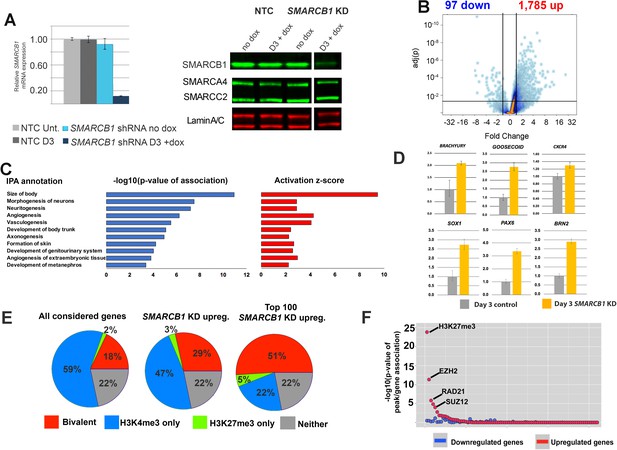
SMARCB1 is a transcriptional repressor of developmental bivalent genes.
(A) qPCR and western blot results showing SMARCB1 KD after 3 days (D3) of doxycycline treatment of inducible SMARCB1 shRNA-expressing H1 hESCs. Band order/spacing was modified from the original gel. qPCR results are relative to untreated NTC hESCs and are normalized to the geometric mean of 18S and GAPDH levels (B) Volcano plot showing the distribution of differentially expressed genes (q < 0.05, FC > 1.5) following 72 hr of SMARCB1. (C) IPA Organismal Development subcategories affected by SMARCB1 KD (q < 0.05, FC > 1.5), with respective activation scores and significance values. (D) qPCR data showing upregulation of early markers for all three germ layers following SMARCB1 KD. (E) Pie charts indicating the percentage of genes with the indicated histone marks, based on data from Pan et al. (2007), in all considered genes (left), those upregulated by SMARCB1 KD (middle), and the top 100 most upregulated genes following SMARCB1 KD (right). (F) Dot plot indicating the significance of intersection between ChIPseq peaks for transcription factors and histone modifications in hESCs and the TSS (±2.5 kb) of genes significantly affected by SMARCB1 KD. The plot is ordered with the ChIPseq peaks most significantly associated with upregulated gene TSS on the left.
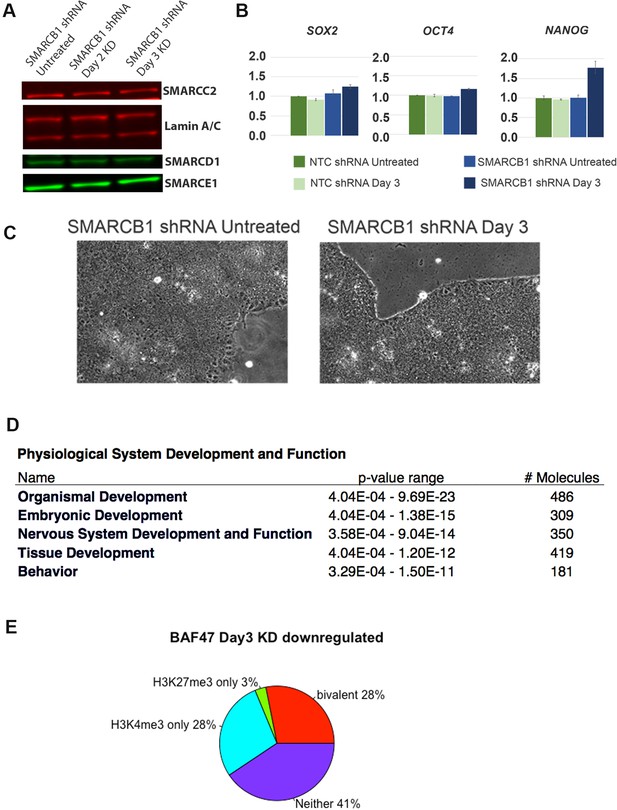
Physical and transcriptional characteristics of SMARCB1 KD hESCs.
(A) Protein levels of SMARCC2, SMARCD1, and SMARCE1 following 2 and 3 days of SMARCB1 KD, with Lamin A/C levels as the loading control). (B) The mRNA levels of SOX2, OCT4, and NANOG are shown for cells carrying shRNAs against a negative control region and SMARCB1, both prior to and following 3 days of treatment with 1 µg/µl doxycycline. (C) Bright-field images of cells carrying shRNAs against SMARCB1 both prior to and following 3 days of treatment with 1 µg/µl doxycycline. (D) Top IPA Physiological System Development and Function categories cells subjected to 3 days of SMARCB1 KD. (E) Percentage of genes downregulated following SMARCB1 KD with the indicated histone marks, based on gene sets defined in Pan et al. (2007).
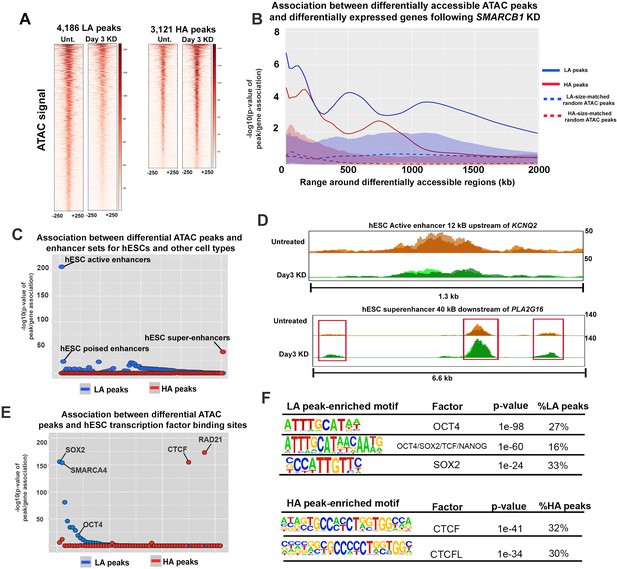
SMARCB1 negatively regulates accessibility at key pluripotency regions in hESCs.
(A) Heatmaps showing the ATAC signal over the peaks with lower (LA) and higher (HA) accessibility (q < 0.05, FC > 1.5) prior to (Unt.) and following 72 hr of SMARCB1 KD. (B) Plot of the significance of the association between SMARCB1 KD lower/higher accessibility peaks (solid lines) and down/up-regulated genes, respectively, by RNAseq (q < 0.05, FC > 1.5) over 2 Mb kb. The blue/red-shaded regions reflect the 5–95% confidence interval (CI) for the significance of the association between down/up-regulated genes and 1000x sets of randomly selected hESC ATAC peaks that were matched in size and number to the lower/higher accessibility ATAC peak sets. The dotted lines indicate the median of the random peak set-based significance range. (C) Dot plot of the significance of the association between SMARCB1 KD lower/higher accessibility peaks and human enhancer regions. The plot is ordered with the enhancer regions most significantly associated with lower accessibility peaks on the left. (D) Top: ATAC signal tracks for untreated and 72 hr SMARCB1 KD cells over an active hESC enhancer 12 kB upstream of KCNQ2. Bottom: ATAC signal tracks for untreated and 72 hr SMARCB1 KD cells over an hESC super-enhancer with three higher accessibility peaks, 40 kB downstream of PLA2G16. (E) Dot plots indicating the significance of intersection between SMARCB1 KD lower/higher accessibility peaks and hESC transcription factor binding sites. The plot is ordered with the ChIPseq peaks most significantly associated with lower accessibility peaks on the left. (F) Top: Pluripotency factor related motifs that are significantly enriched in lower accessibility peaks following SMARCB1 KD, as well as the significance of the association and the percentage of these peaks that contain the motif. Bottom: The significance of the association and the percentage of higher accessibility peaks that contain CTCF and CTCFL motifs following SMARCB1 KD, as well as the significance of the association and the percentage of these peaks that contain the motif.
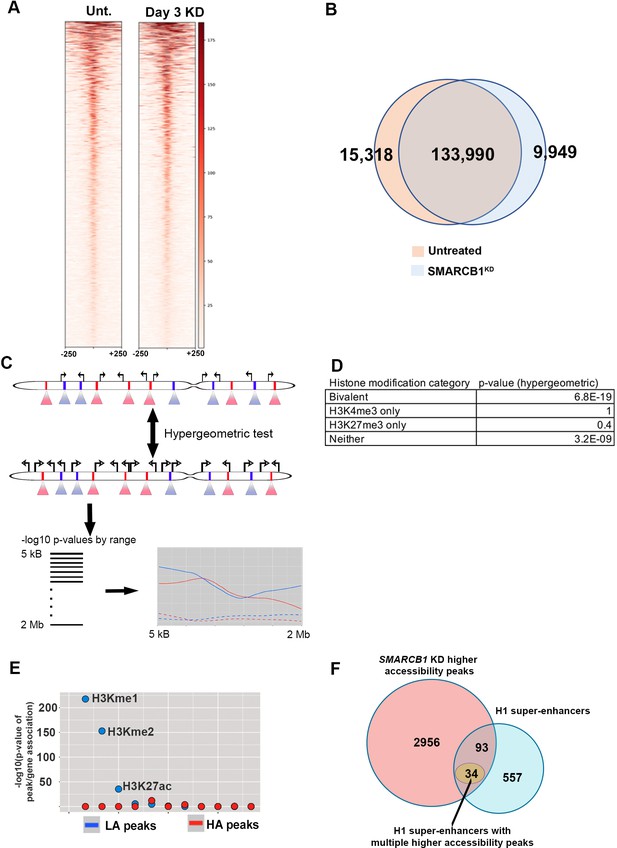
Chromatin accessibility characteristics of SMARCB1 KD hESCs.
(A) ATAC signal over considered peaks for both the control and SMARCB1 KD condition. (B) The number of ATAC peaks called by macs2 for each of the indicated conditions. (C) Visual workflow for differential peak/gene association. Increasing ranges (red/blue triangles) are made around differential ATAC peaks (red/blue bars in chromosome), and the number of DEG TSS (top, solid TSS symbols) is compared with the number of total TSS (bottom, unfilled TSS symbols) that are intersected. A hypergeometric test is used to assess significance, and these p-values are plotted over all ranges considered. (D) Significance of the association between differential peaks and the gene family of the closest TSS (bivalent, H3K27me3 only, H3K4me3 only, or neither marker). (E) The significance of the association between all differential ATAC peaks following SMARCB1 KD and histone modification patterns based on ChIPseq data from ENCODE. (F) Venn diagram showing the intersection between H1 super-enhancers and higher accessibility peaks following SMARCB1 KD. Also indicated are the number of super-enhancers with multiple higher accessibility peaks.
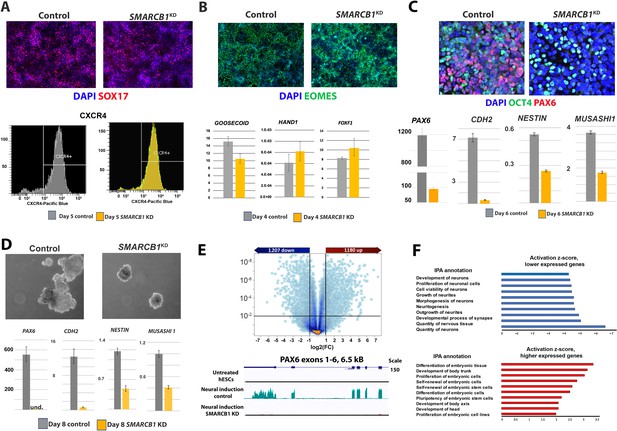
SMARCB1 KD prevents neural induction and upregulation of neural differentiation-related genes.
(A) Top: Control and SMARCB1 KD cells subjected to a 5 day definitive endoderm protocol induction protocol stained for SOX17 (red) and with DAPI (blue). Bottom: Flow cytometry histograms showing similar percentages of CXCR4+ cells in control and SMARCB1 KD cells subjected to this protocol. (B) Top: Control and SMARCB1 KD cells subjected to a 4-day mesodermal induction protocol stained for EOMES (green) and with DAPI (blue). Bottom: qPCR analysis of control and SMARCB1 KD cells subjected to the same protocol for the early mesodermal markers GOOSECOID, HAND1, and FOXF1. All qPCR results are relative to steady state hESCs and are normalized to the geometric mean of 18S and GAPDH levels. (C) Top: Control and SMARCB1 KD cells subjected to a 6 day directed neural induction protocol stained for PAX6 (red), OCT4 (green), and with DAPI (blue). Bottom: qPCR analysis showing a failure of SMARCB1 KD cells to upregulate neural differentiation markers PAX6, CHD2, NESTIN, and MS1. (D) Top: Control and SMARCB1 KD embryoid bodies (EBs) subjected to a neural induction protocol. Bottom: qPCR analysis showing a failure of SMARCB1 KD cells to upregulate the neural differentiation markers PAX6, CHD2, NESTIN, and MS1. (E) Top: Volcano plot illustrating the extent and significance of differential gene expression between control and SMARCB1 KD cells subjected to neural induction protocol as determined by RNAseq (q < 0.01 and FC > 2.0). Bottom: RNAseq tracks PAX6 for steady state hESCs as well as control and SMARCB1 KD cells subjected to a monolayer neural induction protocol. Flat lines indicate undetectable or nearly undetectable expression at the scale used. (F) Top: Activation scores for IPA Nervous System Development sub-categories, considering genes expressed at lower levels in SMARCB1 KD cells compared to controls following the neural induction protocol. Bottom: Activation scores for IPA Embryonic Development sub-categories, considering genes expressed at higher levels in levels in SMARCB1 KD cells compared to controls following the neural induction protocol.
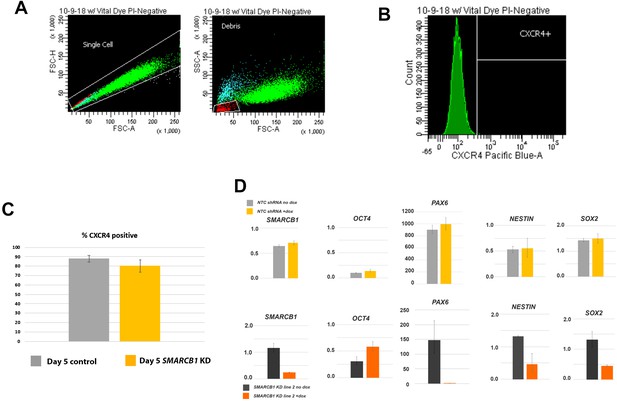
Flow cytometry gating strategy and qPCR data for negative control as well as 2nd SMARCB1 KD line control.
(A) Gating strategy (forward and side scatter) used for flow cytometry analysis of cells subjected to definitive endoderm differentiation. (B) Histogram indicating the data used to define positive CXCR4 expression, representing fluorescence from stained, unmanipulated hESCs. Vertical white line indicates highest level of fluorescence in this cell type. (C) Graph indicating the percentage of CXCR4 positivity in control and SMARCB1 KD cells subjected to the endodermal differentiation protocol. Bar heights indicate the mean of 5 replicates/condition over 3 repeats of experiment. Bars indicate standard deviation. (D) mRNA expression levels of SMARCB1 and neural induction markers for both NTC-shRNA-expressing cells (top row) and a second SMARCB1KD KD line (bottom row) subjected to the monolayer neural induction protocol.
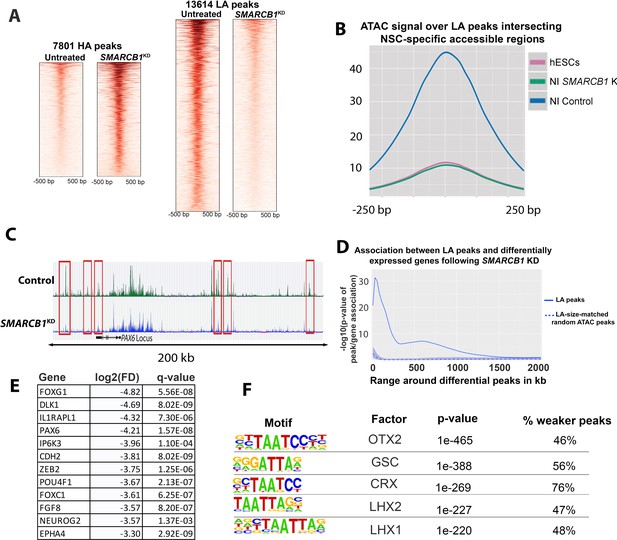
SMARCB1 KD disrupts accessibility dynamics near neural differentiation-related genes.
(A) Heatmaps showing the normalized ATAC signal for control and SMARCB1 KD cells in 7801 peaks with significantly (q < 0.01, FC >2.0) higher (HA) and 13,614 peaks with lower accessibility (LA) peaks following SMARCB1 KD. (B) Accessibility signal over 4107 peaks that are uniquely accessible in NSCs but not iPSCs and which exhibit lower accessibility in SMARCB1 KD cells (Forrest et al., 2017). (C) Accessibility track over the PAX6 locus for steady state hESCs as well as control and SMARCB1 KD cells subjected to a monolayer neural induction protocol. NSC-specific peaks with significantly lower accessibility than the control condition are indicated with red rectangles. (D) Solid line: Significance of the association between lower accessibility peaks and differentially expressed genes (q < 0.01, FD >2.0) over a 2 Mb range. The blue shaded region reflects the 5–95% confidence interval (CI) for the significance of the association between differentially expressed genes and 1000x sets of randomly selected ATAC peaks that were matched in size and number to the lower accessibility ATAC peak set. The dotted line indicates the median of the random peak set-based significance range. (E) Selected differentially expressed genes between the SMARCB1 KD and control condition that have ≥1 lower accessibility region within 500 kb and which are in the IPA Neuron Development Pathway. (F) Motif analysis of the peaks that show stronger or weaker accessibility in SMARCB1 KD cells subjected to the neural induction protocol compared to control cells.
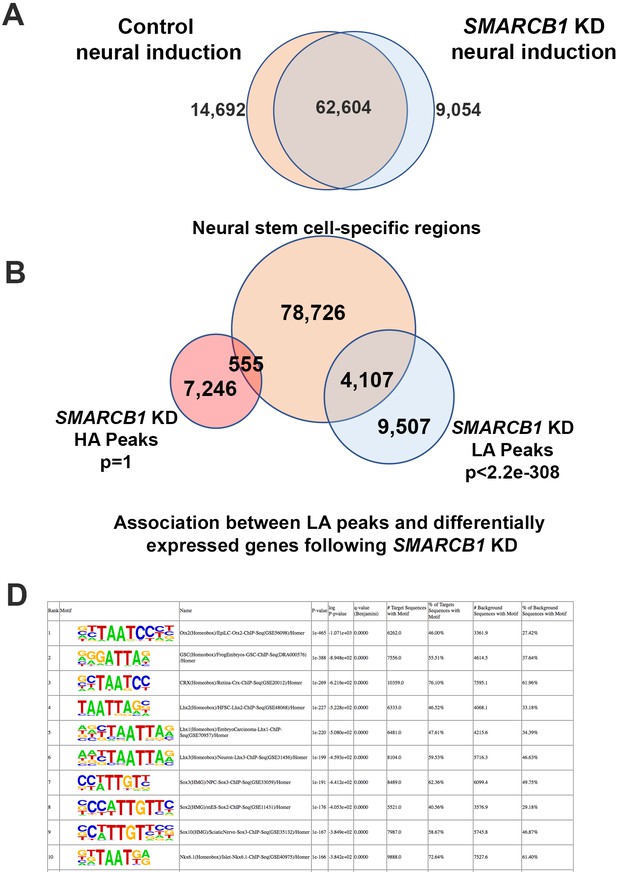
Chromatin accessibility characteristics of SMARCB1 KD cells subjected to neural induction protocol.
(A) The number of ATAC peaks called by macs2 for each the indicated conditions. (B) The number of differentially accessible peaks that overlap with neural stem-cell specific regions, along with significance of overlap. (C) Top 10 HOMER motifs enriched in peaks with lower accessibility in SMARCB1 KD cells following neural induction protocol.
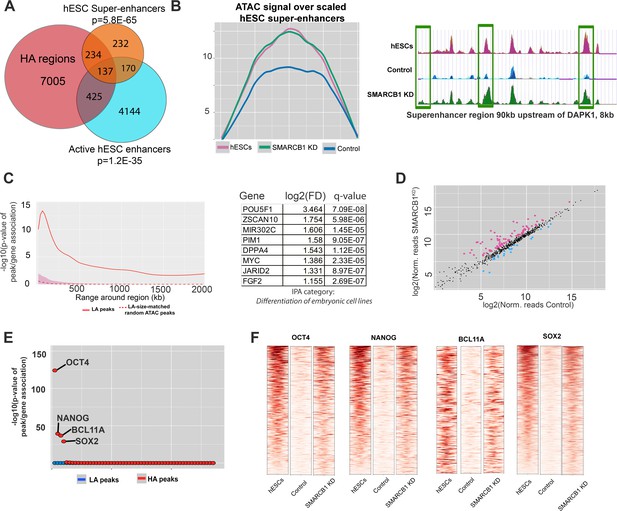
hESC chromatin landscape resists silencing during neural induction protocol in SMARCB1 KD cells.
(A) Venn diagram showing the overlap between higher accessibility peaks in SMARCB1 KD cells subjected to the neural induction protocol and hESC enhancers and super-enhancers. The degree of significance of the overlap is given. (B) Left: Accessibility signals over hESC super-enhancers for steady state hESCs as well as control and SMARCB1 KD cells subjected to neural induction protocol. Right: Track showing normalized accessibility signals for steady state hESCs as well as control and SMARCB1 KD cells subjected to neural induction protocol over an 8 kb portion of a SE near the DAPK1 locus. Differentially accessible peaks are marked with green boxes. (C) Left: Significance of the association between peaks with higher accessibility in hESC super-enhancers and differentially expressed genes by RNAseq over a range of 2 Mb. The red shaded region reflects the 5–95% confidence interval (CI) for the significance of the association between differentially expressed genes and 1000x sets of randomly selected ATAC peaks that were matched in size and number to the set of higher accessibility ATAC peaks in hESC super-enhancers. The dotted line indicates the median of the random peak set-based significance range. Right: Differentially affected genes within 500 kB of an hESC SE with at least one higher accessibility region in SMARCB1 KD cells following the neural induction protocol. (D) Scatterplot showing the number of normalized RNAseq reads over hESC super-enhancers in both control and SMARCB1 KD cells subjected to a neural induction protocol. Pink dots indicate regions with significantly higher levels of RNA, and blue dots indicate regions with lower levels of transcription. (E) Dot plots indicating the significance of intersection between SMARCB1 KD lower/higher accessibility in the neural induction experiments and hESC transcription factor binding sites. The plot is ordered with the ChIPseq peaks most significantly associated with higher accessibility on the left. (F) Heat map indicating the significance of overlap between hESC ChIPseq peaks and higher/lower accessibility in SMARCB1 KD cells following the neural induction protocol. Darker colors indicate a higher degree of significance. Right: Normalized accessibility signals over hESC binding sites for OCT4, NANOG, BCL11A, and SOX2 for steady state hESCs as well as control and SMARCB1 KD cells subjected to the neural induction protocol.
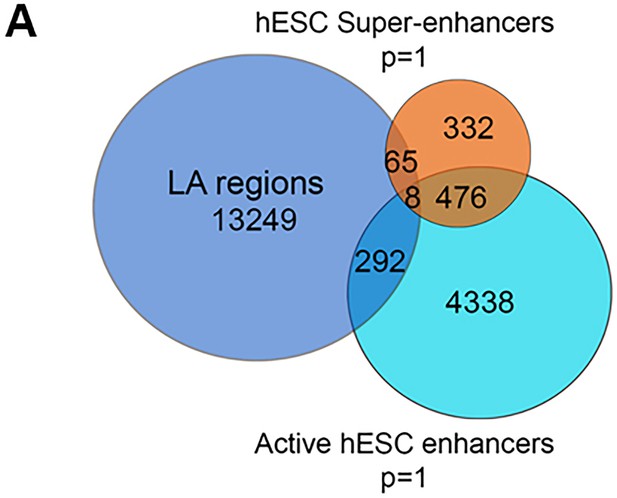
Intersection of LA peaks with hESC enhancer sets.
(A) The numbers of LA peaks that intersect traditional active and superenhancer sets are given. The number of enhancers not intersecting LA regions is also given, with the intersection (476) being in terms of the number of active enhancers (Rada-Iglesias et al., 2011; Hnisz et al., 2013).
Additional files
-
Supplementary file 1
Compilation of shRNAs, differentially regulated genes in SMARCB1 KD cells and the overlap between their TSSs and publicly available genomic features in hESCs.
Sheet 1, The shRNAs used in the present study. Sheet 2, The most strongly upregulated and downregulated genes in SMARCB1 KD cells based on RNAseq analysis. Sheet 3, The degree and significance of the overlap between the TSS of differentially regulated genes and publicaly available ChIPseq datasets for for transcription factors and histone marks in hESCs.
- https://doi.org/10.7554/eLife.45672.013
-
Supplementary file 2
Comparison accessibility peaks in SMARCB1 KD cells and publicly available genomic datasets in hESCs, along with Motif analysis of accessibility peaks.
Sheets 1–2, Degree and significance of overlap between lower and higher accessibility peaks in SMARCB1 KD cells and publicly available ChIPseq datasets for transcription factors and histone marks in hESCs. Sheets 3–4, Motif analysis using HOMER of lower and higher accessibility peaks in SMARCB1 KD cells.
- https://doi.org/10.7554/eLife.45672.014
-
Supplementary file 3
Top differentially affected genes in control in SMARCB1 KD cells subjected to neural induction.
- https://doi.org/10.7554/eLife.45672.015
-
Supplementary file 4
Differentially expressed genes (DEGs) with promoterscontaining differentially accessible peaks and discovered IPA categories in nervous system development and function; along with DEGs in the SMARCB1KD group within 500 kB of an LA peak.
Sheet 1: Differentially expressed genes (q < 0.05, FC > 1.5) with promoters containing differenitally accessible peaks (q < 0.01, FC > 2). Sheet 2: List of IPA categories in Nervous System Development and Function with associated p-values of enrichment and activation scores when considering genes within 500 kb of lower accessibility peaks in SMARCB1KD cells following the neural induction protocol. The category Development of Neurons is highlighted in yellow. Sheet 3: The genes within this category that show significantly different expression in the SMARCB1 KD group and that fall within 500 kB of an LA peak.
- https://doi.org/10.7554/eLife.45672.016
-
Supplementary file 5
Superenhancers with higher expression levels, and those that are near genes with differential expression levels.
Sheet 1: Column A: Tissue/cell line; Column B: Number of defined super-enhancers in tissue/cell line; Column C: Number of all considered accessible regions that overlap super-enhancers; Column D: Percentage of all considered accessible regions that overlap super-enhancers; E: Number of lower accessibility regions that overlap super-enhancers; F: Percentage of lower accessibility regions that overlap super-enhancers; G: p-value of overlap between super-enhancers and lower accessibility regions; H: Number of higher accessibility regions that overlap super-enhancers; I: Percentage of higher accessibility regions that overlap super-enhancers; J: p-value of overlap between super-enhancers and higher accessibility regions. Blue cells indicate significant overlaps with lower accessibility regions, and red cells indicate significant overlaps with higher accessibility regions. Sheet 2: Columns A-C: Location of superenhancer; Column B: q-value of change in super-enhancer RNA expression; Column D: Log2 fold difference between in super-enhancer RNA expression between SMARCB1KD and control cells following the neural induction protocol. Columns E-F: The number of RNAseq reads aligned to the super-enhancer in the control and SMARCB1 KD conditions, respectively. Sheet 3: Column A: Name of differentially expressed gene by RNAseq; Column B: q-value of change in gene expression; Column C: Log2 fold difference in gene expression between SMARCB1KD and control cells following the neural induction protocol. Columns D-F: The location of the super-enhancer with differential eRNA expression. G: Distance from the super-enhancer with differential eRNA expression to the TSS of the gene in column A. Sheet 4: Significant overlap between higher accessibility regions following the neural induction protocol in SMARCB1 KD cells and hESC ChIPseq peaks. The differential accessibility peaks in SMARCB1 KD cells following the neural induction protocol were intersected with available ChIPseq peaks for hESCs. The degree of intersection was assessed for significance using hypergeometric tests. The transcription factor ChIPseq peaks that significantly represented (p<0.05) in higher accessibility peaks are highlighted in in red. No ChIPseq peak set significantly overlapped with lower accessibility peaks.
- https://doi.org/10.7554/eLife.45672.017
-
Supplementary file 6
The qPCR primers used in this study.
- https://doi.org/10.7554/eLife.45672.018
-
Transparent reporting form
- https://doi.org/10.7554/eLife.45672.019





