Homology sensing via non-linear amplification of sequence-dependent pausing by RecQ helicase
Figures
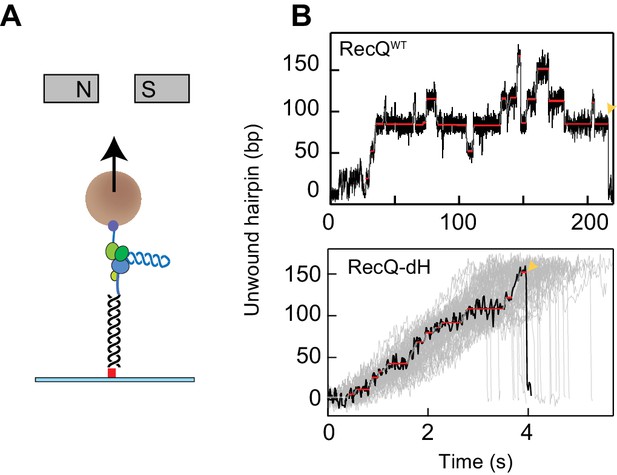
DNA hairpin unwinding activity of RecQ helicase is modulated by the HRDC domain.
(A) Cartoon representation of the experimental scheme (not to scale). The 3′ biotinylated end of the single-stranded poly-dT segment (blue) is attached to a streptavidin-coated 1- or 2.8 µm magnetic bead (brown sphere), whereas the 5′ digoxigenin-labeled double-stranded handle (black line) is attached via anti-digoxigenin (red square) to the surface of the flow-cell. Small magnets above the flow-cell apply a constant upwards force on the magnetic bead. RecQ (purple and green RecA- like domains, yellow zinc binding and winged helix domain, orange HRDC domain) binds at the base of the hairpin (blue helix) and unwinds it, which results in the increase in the extension of the bead. (B) Individual unwinding events of RecQWT and RecQ-dH. Unwound DNA indicates the amount of DNA hairpin opened by RecQ in base pairs. The ends of unwinding events are indicated by a yellow pointer. Pause locations identified from T-test fitting are indicated as solid red lines. Additional RecQ-dH unwinding traces are displayed to show the range of average unwinding rates (gray lines; note that only the region from the beginning to the maximum unwound positions are plotted).
-
Figure 1—source data 1
Source data for Figure 1.
- https://doi.org/10.7554/eLife.45909.004
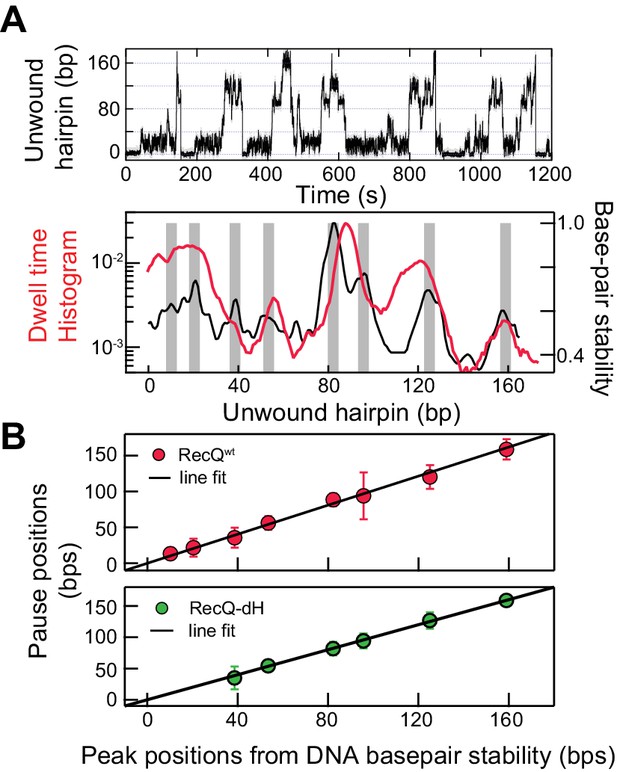
Sequence-dependent pausing of RecQ helicase.
(A) An example trace (top), and dwell-time histogram of RecQWT174 bp hairpin unwinding trajectories (red line) plotted with the exponential of the DNA base-pair melting energy averaged over a 6 bp running window (black line) of the 174 bp DNA hairpin (bottom). The gray bars correspond to peaks in the dwell time histogram associated with RecQ pausing. (B) Linear regression analysis of pause positions of RecQwt (top) and of RecQ-dH (bottom) plotted as function of the peak positions of DNA base-pair stability. The pause positions plotted as a function of energy peak positions were fit with straight lines returning fit values: slope of 0.99 ± 0.03, linear correlation coefficient (Pearson’s r) of 0.97, and χ2 = 0.85 (top); a slope of 0.96 ± 0.06, Pearson’s r = 0.95, and χ2 = 1.1 (bottom). The error bars correspond to the standard error of the mean (SEM).
-
Figure 2—source data 1
Source data for Figure 2—figure supplements 1 and 2.
- https://doi.org/10.7554/eLife.45909.008
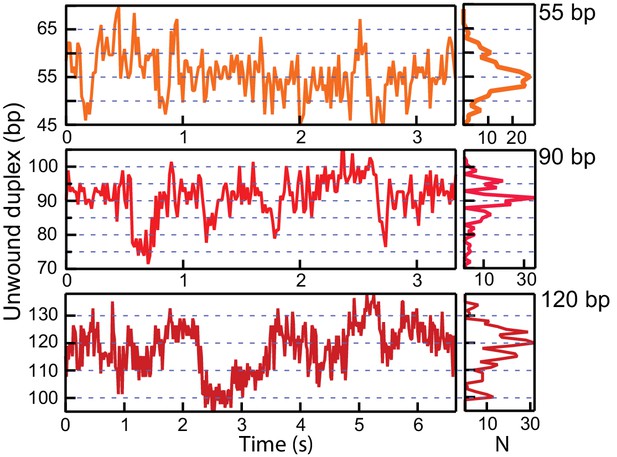
Sequence-dependent pauses are stabilized by the HRDC in RecQWT.
Example traces show long pauses with repetitive rezipping and unwinding of short stretches (<10 bp) around 55 bp, 90 bp, and 120 bp resulting in multiple peaks in the corresponding histograms.
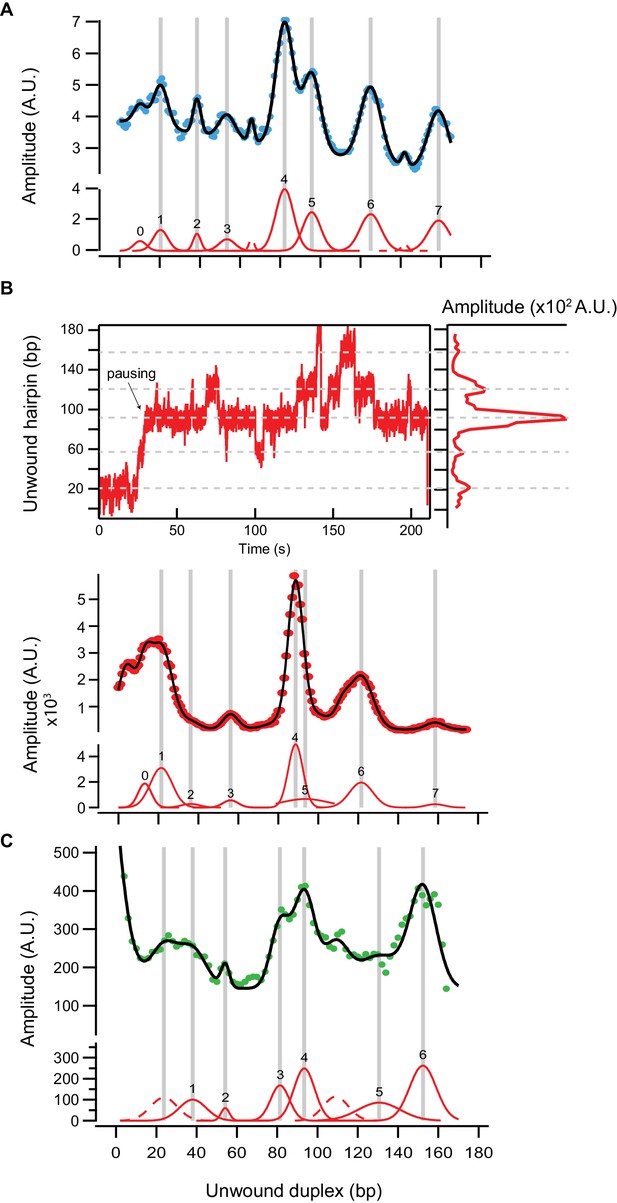
Multi-Gaussian peak fitting of calculated relative pause probability and dwell-time histogram of unwinding traces of RecQwt.
The peak locations in the calculated sequence-dependent pause probability (proportional to the exponential of the DNA hairpin melting energy averaged over 6 bp) and the experimental position dwell-time histogram as a function of hairpin sequence were identified by fitting with a sum of Gaussians. (A) The relative pause probability (blue dots) was globally fitted with a multi-peak Gaussian function (black line). The labeled individual Gaussian peaks are shown below (red lines). (B) Top: The relationship between pausing during unwinding and the peaks in the dwell-time histogram. Bottom: The 1 bp bin width dwell-time histogram (red dots) of RecQwt unwinding traces was fitted with a multi-peak Gaussian function (black line) and individual Gaussian peaks are shown below (red lines). The zero peak corresponds to the base-line extension when the hairpin is fully closed. (C) The 1 bp bin width dwell-time histogram (green dots) for RecQ-dH unwinding traces with the multi-Gaussian fit shown in black and the individual Gaussian peaks in red below.
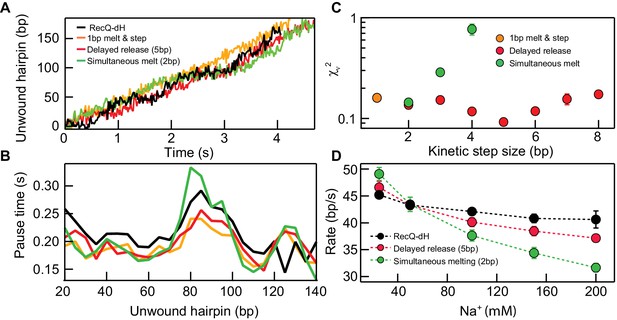
Kinetic modeling of the kinetic step-size.
(A) Example simulated 174 bp DNA hairpin unwinding traces (1 bp melt and step (orange line), 1 bp melt and 5 bp step (red line), and 2 bp melt and step (green line)) overlaid with an example RecQ-dH unwinding trace (black line). Unwinding traces were simulated using Equations 1 and 2. The overall unwinding events are composed of pauses of lifetime (τp) due to melting of the base-pairs, followed by a rapid translocation step in time (τt). τp was calculated based on the sequence stability using nearest neighbor energy parameters (Patten et al., 1984; SantaLucia, 1998; Huguet et al., 2010). The total duration was adjusted to match the mean unwinding rate of RecQ-dH. (B) Pause times plotted as a function of the unwound hairpin for the three example models (with the same marker and line colors) in panel (A). Pause times and positions were obtained by analyzing simulated unwinding traces (100 traces for each condition) using T-test analysis and averaging pause times over a 5 bp window. The experimental pause lifetimes of RecQ-dH are shown in the black solid line. (C) Reduced χ2 (χν2) measure of the correspondence between measured and simulated pause durations as a function of pause position plotted as a function of the kinetic step size for three kinetic stepping models (see main text): 1 bp melt and step (orange filled circles), 1 bp melt and n bp step (red filled circles), and n bp melt and step (green filled circles). χν2 for 1 bp melt and step is significantly larger than the minima of the other two models. The χν2 is minimized for n = 2 bp for the n-bp melt and step model whereas χν2 is minimized for n = 5 bp for the 1 bp melt and n bp step model. (D) Na+ dependent unwinding rates of RecQ-dH (black filled circles and dashed line) and predictions of the two kinetic models with the kinetic step-size, n, that minimizes χν2 for each model: 1 bp melt and 5 bp step (red filled circles and dashed line), and 2 bp melt and step (green filled circles and dashed line). The error bars correspond to the standard error of the mean (SEM).
-
Figure 3—source data 1
Source data for Figure 3 and Figure 3—figure supplement 1.
- https://doi.org/10.7554/eLife.45909.011

Comparison of simulated and experimental traces.
Measured RecQ-dH unwinding traces compared with simulated traces of 10 different pausing models (see main text).
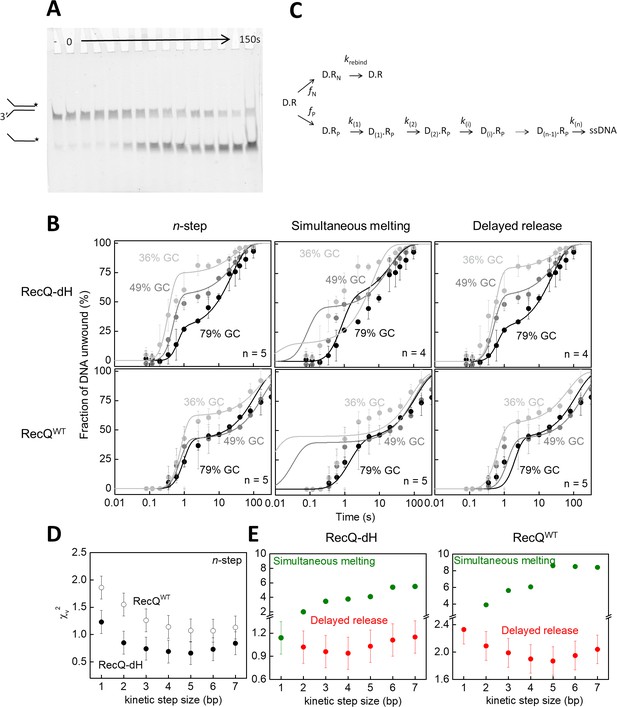
Single-turnover ensemble kinetic experiments.
(A) Electrophoretogram of a single-turnover unwinding experiment. Preincubation of fluorescein-labeled forked duplex DNA (30 nM, gc36) with RecQ-dH (100 nM) was followed by rapid mixing with ATP (3 mM) plus excess ssDNA trap strand (3 µM) (final post-mixing concentrations). Reactions were stopped by the addition of EDTA (40 mM) and loading dye at different time points (0–150 s, cf. panel B) using a quenched-flow instrument or by manual mixing. Amounts of DNA species (forked duplex and ssDNA, depicted by cartoons) labeled with fluorescein (asterisk) were detected by a fluorescence imager. “– “denotes a 150 s control reaction in which ATP was absent. (B) Single-turnover unwinding kinetics of forked DNA substrates with GC contents of 36% (light gray), 48% (gray) and 79% (black) of RecQ-dH and RecQWT. Error bars represent SEM calculated from three experiments. Solid lines show fits based on: the n-step model at n = 5 for both helicase constructs (see scheme on panel C); simultaneous model (Equation 1 and panel C) at n = 4 for RecQ-dH and n = 5 for RecQWT; and delayed release model (Equation. and panel C) at n = 4 for RecQ-dH and n = 5 for RecQWT. (C) Common scheme for the modified n-step and derived simultaneous melting and delayed release models. In the models, unwinding starts from the ssDNA-dsDNA junction. Of all DNA-RecQ complexes (D.R), only a fraction (fP, D.Rp, lower row) unwinds the dsDNA segment in a single run, consisting of n consecutive irreversible kinetic steps (k(1)…k(n)). In the n-step model, the rate constant of each unwinding kinetic step is identical (k(1) = k(n)). In contrast to this, in the simultaneous melting and delayed release models, the rate constant of each unwinding kinetic step depends on the base pair energy of the dsDNA segment to be unwound (segment length according to kinetic step size), as described in Equation 1 and 2, respectively. A fraction of DNA bound helicase molecules (fN, D.RN, upper row) is unable to successfully unwind DNA due to the limited processivity of the enzyme and/or more complex unwinding patterns. After dissociating from the DNA substrate, these enzyme molecules can rebind to the substrate at rate constant krebind and start a new unwinding run (D.R). (D–E) Determined χv2 values from fitting the (D) n-step model for RecQ-dH (filled circles) and RecQWT (open circles) or (E) fitting the simultaneous melting (green) and delayed release models (red) for the indicated helicase construct. Other determined parameters are listed in Supplementary file 1 Table S2.
-
Figure 4—source data 1
Source data for Figure 4.
- https://doi.org/10.7554/eLife.45909.013
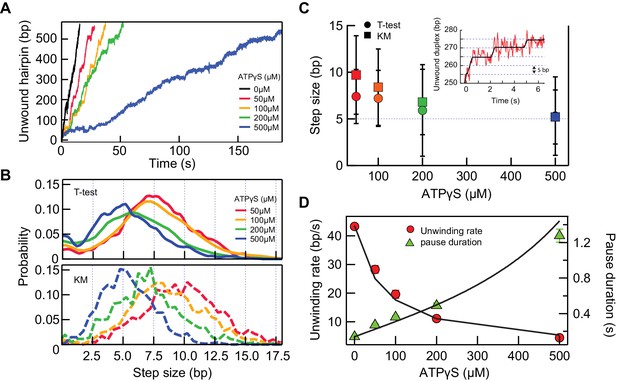
5 bp kinetic step-size and tight mechano-chemical coupling.
(A) Example traces of hairpin unwinding by RecQ-dH with increasing concentration of ATPγS while maintaining the combined concentration of ATP and ATPγS at 1 mM. (B) Step-size distributions were obtained by analyzing unwinding traces collected at each ATPγS concentration with either T-test or Kerssemakers (KM) step analysis algorithms (see main text). (C) The mean step-size obtained by fitting the distributions in panel (B) with Gaussians plotted as a function of ATPγS concentration. The average step sizes from both the T-test and Kerssemakers (KM) analysis converge to 5 bp with increasing ATPγS concentration. Inset: An example trace with 5 bp steps (red line) and the T-test fit (blue line). (D) Global fitting of the mean pause duration (green solid triangles) and average unwinding rate (red solid circles) as a function of ATPγS concentration, using Equations 4 and 5, reveal a tight mechano-chemical coupling ratio of C = 1.0 ± 0.2 bp/ATP.
-
Figure 5—source data 1
Source data for Figure 5 and Figure 5—figure supplements 1–4.
- https://doi.org/10.7554/eLife.45909.019
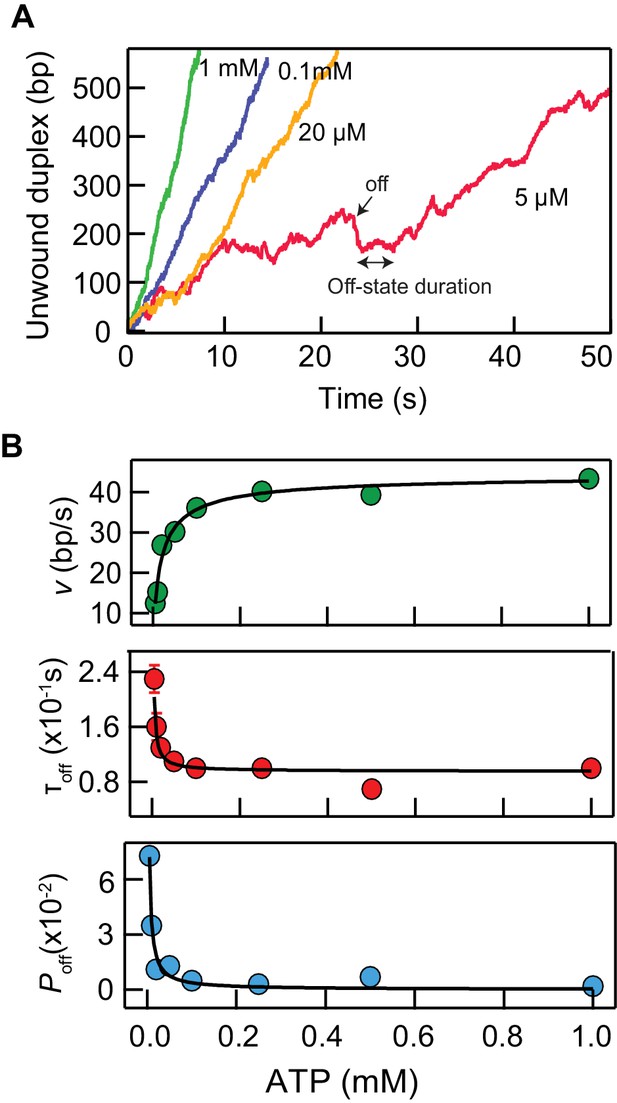
ATP dependence of unwinding, stepping, and backsliding kinetics of RecQ-dH.
(A) Unwinding traces at four different ATP concentrations. At lower ATP concentrations, events of rapid backward transitions (‘off’) followed by a short pause (off-state) and resumption of unwinding occur more frequently, suggesting an ATP-dependent DNA binding stability and entry into an off-pathway weak DNA-binding state of the enzyme. (B) Global fitting of unwinding rates (v), off-state duration (τoff), and off-state probability (Poff) as function of ATP concentration based on the ATP binding and DNA melting scheme (Scheme A in the Appendix 1). See Supplementary file 1 Table S3 for the fitting parameters.
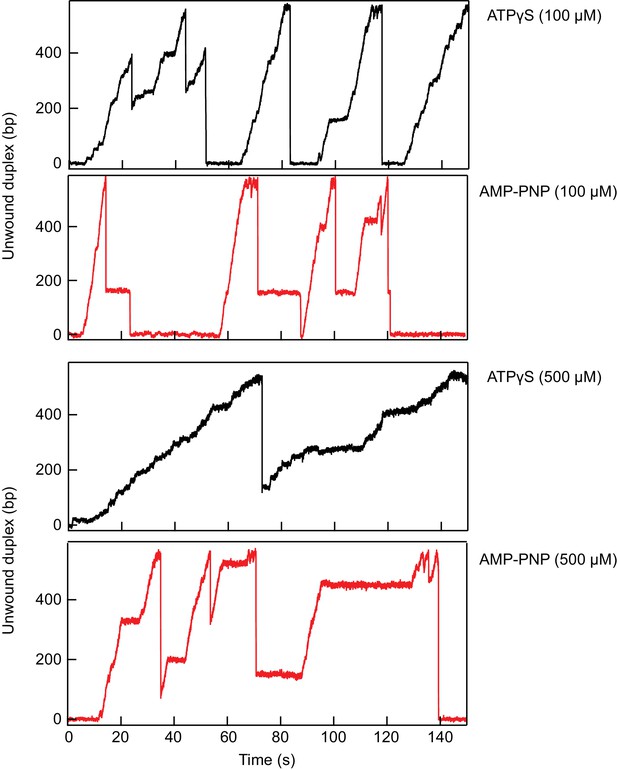
Comparison of RecQ-dH unwinding the 584 bp DNA hairpin in the presence of AMPPNP or ATPγS and ATP.
In comparison to ATPγS, increasing AMPPNP concentration from 100 to 500 µM, with a constant total concentration of ATP and analog of 1 mM, does not gradually slow down the unwinding of RecQ but increases the frequency of very long pauses. These results suggest that AMPPNP is bound significantly less frequently than ATP but it is stably bound for a long time (>5 s) once incorporated by RecQ-dH. In contrast, ATPγS appears to be bound relatively frequently at higher concentrations, but the resulting pauses in unwinding are significantly shorter than the pauses induced by AMPPNP binding.
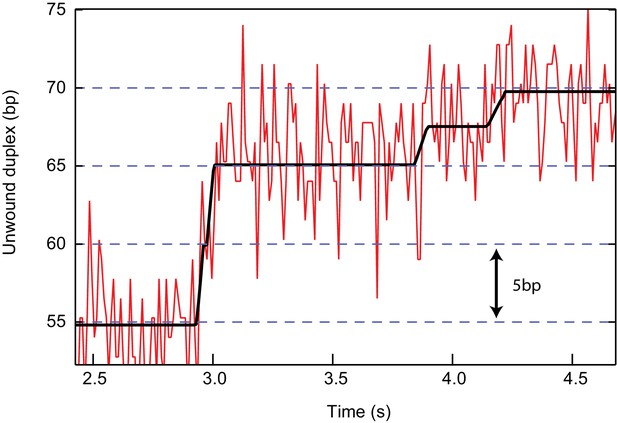
Example trace of RecQ-dH at 500 µM ATPγS showing a 2.5 bp step.
https://doi.org/10.7554/eLife.45909.017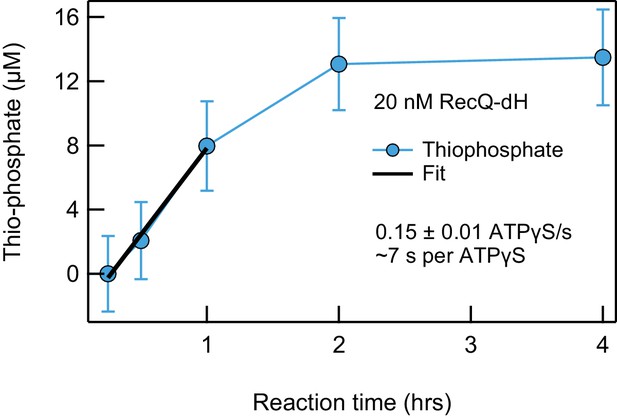
RecQ-dH inefficiently hydrolyzes ATPγS.
The pauses in the unwinding trajectories in the presence of ATPγS could arise from slow hydrolysis of ATPγS, from slow release of ATPγS, or a combination of both hydrolysis and release. Hydrolysis of ATPγS by RecQ-dH produces thiophosphate, which can be detected with a Malachite green assay (Saran et al., 2006). To determine the origin of the ATPγS-induced pauses, we measured the production of thiophosphate (blue circles) by 20 nM RecQ with saturating concentration of ssDNA (200 nM dT54 DNA) in 1 mM ATPγS. We fitted the linear portion of the data with a line (black line, slope = 10.8 ± 0.9 ATPγS·hr−1) and normalized the slope of the line by the enzyme concentration to obtain a rate of 0.15 ± 0.01 ATPγS·s−1 per RecQ-dH (cf. the markedly higher dT54-activated RecQ ATPase activity measured earlier: 86 ± 2 ATP·s−1 per RecQ-dH; Harami et al., 2015). The slow rate of hydrolysis suggests that the vast majority of ATPγS-induced pauses correspond to slow release of ATPγS, which is faster than the hydrolysis rate.
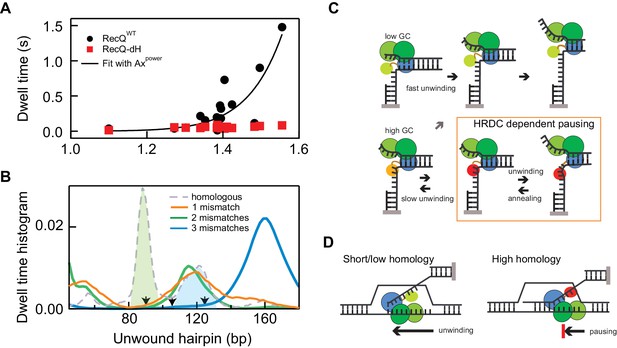
Non-linear amplification of sequence-dependent pausing by HRDC.
(A) The average dwell times of RecQWT and RecQ-dH plotted as a function of the exponential of the average base-pair energy over a 10 bp window. The non-linearity of RecQWT dwell time data was analyzed with a power law function (y = A·xP solid line). The fit returned A = 7 ± 0.9 (x10−4) and Power = 17.2 ± 3.2 (Errors indicate standard deviations of fitting parameters). (B) Dwell-time histograms of RecQ unwinding the 174 bp DNA hairpin with zero mismatches, that is perfect homology, one mismatch (90 bp), two mismatches (90 and 104 bp), and three mismatches (90, 104, and 124 bp). The prominent peak around 90 bp (green shaded region) shown in the dwell time histogram of the intact DNA unwinding by RecQWT was significantly reduced in the hairpin with a single mismatch and the additional peak around 120 bp (blue shaded region) was further suppressed by the third mismatch at 124 bp. The mismatch sites are indicated as arrows. (C) Model for D-loop homology discrimination via HRDC-mediated non-linear amplification of sequence-dependent pausing by RecQ. At regions of low GC content (upper row), RecQ rapidly unwinds duplex DNA and the HRDC remains in a weak binding ssDNA mode (light green HRDC). At regions of high GC content (lower row), RecQ pauses and the HRDC can switch to a strong ssDNA binding mode (orange HRDC). The subsequent binding of the HRDC to the displaced ssDNA (red HRDC) results in stabilization of the GC-induced pauses. As ssDNA is under tension or otherwise constrained, this interaction effectively hinders the movement of the RecQ core, resulting in short-range (5–10 bp) repetitive unwinding and annealing of DNA. (D) HRDC-dependent pausing regulates D-loop disruption in a homology dependent manner. RecQ can quickly unwind an invading strand of short or only partially homologous DNA, whereas HRDC-dependent pausing slows down unwinding and prevents disruption of an invading strand with an extended homology.
-
Figure 6—source data 1
Source data for Figure 6 and Figure 6—figure supplements 1–2.
- https://doi.org/10.7554/eLife.45909.023
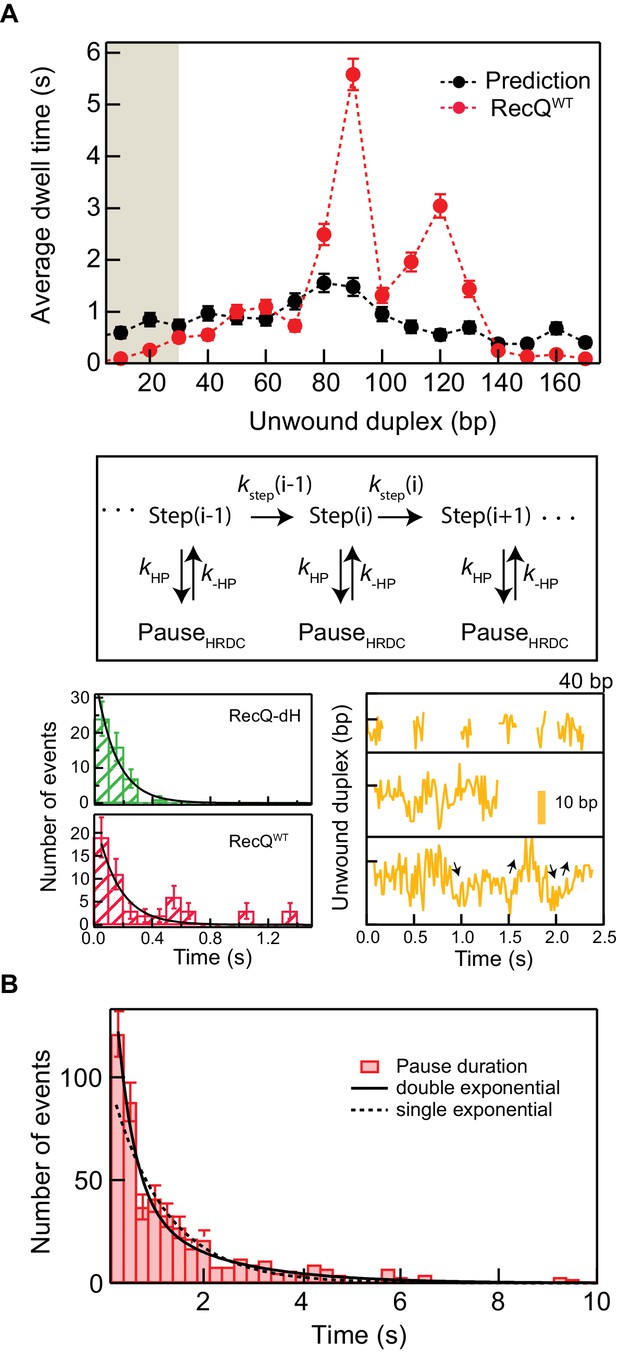
HRDC-dependent pausing kinetics are inconsistent with a simple kinetic competition mechanism of pausing.
(A) Top: Comparison between the dwell time histograms of RecQWT (red circles, 5 bp binning) and the simulation (black circles, 5 bp binning) based on a simple kinetic competition model. The pause durations of RecQWT for less than 30 bp of unwound duplex (brown shaded region) were not analyzed since long and frequent shuttling activity was difficult to determine from the step finding analysis in this region. Middle: Schematic of the kinetic competition model for HRDC-stabilization of sequence-dependent pauses. HRDC-dependent pausing occurs via kinetic competition between the stepping rate (kstep(i)) and rate of entering into a pause state (kHP), of duration 1/k-HP. Bottom: The rates were estimated from analysis of the pausing around the region of 40 bp opening of the hairpin Pause duration histograms of RecQ-dH and RecQWT with single exponential fits (left). The pauses around 40 bp exceeding 0.4 s, that were not observed in RecQ-dH, show repetitive rezipping and unwinding (black arrows) by RecQWT (right; middle and bottom). There was no apparent shuttling at short pauses less than ~0.4 s (right; top). (B) Pause duration distribution of RecQWT from T-test analysis fit with single and double exponential functions. The single exponential fit provides an average pause time of 1.1 ± 0.1 s with χ2 = 67.1; the double exponential fit yields two different time constants: 1.8 ± 0.3 s and 0.4 ± 0.1 s with χ2 = 40.3. Note: both of these pause durations are longer than the average pause duration (0.14 ± 0.03 s, see above) for the core RecQ (RecQ-dH), indicating that there are multiple HRDC-dependent pause states.

Example traces of intact (homologous) hairpin DNA, hairpin with one mismatch, hairpin with two mismatches, and hairpin with three mismatches by RecQWT.
https://doi.org/10.7554/eLife.45909.022
Cartoon of DNA haripin used for single-molecule measurements.
https://doi.org/10.7554/eLife.45909.027Tables
| Reagent type (species)or resource | Designation | Source/reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | ER2566 | New England Biolabs | NEB Cat. #: E4130 | |
| Recombinant DNA | Modified pTXB vector | PMID: 26067769 | Transformation and expression of RecQ constructs | |
| Recombinant DNA | pKZ1 | PMID: 28069956 | Template for hairpin DNA substrate | |
| Antibody | Anti-digoxigenin (Sheep Polyclonal) | Roche | Roche Cat# 11333089001, RRID:AB_514496 | Reconstituted in 1x Phosphate buffered saline (0.6 µg) |
| Commercial assay or kit | IMPACT purification system | New England Biolabs | NEB Cat. #: E6901S | |
| Commercial assay or kit | PCR DNA purification kit | Qiagen | Qiagen Cat. #: 28104 | |
| Chemical compound | Streptavidin coated magnetic beads (ø: 1 and 2.8 µm) | Invitrogen | Invitrogen Cat. #: 65602 and 65305 | |
| Chemical compound | Phusion high-fidelity DNA polymerase | New England Biolabs | NEB Cat. #: M0530 | |
| Chemical compound | T4 DNA ligase | Promega | Promega Cat. #: M1801 | |
| Chemical compound | Nt.BbvcI | New England Biolabs | NEB Cat. #: R0632 | |
| Chemical compound | BsaI-HF | New England Biolabs | NEB Cat. #: R3535 | |
| Software, algorithm | LabVIEW, Instrument control software | National Instruments | NI Cat. #: 776678–35 | |
| Software, algorithm | Igor pro 7, Data analysis | Wavemetrics PMID: 28069956 | ||
| Software, algorithm | MATLAB, Data analysis | MathWorks PMID: 16799566 | ||
| Software, algorithm | KinTek Global Kinetic Explorer 4.0, Data analysis | KinTek |
Additional files
-
Supplementary file 1
Supplementary Tables including DNA sequences and fitting paramters.
Table S1: Sequence (5' to 3') of ssDNA strands composing gc36, gc46 and gc79 forked dsDNA substrates (complementary regions are bold; Flu represents 3’ fluorescein labeling). Table S2: Parameters determined from global fitting ensemble unwinding data with the n-step and delayed release models a Parameters determined from fitting ensemble unwinding data with the n-step model with n = 5 (cf. Figure 4B). b Parameters determined from fitting ensemble unwinding data with the delayed release model with n = 5 for RecQWT and n = 4 for RecQ-dH (cf. Figure 4B). Table S3 Parameters from fitting of Poff, τoff, and v as a function of ATP with three schemes. Table S4 Number of events in the analysis a: Number of events (Na+ mM), b: Number of events (ATPγS µM), c: Number of events (ATP µM)
- https://doi.org/10.7554/eLife.45909.024
-
Transparent reporting form
- https://doi.org/10.7554/eLife.45909.025