Temporal regulation of axonal repulsion by alternative splicing of a conserved microexon in mammalian Robo1 and Robo2
Figures
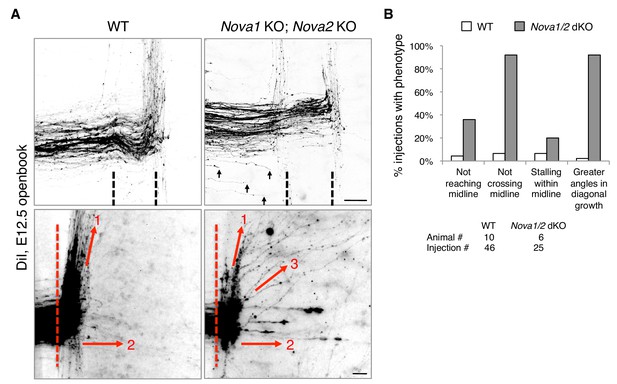
Nova1/2 double knockout (dKO) disrupts commissural axon midline crossing and postcrossing guidance.
(A) DiI labeling of E12.5 mouse spinal cords in openbook preparations. The top panel shows confocal micrographs of axons at the midline. Nova1/2 dKO partially blocked midline crossing. Some axons had not yet reached the midline in Nova1/2 dKO embryos (arrows), consistent with a reduction in DCC-mediated attraction as previously reported (Leggere et al., 2016). The bottom panel shows postcrossing axons that projected away from the midline. Some axons projected diagonally (arrow 1) and some projected straight away (arrow 2). In Nova1/2 dKO embryos, some axons projected diagonally at greater angles from the midline than normal (arrow 3). Dashed lines indicate midline boundaries. Scale bars, 50 μm. (B) Quantification of the guidance defects from DiI tracing. Data are shown as the percentage of injection sites with the stated phenotypes. Midline recrossing was not observed in WT or Nova1/2 dKO embryos. The trajectories of postcrossing axons along the rostrocaudal axis were comparable between WT and Nova1/2 dKO embryos.
-
Figure 1—source data 1
DiI tracing.
- https://doi.org/10.7554/eLife.46042.003
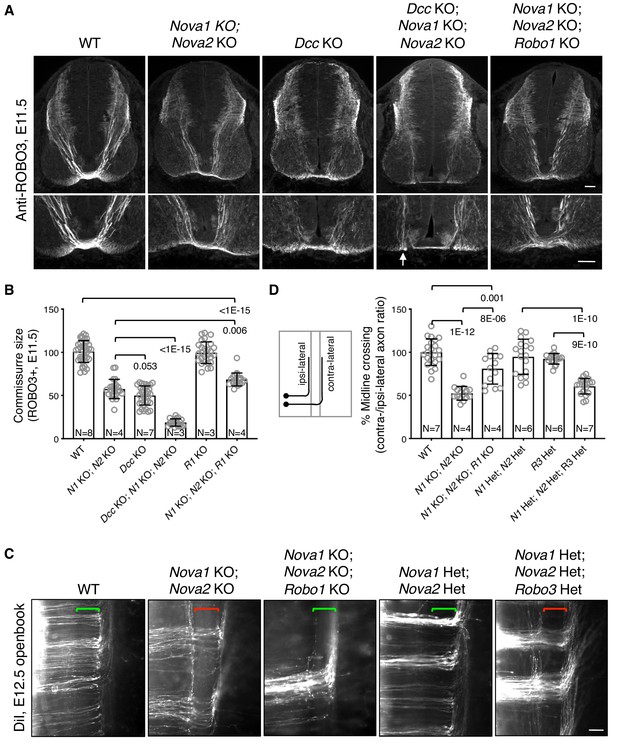
Midline crossing defect in Nova1/2 dKO embryos results from elevated ROBO repulsion.
(A) Anti-ROBO3 staining of commissural axons in transverse sections of E11.5 spinal cords. The bottom panel shows closeup images of the floorplate. Nova1/2 dKO and Dcc KO embryos had reduced ventral commissures. Dcc; Nova1; Nova2 triple KO mutants had even thinner commissures than Dcc KO embryos, and some axons appeared to remain on the ipsilateral side without entering the midline (arrow) in the triple KO mutants. In contrast, Nova1; Nova2; Robo1 triple KO mutants had a slightly increased ventral commissure size compared to Nova1/2 dKO embryos. Scale bars, 50 μm. (B) Quantification of the ventral commissure size in A. Data are normalized to the WT and are represented as the mean ± SD (one-way ANOVA and Bonferroni post test; animal numbers and p values are indicated). (C) DiI labeling of E12.5 mouse spinal cords in openbook preparations. In Nova1/2 dKO embryos, about half of the axons arriving at the midline did not project across. This defect was alleviated by Robo1 KO. Nova1/2 dHet and Robo3 Het, which were phenotypically normal on their own, synergistically blocked midline crossing. Brackets indicate the midline. Scale bar, 50 μm. (D) Quantification of midline crossing in C. Data are normalized to the WT and are represented as the mean ± SD (one-way ANOVA and Bonferroni post test; animal numbers and p values are indicated).
-
Figure 2—source data 1
Commissure size and midline crossing.
- https://doi.org/10.7554/eLife.46042.005
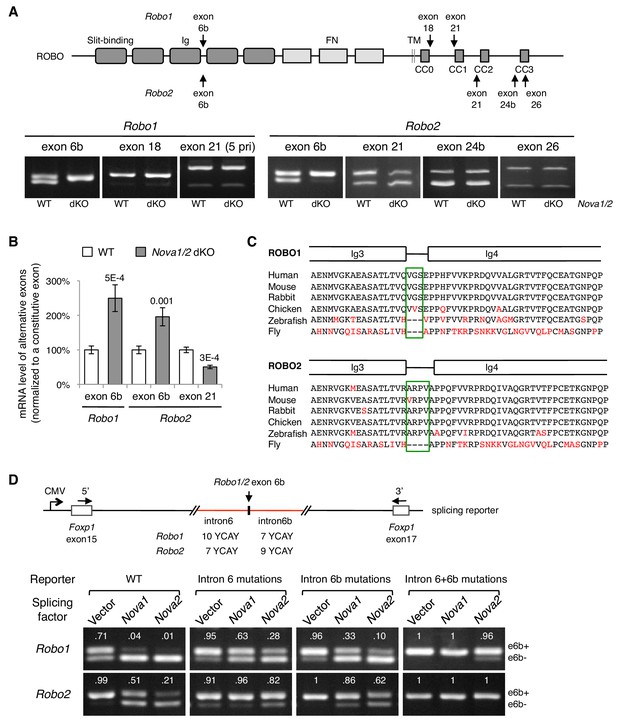
Nova1/2 regulate Robo1/2 alternative splicing.
(A) Alternative splicing of Robo1/2 exons assessed by semi-quantitative RT-PCR using E11.5 dorsal spinal cord (where commissural neurons are largely located). Schematic shows ROBO1/2 protein domains and the locations of alternatively spliced sequences. Ig, immunoglobulin domain; FN, fibronectin domain; TM, transmembrane domain; CC0-CC3, conserved cytoplasmic domain 0–3. The first Ig domain interacts with SLIT (Morlot et al., 2007). (B) Quantitative RT-PCR analyses of Robo1/2 alternative exons whose expression was altered by Nova1/2 dKO. Data are normalized to the WT and are represented as the mean ± SD (Student’s t-test, two-tailed and unpaired; n = 3 animals; p values are indicated). (C) Alignment of peptide sequences encoded by Robo1/2 exon 6b. Exon 6b encodes a short linker (boxed sequences) between the Ig3 and Ig4 domains. Identical residues are shown in black and non-identical ones in red. Drosophila ROBO1 was used for alignment, as it is homologous to both ROBO1 and ROBO2 in vertebrates. (D) Splicing assays of Robo1/2 exon 6b in COS-1 cells, where a splicing reporter was coexpressed with Nova1 or Nova2 and the levels of splice variants were assessed by RT-PCR. Flanking introns of exon 6b in the reporter (in red) contain candidate NOVA-binding sites (7–10 YCAY repeats; Y = C/U; mutations created YAAY repeats; see Materials and methods and Figure 3—figure supplement 1E). Alternatively spliced sequences were detected by semi-quantitative RT-PCR using 5’ and 3’ primers as indicated. Numbers in the electrophoresis images indicate the e6b+ level normalized to the total amount of both isoforms. Nova1/2 normally repressed the e6b+ levels of both Robo1 and Robo2. Mutating candidate NOVA-binding sites in either flanking intron partially blocked NOVA activity. Eliminating all NOVA-binding sites caused e6b+ to be exclusively expressed.
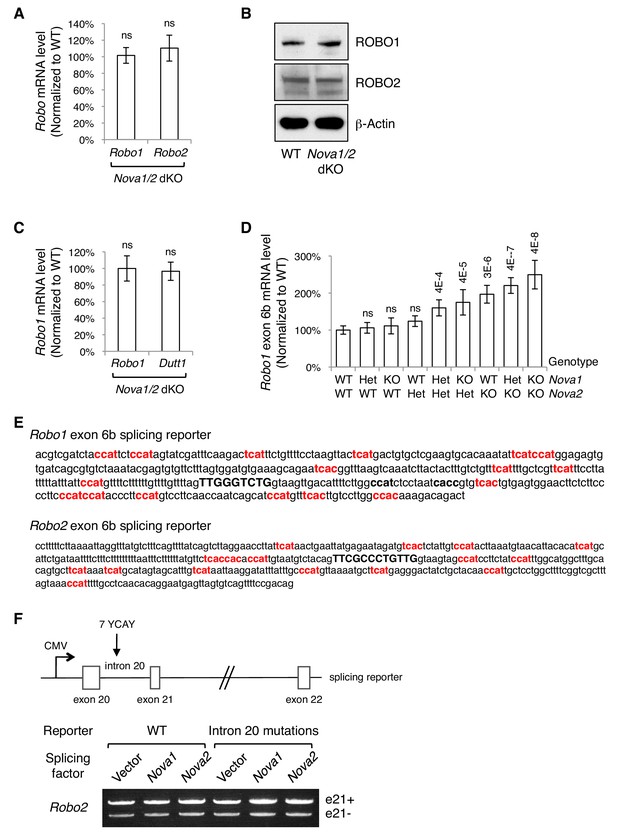
Alternative splicing of Robo1/2.
(A) Quantitative RT-PCR of a constitutive exon in Robo1/2 showing the total Robo mRNA levels in Nova1/2 dKO dorsal spinal cord at E11.5. (B) Western blotting analysis of ROBO1/2 protein levels in E11.5 spinal cords. The antibodies do not distinguish between the alternative isoforms. (C) Quantitative RT-PCR analysis of Robo1 and Dutt1 levels in Nova1/2 dKO dorsal spinal cord, which are two alternative mRNAs produced from two different promoters (Nural et al., 2007),. Other alternative 5’ sequences of Robo1 and Robo2 were also tested and no significant difference was observed between WT and Nova1/2 dKO spinal cords. (D) Quantification of Robo1 exon 6b expression in nine different genotype combinations of Nova1 and Nova2. Robo1 exon 6b level was sensitive to Nova1/2 gene copy number. Robo2 exon 6b level was regulated in a similar dose-sensitive manner. Data are normalized to the WT and are represented as the mean ± SD (Student’s t-test in A,C; one-way ANOVA and Bonferroni post test in D; n = 3 for all genotypes; p value are indicated; ns, not significant). (E) Genomic DNA included in exon 6b splicing reporters. Exon 6b sequences are capitalized and in bold. Potential NOVA-binding sites (YCAY repeats; Y = C/U) that were mutated in the splicing assays are in bold and in red. Two additional candidate NOVA-binding sites (in black, bold, and lower case) are present in Robo1 intron 6b but were not mutated, as the mutations themselves constitutively repressed exon 6b inclusion. (F) Splicing assays of Robo2 exon 21. Schematic shows the Robo2 exon 21 splicing reporter, which contains the complete genomic sequences between exons 20 and 22 (5.8 kb). Candidate NOVA-binding sites (7 YCAY repeats) are located in intron 20. Expression of e21- and e21+ isoforms was detected by semi-quantitative RT-PCR. NOVAs had no direct effect on Robo2 exon21 alternative splicing.
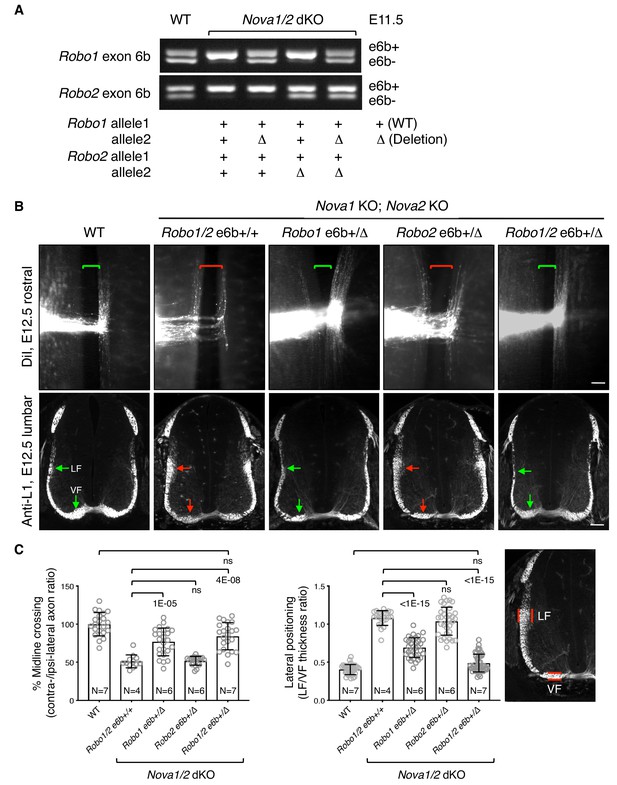
Restoring Robo1/2 exon 6b levels rescues axon guidance defects in Nova1/2 dKO embryos.
(A) Alternative splicing of exon 6b in animals with or without the exon 6b deletion allele. Semi-quantitative RT-PCR was performed using RNA from E11.5 dorsal spinal cord. Deleting one copy of exon 6b from Robo1/2 genomic DNA in Nova1/2 dKO embryos restored e6b+ expression to a comparable level as in the WT. (B) DiI labeling of spinal cord openbooks (top panel) and anti-L1 staining of transverse spinal cord sections (bottom panel) in E12.5 animals with or without exon 6b deletion. As it is difficult to obtain a large number of compound mutants, the rostral half of the spinal cord was used for DiI labeling and the lumbar level was used for anti-L1 staining. Brackets indicate the midline. Arrows indicate the lateral funiculus (LF) and ventral funiculus (VF). Reducing Robo1(e6b+) partially rescued both midline crossing and lateral positioning defects in Nova1/2 dKO embryos, while reducing Robo2(e6b+) alone did not rescue. Reducing Robo1/2(e6b+) together further rescued both defects. Scale bars, 50 μm. (C) Quantification of axon midline crossing and lateral positioning in B. As Nova deficiency reduces the number of postcrossing axons due to fewer axons reaching the midline, we compared the ratio between the thickness of the lateral and ventral funiculi within the same section. Data are represented as the mean ± SD (one-way ANOVA and Bonferroni post test; animal numbers and p values are indicated; ns, not significant).
-
Figure 4—source data 1
Phenotypic rescues by exon 6b deletion.
- https://doi.org/10.7554/eLife.46042.012
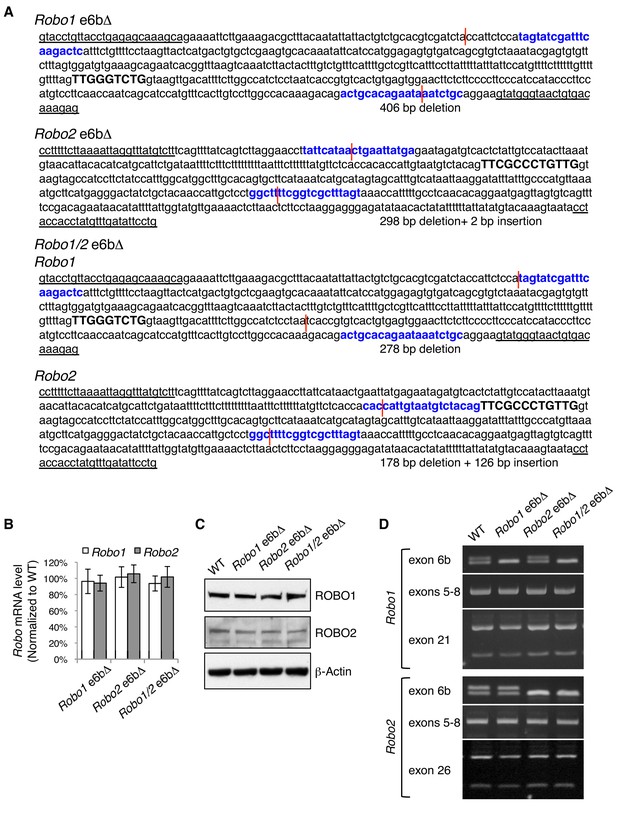
Generation of Robo1/2 exon 6b deletion alleles by CRISPR/Cas9.
(A) Exon 6b deletion mutations in Robo1 alone, in Robo2 alone, and in both Robo1 and Robo2. Robo1/2 are linked on mouse chromosome 16. Single guide RNAs (in bold and in blue) targeted both 5’ and 3’ of exon 6b (capitalized and in bold). Genomic DNA breakpoints are indicated by red lines. Forward and reverse genotyping primers are underlined. (B) Quantitative RT-PCR of a constitutive exon showing the total Robo mRNA levels in E11.5 dorsal spinal cord in homozygous exon 6b deletion mutants. Data are normalized to the WT and are represented as the mean ± SD (one-way ANOVA and Bonferroni post test; n = 3 for all genotypes; not significant at all data points). (C) Western blotting analysis of ROBO1/2 proteins in E11.5 spinal cord in homozygous exon 6b deletion mutants. The antibodies do not distinguish between alternative isoforms. (D) Semi-quantitative RT-PCR showing the alternative splicing of exon 6b, exons 5 to 8, and 3’ alternative exons (exon 21 in Robo1 and exon 26 in Robo2) in homozygous exon 6b deletion mutants. The RT-PCR products from exons 5–8 were cloned and sequenced, and no change other than at exon 6b was detected. Overall, removing exon 6b does not affect the splicing of nearby exons or of other alternative exons.
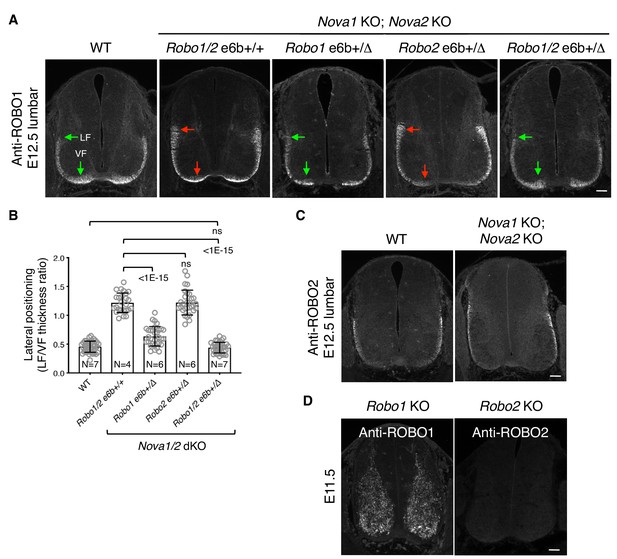
Lateral positioning of ROBO1-expressing longitudinal axons in Nova1/2 dKO embryos.
(A) Anti-ROBO1 staining of transverse sections of E12.5 lumbar spinal cords. The antibodies do not distinguish between isoforms. ROBO1 was expressed at a low level on precrossing axons and was highly upregulated on postcrossing axons in both WT and Nova1/2 dKO embryos. ROBO1-expressing axons displayed the same lateral positioning defect as anti-L1 labeled axons (compare with Figure 4B). Reducing Robo1(e6b+) partially rescued the lateral positioning defect in Nova1/2 dKO embryos, while reducing Robo2(e6b+) alone did not rescue. Reducing Robo1/2(e6b+) together further rescued the defect. Arrows indicate the lateral funiculus (LF) and ventral funiculus (VF). Scale bar, 50 μm. (B) Quantification of the lateral positioning of ROBO1-expressing axons in A. Data are represented as the mean ± SD (one-way ANOVA and Bonferroni post test; animal numbers and p values are indicated; ns, not significant). (C) Anti-ROBO2 staining of transverse sections of E12.5 lumbar spinal cords. The antibodies do not distinguish between isoforms. ROBO2 was primarily expressed by axons in the lateral funiculi, and the overall patterns were comparable between the WT and Nova1/2 dKO embryos. (D) Anti-ROBO1 and anti-ROBO2 staining in the corresponding KO embryos, showing the specificity of the antibodies. The Robo1 KO was generated by trapping the protein product in intracellular compartments (Friedel et al., 2005). ROBO1 expression was seen in neuronal cell bodies but not in axons in Robo1 KO spinal cords, as previously reported (Long et al., 2004).
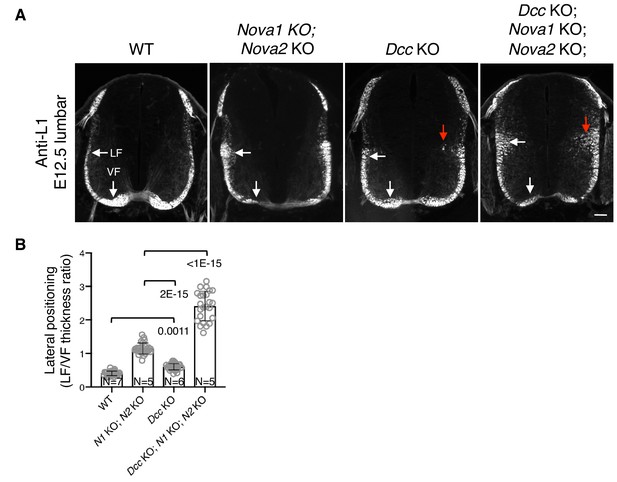
Nova1/2 and Dcc mutants have distinct postcrossing guidance defects.
(A) Anti-L1 staining of transverse sections of E12.5 lumbar spinal cords. Nova1/2 dKO embryos had enlarged lateral funiculi. There was a small increase in the lateral funiculi in Dcc KO mutants and some lateral axons projected into the medial spinal cord (red arrow). In Dcc; Nova1; Nova2 triple KO mutants, the lateral funiculi were enlarged and many lateral axons entered the medial spinal cord. Scale bar, 50 μm. (B) Quantification of axon lateral positioning in A. Data are represented as the mean ± SD (one-way ANOVA and Bonferroni post test; animal numbers and p values are indicated).
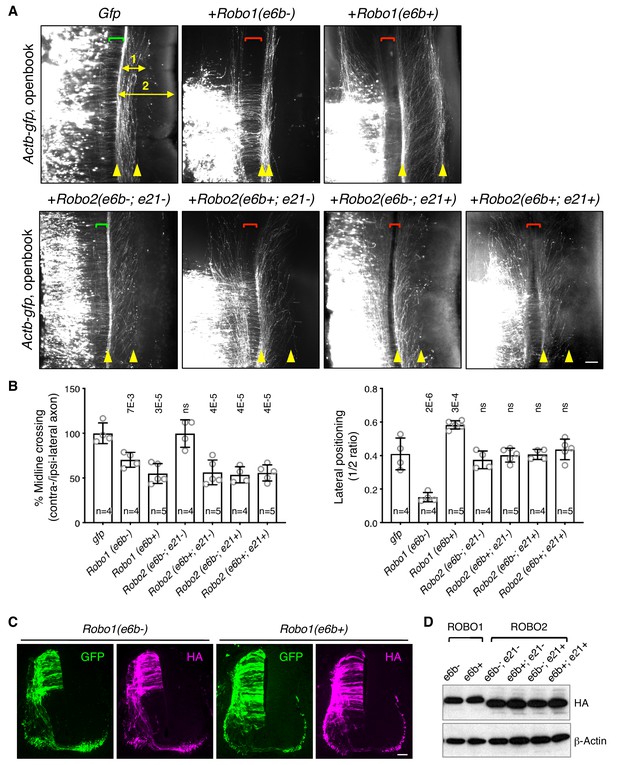
ROBO1/2 splice variants have distinct guidance activities.
(A) Openbook preparations of chicken spinal cords electroporated with Actb-gfp or with different Robo1 or Robo2 isoforms. Transfected neuronal cell bodies are oriented to the left, and the rostral end of the spinal cord is pointing up. Brackets indicate the midline. Arrowheads indicate the medial and lateral positions of postcrossing axons. Distance one is between the midline and the dorsolateral-most longitudinal axons and distance two is the total height of the spinal cord. Scale bar, 50 μm. (B) Quantification of midline crossing and lateral positioning of commissural axons in A. Lateral positioning is quantified as the ratio between distances one and two. Data are represented as the mean ± SD (one-way ANOVA and Bonferroni post test; animal numbers and p values are indicated; ns, not significant). (C) Immunohistochemistry of transverse sections of chicken spinal cords electroporated with HA-tagged Robo1 isoforms. Scale bar, 50 μm. (D) Western blotting analysis of lysates from chick spinal cords electroporated with HA-tagged Robo1/2 isoforms.
-
Figure 5—source data 1
Overexpression of Robo1/2 isoforms.
- https://doi.org/10.7554/eLife.46042.015
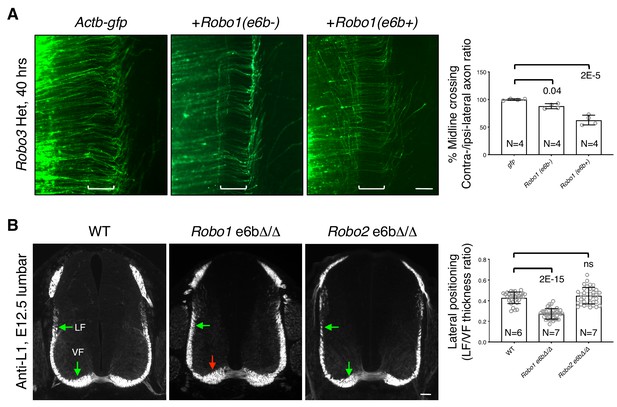
ROBO1/2 splice variants have distinct guidance activities.
(A) Openbook preparations of Robo3 heterozygous spinal cords electroporated with Actb-gfp, Robo1(e6b-), or Robo1(e6b+). Robo3 Het embryos were used for the overexpression in order to reduce the inhibition of exogenously expressed Robo1 cDNA and thus to potentiate the effect. Brackets indicate the midline. Actb-gfp predominantly labels ROBO3-positive commissural axons in cultured mouse embryos (Leggere et al., 2016). (B) Anti-L1 staining of spinal cord transverse sections in WT and homozygous exon 6b deletion embryos. Arrows indicate the lateral funiculus (LF) and ventral funiculus (VF). Robo1 e6bΔ/Δ mutants had enlarged VF (red arrow), while Robo2 e6bΔ/Δ mutants did not have any significant defect. Scale bars, 50 μm. Data are normalized to gfp in A and are represented as the mean ± SD (one-way ANOVA and Bonferroni post test; animal numbers and p values are indicated; ns, not significant).
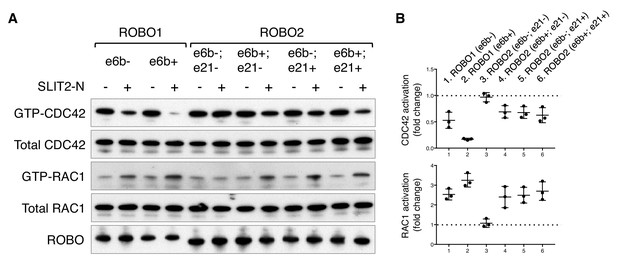
ROBO1/2 splice variants have distinct signaling activities.
(A) Regulation of CDC42/RAC1 activation by ROBO1/2 isoforms in COS-1 cells. SLIT2 N-term was added at 500 ng/ml for 10 min at 37°C. GTP-bound/activated CDC42 or RAC1 was pulled down by PAK-GST beads and detected by western blotting. (B) Data from three independent assays showing fold changes in CDC42/RAC1 activation upon SLIT2 N-term stimulation, represented as the mean ± SD.
-
Figure 6—source data 1
Activation of small GTPases by ROBO1/2 isoforms.
- https://doi.org/10.7554/eLife.46042.018
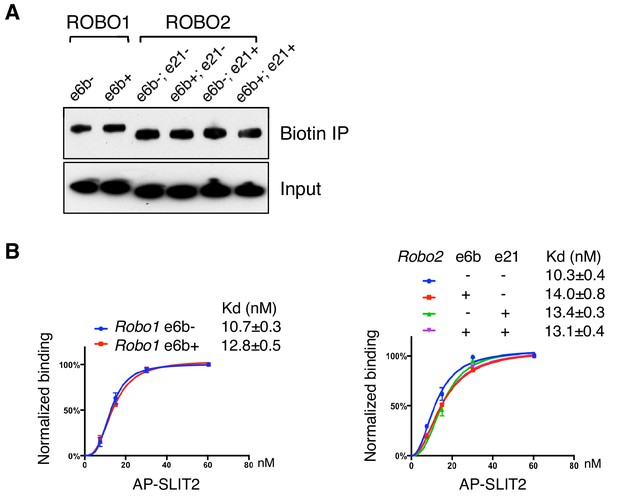
ROBO1/2 splice variants bind to SLIT2 with similar affinities.
(A) Cell surface biotinylation of ROBO1/2 variants expressed in COS-1 cells. Similar levels of the isoforms were present on the surface. (B) Binding between AP-SLIT2 (N-term) and ROBO1/2 isoforms expressed in COS-1 cells. Data are represented as the average of three independent binding assays, with binding normalized to the saturating level from each test, and as the mean ± SD.
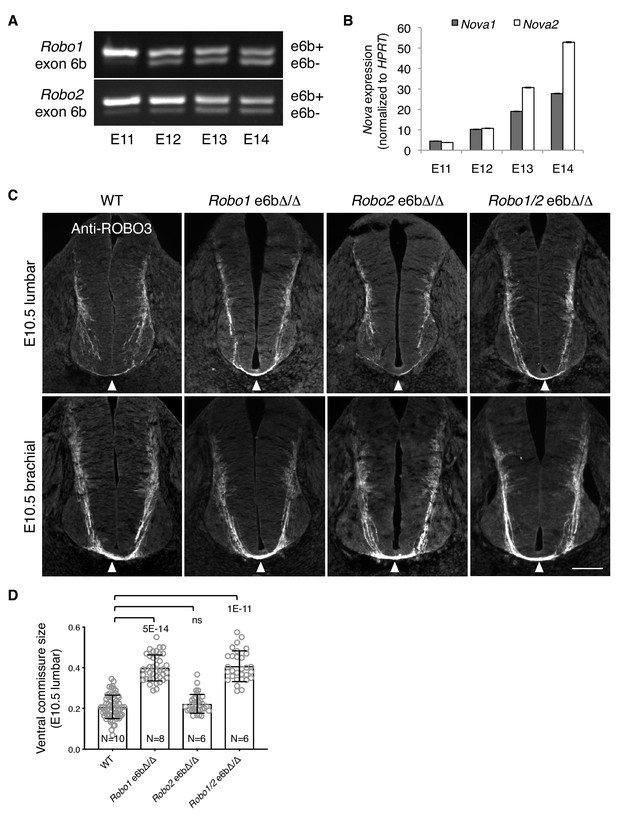
Alternative splicing of Robo1/2 is developmentally regulated to ensure proper timing of midline crossing.
(A,B) The expression levels of Robo1/2 exon 6b and Nova1/2, respectively, in dorsal spinal cord from E11 to E14 rats (equivalent of E9.5 to E12.5 in mice), as measured by RT-PCR. Rat embryos were used to allow separation of the dorsal and ventral halves of the spinal cord. (C) Anti-ROBO3 staining of transverse sections of WT and Robo e6bΔ/Δ spinal cords at E10.5 lumbar (top) and brachial (bottom) levels. Arrowheads indicate the ventral commissure consisting of midline-crossing axons. The ventral commissures in Robo1 e6bΔ/Δ and Robo1/2 e6bΔ/Δ mutants were thicker than in the WT controls at the lumbar level. All genotypes had comparable ventral commissure thickness at the brachial level. Scale bar, 50 μm. (D) Quantification of the ventral commissure size in C. Data are represented as the mean ± SD (one-way ANOVA and Bonferroni post test; animal numbers and p values are indicated; ns, not significant).
-
Figure 7—source data 1
Early projection of commissural axons in exon 6b deletion mutants.
- https://doi.org/10.7554/eLife.46042.021
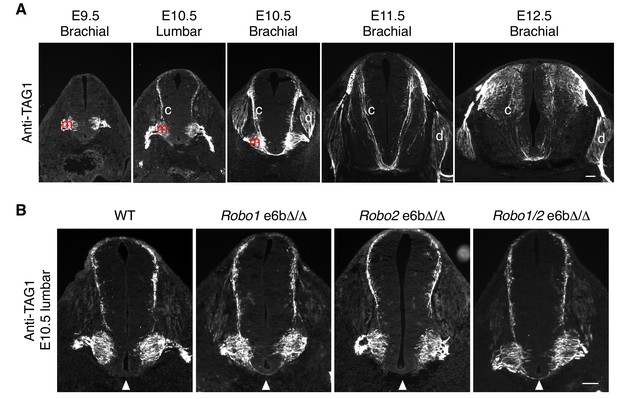
Developmental expression of TAG1 in mouse embryos.
(A) TAG1 expression in motor (m), commissural (c), and dorsal root ganglion (d) neurons from E9.5 to E12.5 in mice. TAG1 expression in motor columns was first observed at E9.5. By E11.5, the expression was no longer seen on motor neuron cell bodies, and has been shown to become restricted to the axons until E14 (Dodd et al., 1988). TAG1 expression in commissural neurons started at E10.5 at the lumbar level and receded by E12.5. In DRG neurons, TAG1 expression initiated at E10.5 at the brachial level, and has been reported to disappear in most DRG populations during late embryonic stages (Dodd et al., 1988). (B) Anti-TAG1 staining of transverse sections of WT and Robo e6bΔ/Δ spinal cords at E10.5 lumbar level. Arrowheads indicate the ventral commissure. TAG1 expression and thus the developmental stage of the embryos were comparable between genotypes. Scale bars, 50 μm.
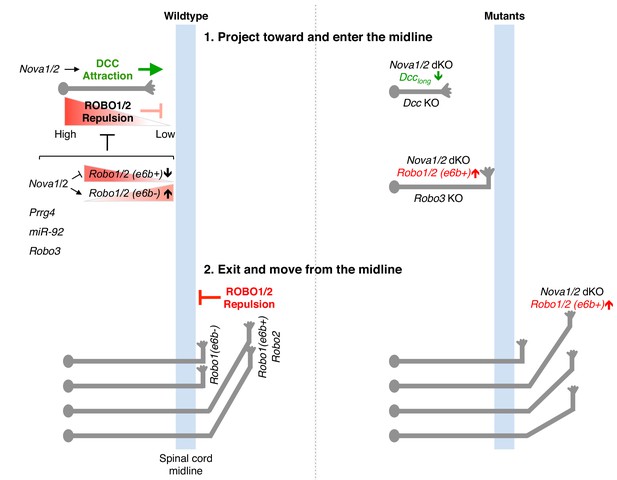
Model of NOVA1/2 regulation of commissural axon guidance.
As commissural axons approach and enter the midline, Netrin/DCC signaling promotes axonal outgrowth and midline attraction. Nova1/2 regulate Dcc alternative splicing by promoting the production of the full-length Dcclong isoform. In Nova1/2 dKO embryos, Dcclong is reduced and fewer axons are able to reach the midline, phenocopying Dcc KO mutants. In precrossing axons, SLIT/ROBO repulsion is repressed by multiple genes including Prrg4, miR-92, and Robo3 to allow midline entry. We show here that ROBO repulsion is not uniformly repressed precrossing, but is developmentally controlled such that ROBO repulsion is active early on to prevent premature crossing and is sufficiently inhibited later to allow crossing. This is achieved through the regulation of Robo1/2 alternative splicing at microexon 6b by the NOVA splicing factors. As NOVAs increase during development, the Robo1/2(e6b+) isoform, which has a greater repulsive activity, is reduced, whereas the Robo1/2(e6b-) isoform is upregulated. In Nova1/2 dKO embryos, e6b+ is abnormally elevated and the axons that are able to reach the midline often fail to enter. A similar and more complete defect is present in Robo3 KO embryos and partial loss of function in Robo3 and Nova1/2 synergistically blocks midline entry. Upon entering the midline, SLIT/ROBO repulsion is upregulated, which facilitates axonal exit, blocks recrossing, and guides postcrossing axons to appropriate lateral positions. Robo1(e6b-) directs the axons medially, while Robo1(e6b+) and Robo2 guide the axons dorsolaterally. Double knockout of Nova1/2 reduces the medial tract and increases the dorsolateral tract.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Mus musculus) | CD-1 | Charles River Laboratories | Crl:CD-1(ICR) | |
| Genetic reagent (Mus musculus) | Dcc knockout | Fazeli et al., 1997 | ||
| Genetic reagent (Mus musculus) | Nova1 knockout | Jensen et al., 2000 | ||
| Genetic reagent (Mus musculus) | Nova2 knockout | Saito et al., 2016 | ||
| Genetic reagent (Mus musculus) | Robo1 knockout | Long et al., 2004 | ||
| Genetic reagent (Mus musculus) | Robo3 knockout | Sabatier et al., 2004 | ||
| Genetic reagent (Mus musculus) | Robo1 e6b del | This paper | Figure 4—figure supplement 1 | |
| Genetic reagent (Mus musculus) | Robo2 e6b del | This paper | Figure 4—figure supplement 1 | |
| Genetic reagent (Mus musculus) | Robo1; Robo2 e6b del | This paper | Figure 4—figure supplement 1 | |
| Genetic reagent (Gallus gallus) | Chicken embryos | Charles River Laboratories | 10100326 | Specific pathogen-free fertile eggs |
| Genetic reagent (Rattus norvegicus) | Sprague Dawley rats | Charles River Laboratories | Crl:SD | |
| Cell line (Cercopithecus aethiops) | COS-1 | American Type Culture Collection | CRL-1650 | Free of mycoplasma contamination |
| Recombinant DNA reagent | Robo1 (e6b-) | This paper | HA tag at C-term; pCAGGS vector | |
| Recombinant DNA reagent | Robo1 (e6b+) | This paper | HA tag at C-term; pCAGGS vector | |
| Recombinant DNA reagent | Robo2 (e6b-; e21-) | This paper | HA tag at C-term; pCAGGS vector | |
| Recombinant DNA reagent | Robo2 (e6b-; e21+) | This paper | HA tag at C-term; pCAGGS vector | |
| Recombinant DNA reagent | Robo2 (e6b+; e21-) | This paper | HA tag at C-term; pCAGGS vector | |
| Recombinant DNA reagent | Robo2 (e6b+; e21+) | This paper | HA tag at C-term; pCAGGS vector | |
| Recombinant DNA reagent | Robo1 exon 6b splicing reporter | This paper | Figure 3; Figure 3—figure supplement 1E | |
| Recombinant DNA reagent | Robo2 exon 6b splicing reporter | This paper | Figure 3; Figure 3—figure supplement 1E | |
| Recombinant DNA reagent | Nova1-V5 | Leggere et al., 2016 | V5 tag at C-term; pCAGGS vector | |
| Recombinant DNA reagent | Nova2-V5 | Leggere et al., 2016 | V5 tag at C-term; pCAGGS vector | |
| Antibody | ROBO3 rabbit polyclonal | Sabatier et al., 2004 | IHC (1:1000) | |
| Antibody | L1 rat monoclonal | MilliporeSigma | MAB5272 | IHC (1:1000) |
| Antibody | ROBO1 rabbit polyclonal | Novus Biologicals | NBP2-20195 | IHC (1:1000); Western blot (1:500) |
| Antibody | ROBO2 rabbit polyclonal | Novus Biologicals | NBP1-81399 | IHC (1:1000) |
| Antibody | ROBO2 mouse monoclonal | R and D Systems | MAB3147 | Western blot (1:500) |
| Antibody | TAG1 mouse monoclonal IgM | Developmental Studies Hybridoma Bank | 4D7 | IHC (1:1000) |
| Antibody | HA rat monoclonal | Roche | 3F10 | IHC (1:1000); Western blotting (1:1000, HRP conjugated) |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.46042.023





