Cryo-EM structures of remodeler-nucleosome intermediates suggest allosteric control through the nucleosome
Figures
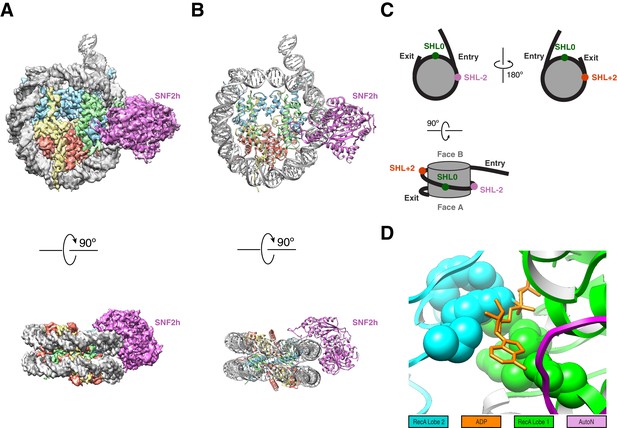
High resolution structure of SNF2h bound to a nucleosome with 60 bp of flanking DNA in the presence of ADP-BeFx and 140 mM KCl.
(A) Cryo-EM density map of SNF2h bound to the nucleosome at 3.4 Å from data recorded with a K2-summit camera. (B) Model built using the density in (A). (C) Cartoon representation of a nucleosome with asymmetric flanking DNA as in our structures. Super Helical Location (SHL) ± 2 as well as the entry and exit site DNAs are labeled. The SHL0 location is also labeled and is defined as the dyad. Faces A and B of the histone octamer are labeled in gray. (D) Zoom into the ATP-binding pocket of SNF2h with ADP in orange and represented with sticks. In spheres are the SNF2h residues that bind nucleotide with the helicase motif I in green and helicase motif VI in blue (Figure 3—figure supplement 4).
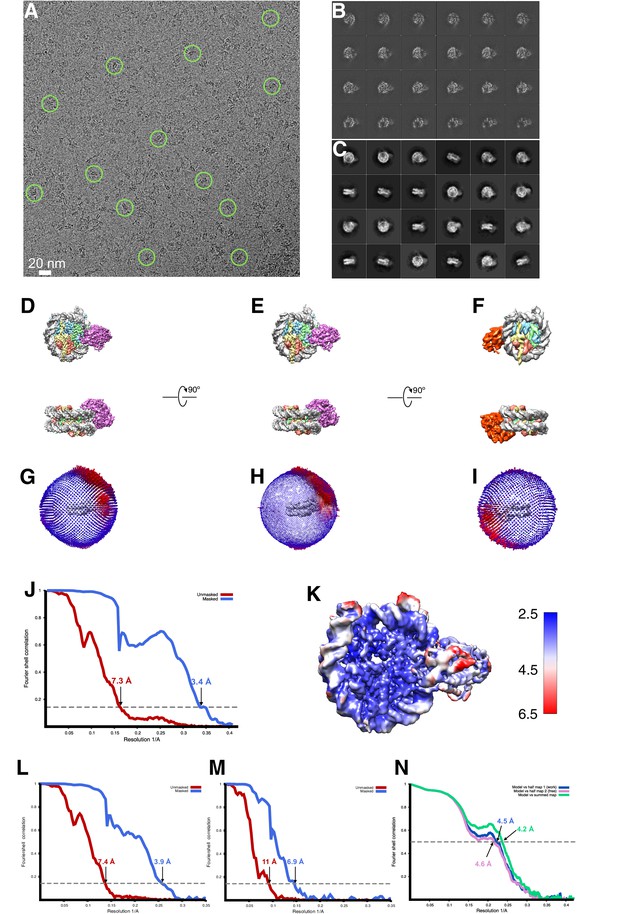
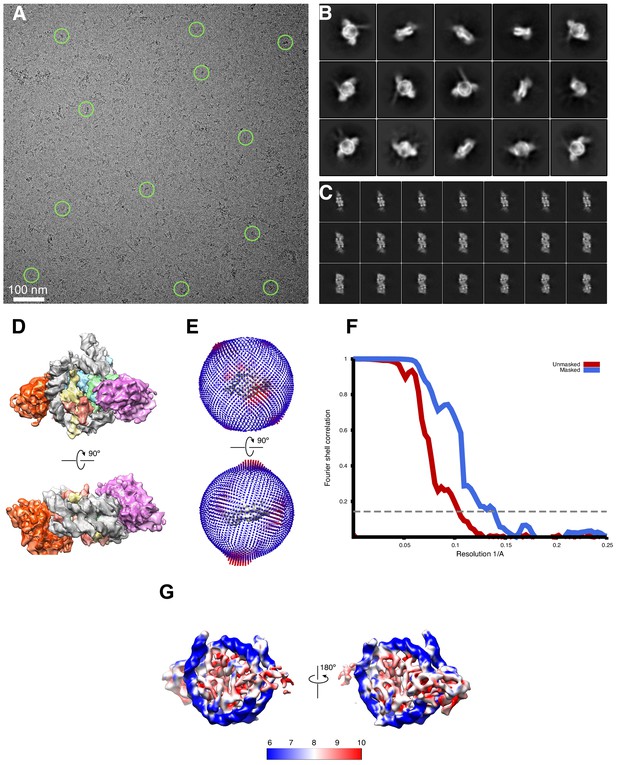
Cryo-EM analysis of singly bound SNF2h-nucleosome complexes (140 mM KCl) .
(A) A raw cryo-EM micrograph of single bound SNF2h-nucleosome complex recorded as described in Materials and methods. Examples of monodisperse particles are circled. (B) Slices through the unsharpened density of the 3.9 Å resolution map, which has SNF2h bound to the SHL-2 position, at different levels along the top view. (C) Typical 2D class averages of selected particles. The box size is 256 pixels. (D) Representative views of the 3D reconstruction of the 3.4 Å resolution . (E) Representative views of the 3.9 Å resolution map with SNF2h bound to the SHL-2 position. (F) Representative views of the 6.9 Å resolution map with SNF2h bound to the SHL+2 position. (G) Euler angle distribution of all particles used for the final 3D reconstruction of the 3.4 Å resolution. The length of each cylinder is proportional to the number of particles visualized from this specific orientation. (H) Euler angle distribution of all particles used for the 3.9 Å 3D reconstruction. The length of each cylinder is proportional to the number of particles visualized from this specific orientation. (I) Euler angle distribution of all particles used for the 6.9 Å resolution 3D reconstruction. (J) FSC curves of the 3.4 Å complex between two independently refined half maps before (red) and after (blue) masking in Cryosparc v2, indicated with resolutions corresponding to FSC = 0.143. (K) Final unsharpened 3.4 Å 3D density map of SNF2h-nucleosome complex colored with local resolution. (L) FSC curves of the 3.9 Å reconstruction between two independently refined half maps before (red) and after (blue) masking in Cryosparc, indicated with resolutions corresponding to FSC = 0.143. (M) FSC curves of the 6.9 Å complex (N) Cross-validation using FSC curves of the density map calculated from the refined model versus half map 1 (‘work’, blue), versus half map 2 (‘free’, pink) and versus summed map (green).
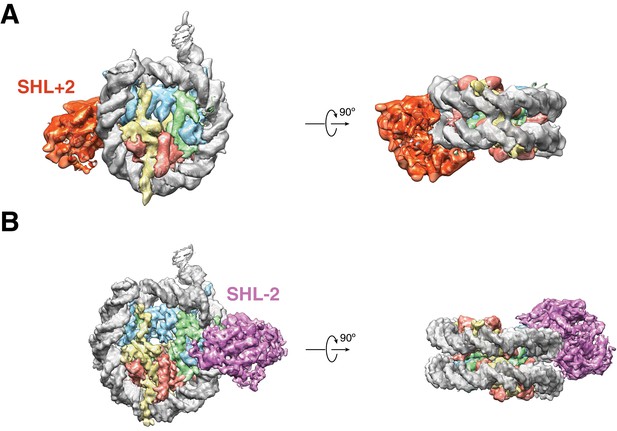
Cryo-EM Densities of SHL+2 and SHL-2 SNF2h-Nucleosome complexes obtained at 140 mM.
KCl Cryo-EM reconstructions of single-SNF2h bound nucleosomes from data recorded on a K2-summit direct electron bound at (A) SHL +2 at 6.9 Å resolution and (B) SHL-2 at 3.9 Å resolution.
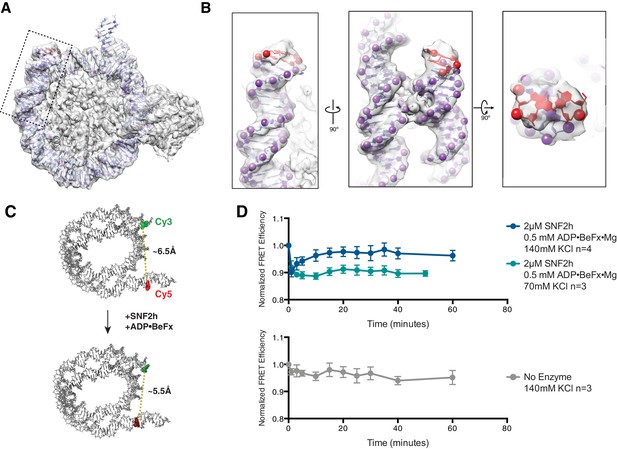
Cryo-EM reconstructions of the SNF2h-Nucleosome complexes at 140 mM KCl are translocated ~2 bp.
(A) High resolution structure from Figure 1—figure supplement 2B with Cryo-EM density fit with an atomic model. The additional 2 bp of DNA exiting the canonical nucleosome structure are highlighted in red. The box indicates the region zoomed to show greater detail in (B). (B) Zooms of the boxed region in A to show greater detail. The phosphates of the phosphodiester backbone are shown in spheres and fit precisely into the Cryo-EM density. (C) Schematic of the ensemble FRET experiment performed in (D). Nucleosomes were reconstituted on identical DNA sequence used to obtain Cryo-EM reconstructions and labeled with two fluorophores: cy3 on the 5’ phosphate at the DNA exit side of the nucleosome with a three carbon flexible linker and with cy5 on C8 of the cytosine base at position 149 near the entry side of the nucleosome via a six carbon flexible linker. Upon addition of SNF2h and ADP-BeFx, if the nucleosome is translocated as in the Cryo-EM reconstruction, the two labeled bases will be brought closer together in 3D space. This change in DNA conformation could be read out as a very small increase in FRET efficiency between the two labels. (D) Ensemble FRET experiment described in (C). FRET efficiency values were normalized to the FRET efficiency of the nucleosome prior to adding SNF2h and nucleotide. Points represent the mean of the indicated number independent experiments and error bars represent the standard error of the mean. Addition of SNF2h and ADP-BeFx produces an instantaneous drop in FRET efficiency that slowly increases in the presence 140 mM KCl but not in the presence of 70 mM KCl (top). The instantaneous drop in FRET efficiency may be due to subtle changes in the conformation of DNA immediately upon SNF2h binding or due to previously reported changes in the photophysics of the cy3 dye labeled at this position (Racki et al., 2009). The subsequent increase in FRET reflects structural changes in nucleosomal DNA which, in principle, could reflect many types of motions such as subtle changes in DNA trajectory. However, the increase observed is consistent with the small ~2 bp translocation captured in our cryo-EM reconstructions. Changes in the photophysical properties of the cy5 label were ruled out as contributing to the change in FRET efficiency as there was no change in fluorescence intensity when cy5 was directly excited (data not shown). No increase in FRET signal was seen when only SNF2h buffer was added (bottom).
-
Figure 1—figure supplement 3—source data 1
Values used to obtain plots in D.
- https://doi.org/10.7554/eLife.46057.006
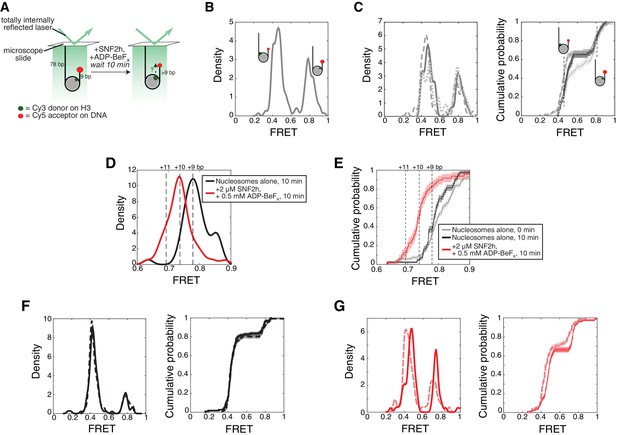
By a single molecule assay, SNF2h induces a change in FRET under the 140 mM KCl conditions, consistent with a movement of the nucleosomal DNA.
(A) Setup of the single molecule assay. Nucleosomes with 78 bp flanking DNA on one side are attached to the surface of a microscope slide and imaged using total internal reflection fluorescence microscopy. The nucleosomes have 9 bp flanking DNA on the other side, with a Cy5 dye at the end of the 9 bp. Movement of the DNA will change the distance between the Cy3 and Cy5 dyes, changing the observed FRET. (B) Kernel density estimation (KDE) plot of the starting FRET values of 179 nucleosomes, without SNF2h, combined from the three independent replicates shown in (C). FRET values cluster into two peaks, near 0.8 FRET and 0.45 FRET, corresponding to two possible labeling schemes: a Cy3 dye on the histone H3 closer to the Cy5-labeled DNA end (proximal labeling), or a Cy3 dye on the other copy of histone H3 (distal labeling). As demonstrated in Zhou et al. (2018), the distally-labeled population is relatively insensitive to changes in the length of the DNA separating the Cy5 from the edge of the nucleosome, and so is not shown in (D) and (E). (C) KDEs (left) and empirical cumulative distribution functions (CDFs; right) of starting nucleosomal FRET values from three independent replicates. Note that the FRET value of the midpoint of a peak in a KDE corresponds to the FRET value where the slope of the CDF is steepest. Thus the distal and proximal peaks in the KDEs appear as two regions of steep slopes in the CDFs. For two data sets with similar peak locations, the FRET value (position along the x-axis) where the slope of the CDF is steepest will be similar. (D) (black) KDE of FRET values for 43 proximally-labeled nucleosomes after 10 min of incubation without SNF2h, and (red) 48 proximally-labeled nucleosomes after a 10 min incubation in the presence of 2 µM SNF2h and 0.5 mM ADP-BeFx. Dashed lines labeled ‘+11’, ‘+10’, and ‘+9’ indicate expected FRET values for nucleosomes with 11, 10, or 9 bp of flanking DNA between the Cy5 and the edge of the nucleosome, based on the calibration curve derived in Zhou et al. (2018). (E) Empirical cumulative distribution functions (CDFs) of the data in B and C. (F) Same as (C) but for the two replicates of nucleosomes alone after a 10 min incubation. The two replicates have 115 and 117 nucleosomes each (with the distally labeled population included). (G) Same as (C) but for the two replicates of nucleosomes plus SNF2h and ADP-BeFx after 10 min. The two replicates have 72 and 83 nucleosomes each. Shaded regions in CDFs in all panels represent an estimate of the error based on a bootstrapping method (Gamarra et al., 2018; Zhou et al., 2018).
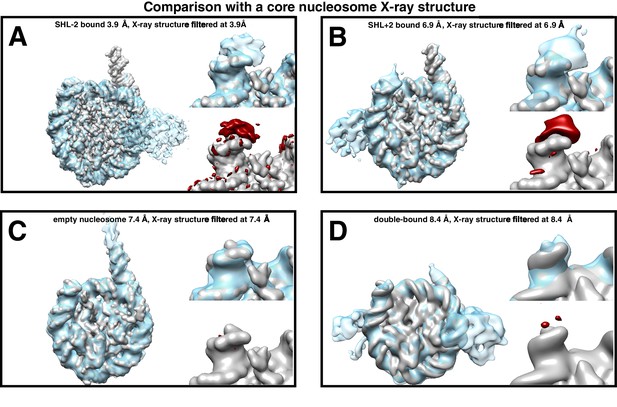
Difference maps to test for extra density of DNA at exit side of SNF2h-nucleosome complexes.
(A–C) Cryo-EM density map of the nucleosome either with SNF2h bound at (A) SHL-2 and (B) SHL+2 with 140 mM KCl with the simulated density map of nucleosome without two additional base-pairs filtered to a common resolution (A: 3.9 Å, B: 6.9 Å). (C) Cryo-EM density map of nucleosome without added SNF2h with the simulated density map of nucleosome without two additional base-pairs filtered to a common resolution (7.4 Å). For A–C), left: each panel shows an overview of the EM density (light blue) overlaid onto the simulated nucleosome map. Top right: enlarged view centered on the DNA exit site. Bottom right: enlarged view of the DNA exit site showing the difference density between the simulated map and the EM density at σ = 7. (D) Left: cryo-EM density map of doubly-bound nucleosome with 70 mM KCl is overlaid with the cryo-EM density map of the simulated nucleosome map. Both maps are filtered to a resolution of 8.4 Å. Top right: an enlarged view of the DNA end. Bottom right: density map of nucleosome alone is overlaid with the difference density calculated between the two, and visualized at σ = 7.
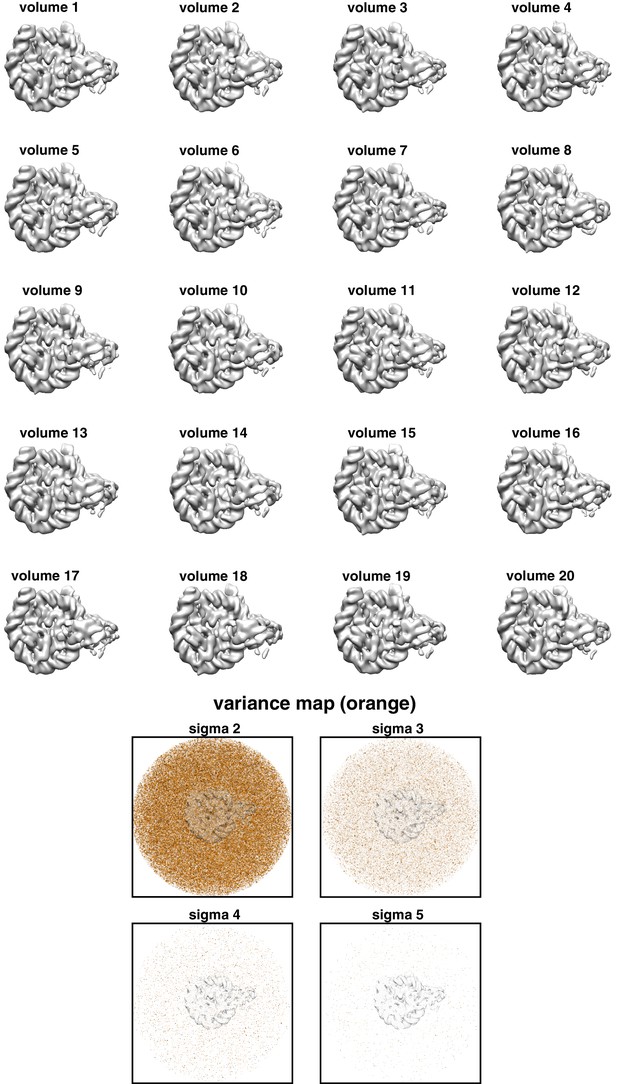
Bootstrapped maps of SNF2h-nucleosome complex.
20 bootstrapped maps were calculated by using 20 subsets of 5000 particles bootstrapped from the particles that were used to calculate the 3.9 Å reconstruction. Slices through the variance map at different sigma levels (2, 3, 4 and 5) are shown, and reveal no significant variance. Transparent overlay of the SHL-2 side 3.9 Å density is shown for orientation.
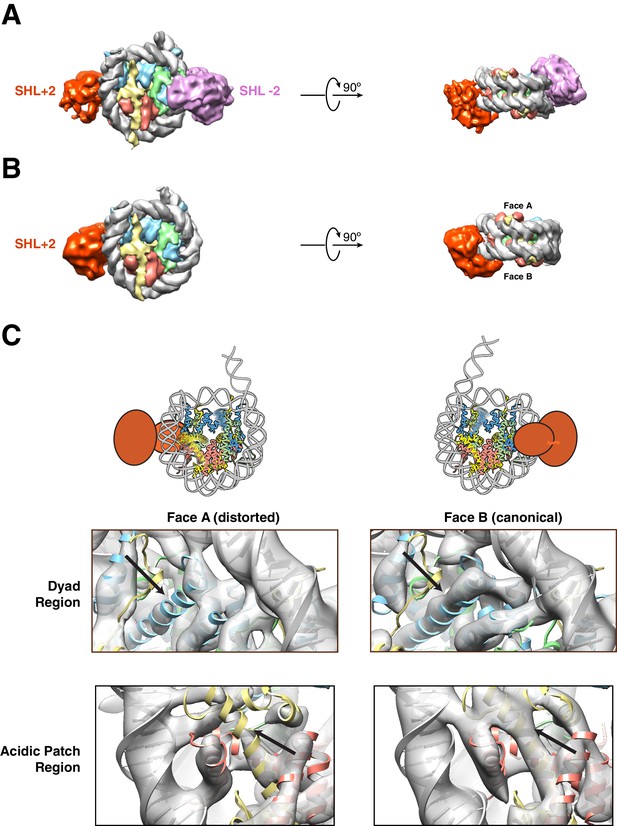
Structures of SNF2h bound to a nucleosome with 60 bp of flanking DNA in the presence of ADP-BeFx and 70 mM KCl.
(A–C) Cryo-EM density maps of SNF2h bound to the nucleosome recorded with a scintillator-based camera (A) Doubly bound SNF2h-nucleosome complex at 8.4 Å resolution. (B) Singly bound SNF2h at SHL+2. (C) Comparison of the Cryo-EM density on the two faces of the nucleosome. Face A of the nucleosome (left column) has weaker EM density at the histone H2A acidic patch (bottom row) and the α2 helix of H3 (top row) when compared to face B (right column) at the same contour level. The black arrows point to the helices that show altered densities in Face A vs. Face B. The regions of increased dynamics are also shown schematically as blurry helices in cartoons of the nucleosome above the densities for Face A and Face B.
Cryo-EM analysis of doubly bound SNF2h-nucleosome complexes obtained at 70 mM KCl.
(A) A raw cryo-EM micrograph of doubly bound SNF2h-nucleosome complex recorded as described in Materials and Methods. (B) Typical 2D class averages of selected particles. The box size is 300 pixels. (C) Slices through the unsharpened density at different levels along the side view. (D) Representative views of the doubly bound complex. (E) Euler angle distribution of all particles used for the final 3D reconstruction. The length of each cylinder is proportional to the number of particles visualized from this specific orientation. (F) FSC curves between two independently refined half maps before (red) and after (blue) masking in Cryosparc, indicated with resolutions corresponding to FSC = 0.143. (G) Final unsharpened 8.4 Å 3D density map of the doubly-bound complex colored with local resolution.
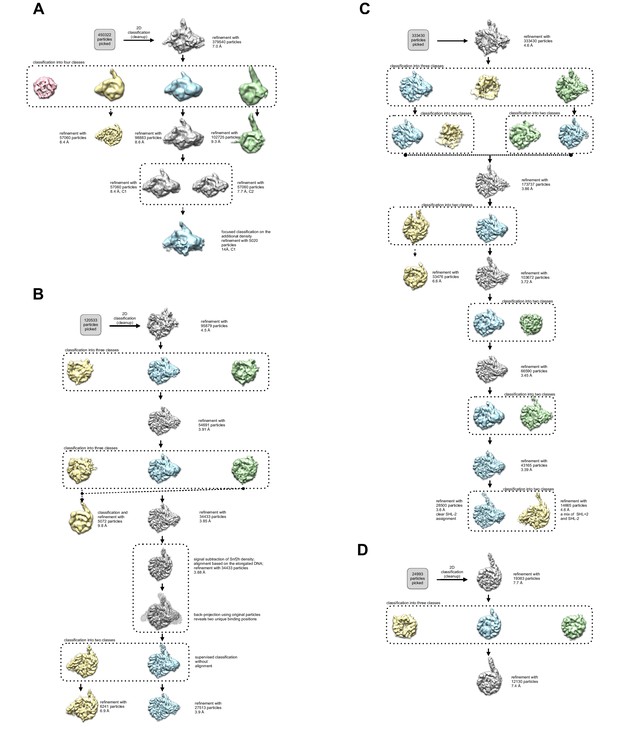
3D Classification and refinement.
(A–B, D). The flowchart of classification and refinement procedures using RELION and Cryosparc is shown. Following 1 round of 2D classification, each dataset was subjected to 3D refinement and subsequent classification. (A) Scintillator-based doubly-bound dataset. (B) Direct electron detector-based SNF2h-nucleosome complex dataset that resulted in single-bound flanking DNA proximal and distal complexes as well as unbound nucleosomes. (C) Recalculated direct electron detector-based SNF2h-nucleosome complex dataset, resulting in the 3.4 Å consensus Coulomb potential map. (D) Direct electron detector-based data set for core nucleosome with flanking DNA without added SNF2h.
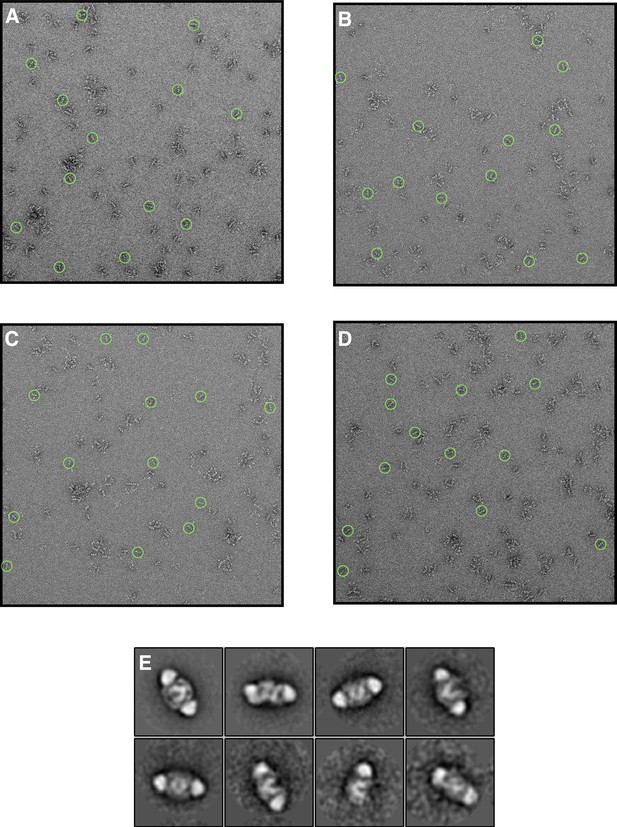
Negative stain EM of SNF2h in the presence of ADP-BeFx and 140 mM KCl.
(A–D) Raw micrographs of negatively stained sample performed under the same buffer conditions as the dataset in Figure 1. Complexes were assembled identically to the high resolution EM condition and then diluted 40-fold in buffer immediately before applying the sample to the grid to permit visualization by negative stain EM. The final concentration applied to the grid was 15 nM nucleosomes and 31 nM SNF2h. Examples of monodisperse particles are circled; (E) Selected class averages from (A–D) showing that in these conditions, the majority of nucleosomes are doubly-bound.
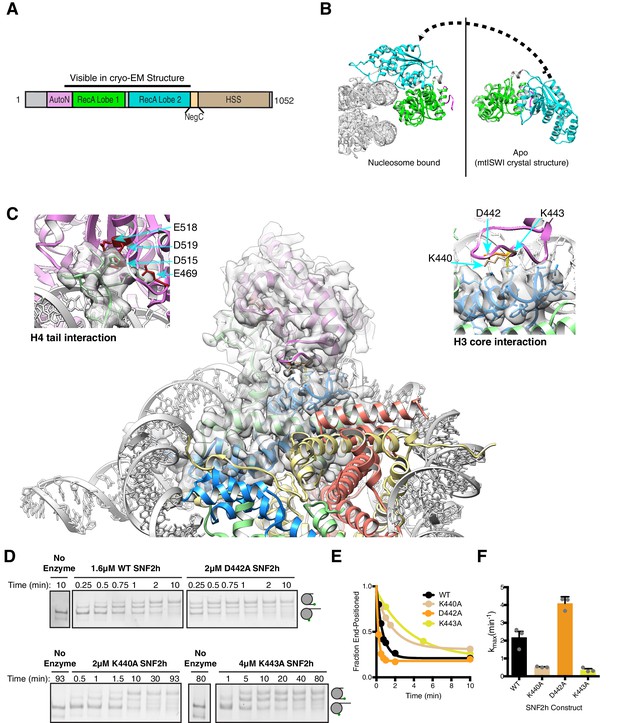
Interactions of SNF2h with the histone proteins.
(A) Domain diagram of SNF2h. (B) Conformational changes in SNF2h associated with nucleosome binding. SNF2h is colored according to the domain diagram. The apo structure is the Myceliophtora thermophila ISWI crystal structure (PDBID: 5JXR). (C) Middle. High resolution SNF2h-nucleosome structure from Figure 1 enlarged to show details of the interactions with the histone proteins. Colored in red on SNF2h are the acidic residues contacting the histone H4 tail. Colored in orange, tan, and yellow are the residues mutated in this study. Left. Enlarged to show details of the H4 tail interaction. Right. Enlarged to show details of the H3 core interaction. (D) Native gel remodeling assay of SNF2h constructs. Cy3-DNA labeled nucleosomes were incubated with saturating concentrations of enzyme and ATP and resolved on a native 6% polyacrylamide gel. (E) Quantifications of the data in (D) zoomed on the x-axis to show effects more clearly and fit to a single exponential decay. Un-zoomed plots are in Figure 3—figure supplement 6. (F) Rate constants derived from remodeling assays. Bars represent the mean and standard error from three experiments.
-
Figure 3—source data 1
Values plotted in E and F.
- https://doi.org/10.7554/eLife.46057.022
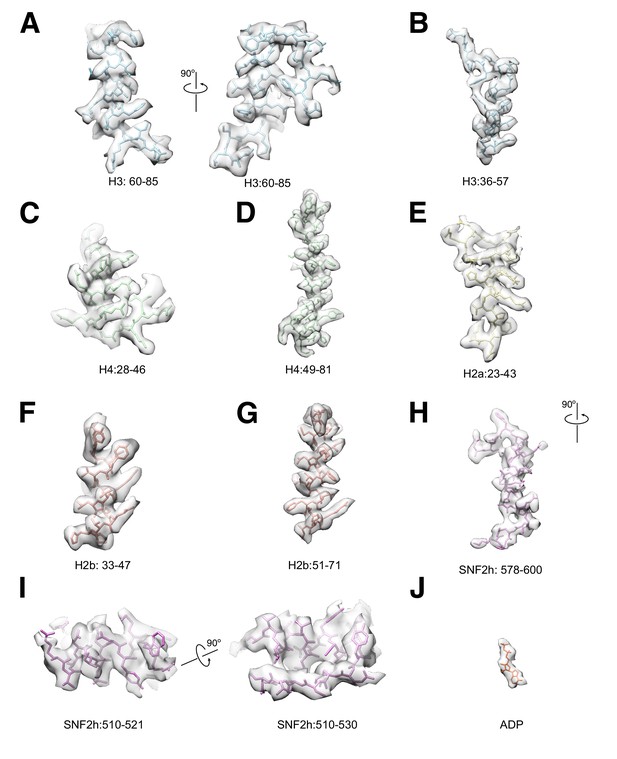
Selected Cryo-EM protein densities.
Representative 3.4 Å resolution cryo-EM densities of SNF2h and core histones superimposed on the atomic model. The density maps are shown as semi-transparent, and the model is colored according to the color scheme in the main figures ((A–B) H3: blue, (C–D) H4: green, (E) H2a: yellow, (F–G) H2b: red, (H–I) SNF2h: purple, (J) ADP: orange).
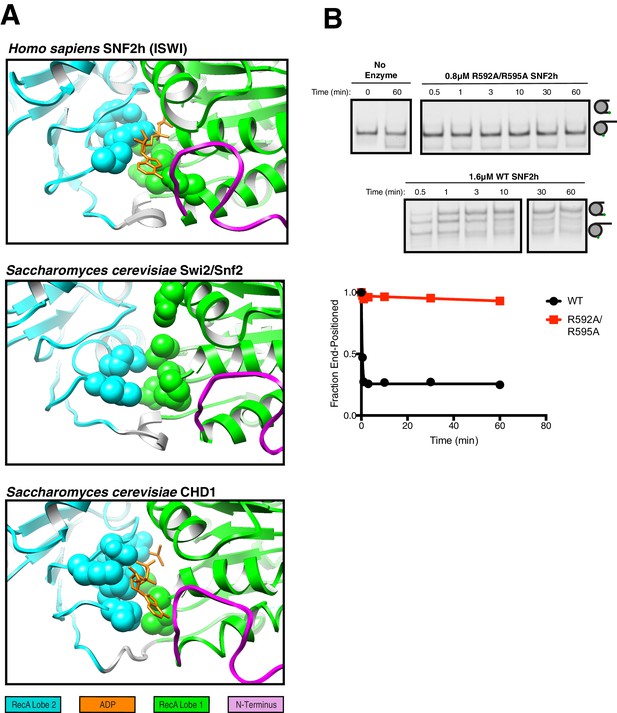
Comparison of ATP-binding pockets of SNF2h with CHD1 and Swi2/Snf2 and functional validation of SNF2h ATP-binding pocket.
(A) Top. Close up view of the SNF2h ATP-binding pocket from the 3.4 Å structure with ADP fit in the canonical binding site. Middle. Close up view of the ATP-binding pocket from S. cerevisiae Swi2/Snf2 Cryo-EM structure (Liu et al., 2017). Bottom. Close up view of the ATP binding pocket from S. cerevisiae CHD1 Cryo-EM structure with ADP-BeFx fit in the canonical binding site (Sundaramoorthy et al., 2018). The conserved helicase motif I and helicase motif VI that are required for ATP hydrolysis are shown in space fill. (B) Mutation of the putative arginine fingers to alanine abolishes sliding activity. Top. Native gel remodeling assay with saturating concentrations of SNF2h. Bottom. Quantification of the gel fit to a single exponential decay.
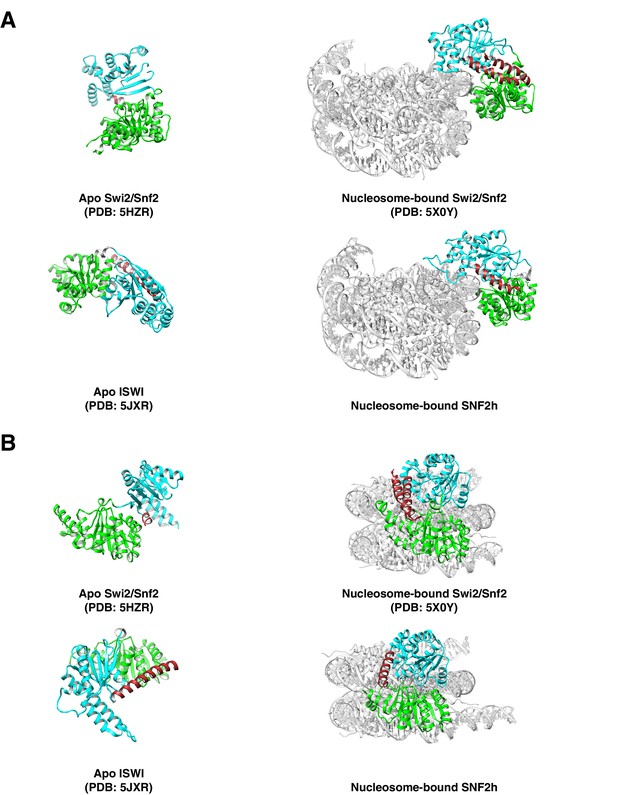
Brace helix comparisons.
(A, B). Comparison of bound and unbound conformations of Swi2/Snf2 (top A, (B) and ISWI (bottom A, (B) remodelers in two views. Left: Apo structures of Myceliophthora thermophila Swi2/Snf2 ATPase and ISWI ATPase (Xia et al., 2016; Yan et al., 2016). Right: nucleosome-bound S. cerevisiae Swi2/Snf2 and H. sapiens Snf2h (Liu et al., 2017). Brace helix is shown in brown.

Multiple sequence alignment of the ATPase domains of selected members of chromatin remodeling families.
Alignment of the ATPase domains of members of the ISWI (blue), SWI/SNF (red), and CHD families (yellow). Below the sequence, canonical SF2 nucleic acid helicase motifs are annotated (Flaus et al., 2006). Residues mutated in this study are highlighted in yellow. Red (•) denotes K443.
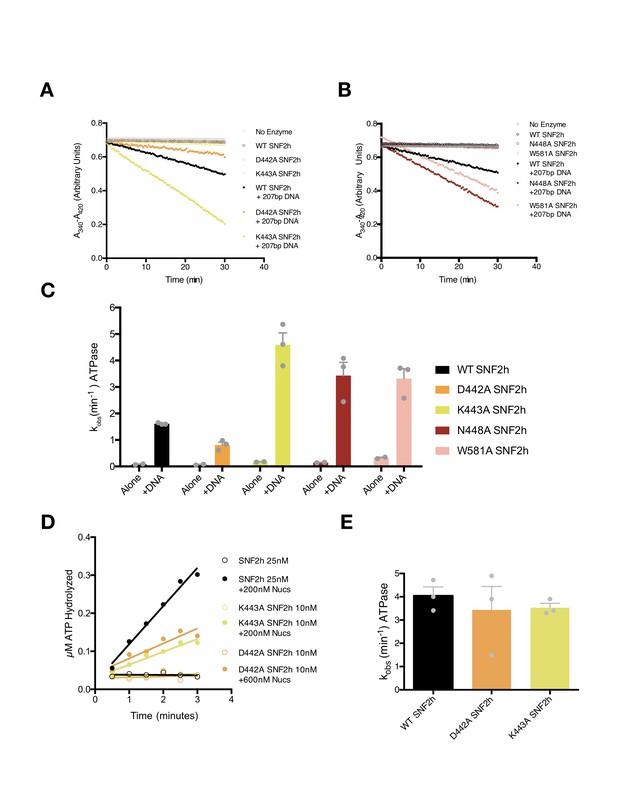
ATPase activities of point mutants in this study.
(A–B) Example absorbance plots of the NADH coupled ATPase assay to measure DNA-stimulated ATPase activity. Reactions were performed with 800 nM of the indicated SNF2h constructs either without DNA (open circles) or with saturating concentrations (208 nM) of the 207 bp DNA used to assemble nucleosomes in our Cryo-EM reconstructions. (C) Mean and standard error of DNA-stimulated ATPase rate measurements for n = 3 experiments. All mutations tested had modest effects on DNA-stimulated hydrolysis rates (~2 fold or less). (D) Example plot of radioactive ATP hydrolysis reactions used to measure nucleosome-stimulated ATPase activity. Reactions were performed with the indicated concentrations of SNF2h and nucleosomes without flanking DNA. The concentrations of nucleosomes used were confirmed to be saturating by this assay while the concentration of ATP-MgCl2 used was subsaturating (20 µM). (E) Mean and standard error of nucleosome-stimulated ATPase rate measurements from n = 3 measurements. All rate constants measured were within error of each other.
-
Figure 3—figure supplement 5—source data 1
Values used to obtain plots.
- https://doi.org/10.7554/eLife.46057.020
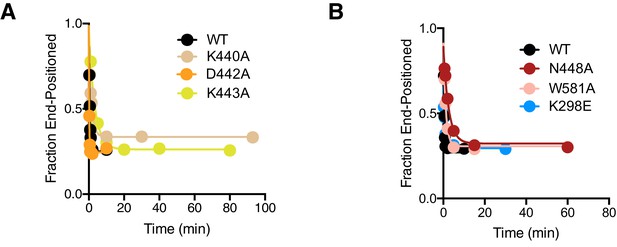
Full fits of native gel remodeling assays.
https://doi.org/10.7554/eLife.46057.021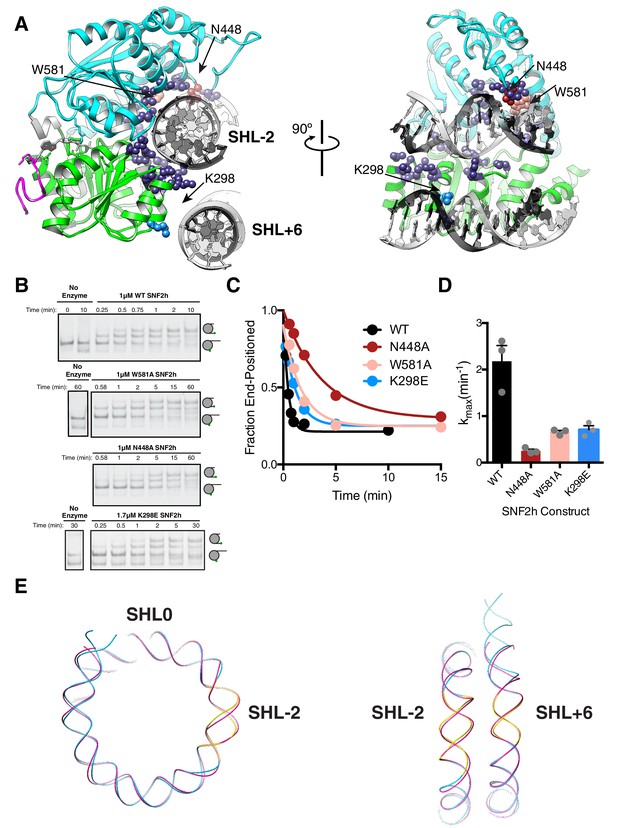
DNA contacts in the SNF2h-Nucleosome structure.
(A) SNF2h residues contacting the nucleosome are shown in spheres. In light blue is a contact with the second gyre near SHL + 6. In red, pink, and light blue are residues mutated in this study. (B) Native gel remodeling assay of DNA contact mutants. (C) Quantification of gels in (B). The x-axis is zoomed to show detail. The un-zoomed plots are in Figure 3—figure supplement 6. (D) Mean and standard error of rate constants derived from three experiments. (E) Comparison of the nucleosomal DNA in the SNF2h-nucleosome structure (blue) with the unbound structure (magenta) (PDBID: 1K × 5). Contacts with the remodeler at SHL-2 and SHL+6 are highlighted in yellow.
-
Figure 4—source data 1
Values plotted in C and D.
- https://doi.org/10.7554/eLife.46057.024
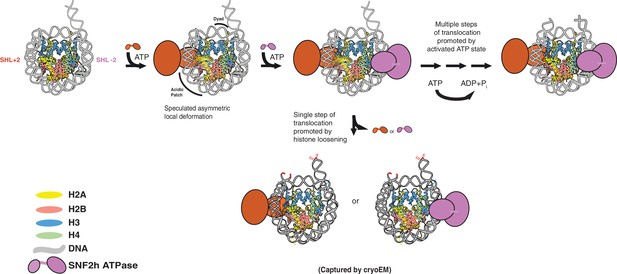
Speculative model that places SNF2h-nucleosome Cryo-EM structures within SNF2h reaction cycle .
Two protomers of SNF2h bind to the nucleosome along with ATP. Based on previous work, the directionality of nucleosome sliding is determined by the motor that engages the longer flanking DNA (Leonard and Narlikar, 2015). By this model, the SNF2h motor bound at SHL+2 (orange protomer) will be the active motor and determine the direction of sliding because it would contact the 60 bp flanking DNA with its HSS domain (Leonard and Narlikar, 2015). For simplicity, the HSS domain is not shown. Binding of the SHL+2 protomer asymmetrically deforms the acidic patch and histone H3 near the dyad on the opposite face of the histone octamer. The second protomer can bind at SHL-2, but cannot act because deformation of the acidic patch inhibits its ability to slide nucleosomes. The SNF2h complex with the deformed octamer represents an intermediate that is poised for translocation. Processive DNA translocation is enabled by successive ATP hydrolysis cycles from this activated intermediate, moving DNA in 1–2 bp fundamental increments. We speculate that the cryo-EM structure captured at low salt represents the deformed intermediate, while the structure captured at high salt represents a collapsed product state in which the nucleosome is translocated by 2 bp (translocated bases are highlighted in red).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | Rosetta (DE3) | Millipore sigma | 70954 | Chemically competent cells |
| Strain, strain background (Escherichia coli) | BL1 (DE3) pLysS | Agilent Technologies | 200132 | Chemically competent cells |
| Recombinant DNA reagent | core Widom 601 (bold) andflanking DNAsequences | (Lowary and Widom, 1998) | 5’-CGGCCGCCCTGGAGAATCCCGGTGCCGAGGCCGCTCAATTGGTCGTAGACAGCTCTAGCACCGCTTAAACGCACGTACGCGCTGTCCCCCGCGTTTTAACCGCCAAGGGGATTACTCCCTAGTCTCCAGGCACGTGTCAGATATATACATCCTGTGCATGTATTGAACAGCGACCTTGCCGGTGCCAGTCGGATAGTGTTCCGAGCTCCCACTCTAGAGGATCCCCGGGTACC-3’ | |
| Recombinant DNA reagent | 601 plasmid | (Lowary and Widom, 1998) | PCR template | |
| Recombinant DNA reagent | pBH4-SNF2h | (Leonard and Narlikar, 2015) | Expression plasmid | |
| Recombinant DNA reagent | Pet3a-H2A | (Yang et al., 2006) | Expression plasmid | |
| Recombinant DNA reagent | Pet3a-H2B | (Yang et al., 2006) | Expression plasmid | |
| Recombinant DNA reagent | Pet3a-H3 | (Yang et al., 2006) | Expression plasmid | |
| Recombinant DNA reagent | Pet3a-H4 | (Yang et al., 2006) | Expression plasmid | |
| Sequence-based reagent | 601 core forward primer | IDT | 5’-CTGGAGAATCCCGGTGCCG-3’ | |
| Sequence-based reagent | 601 + 60 reverse primer | IDT | 5’-AGAGTGGGAGCTCGG AACAC-3’ | |
| Sequence-based reagent | Cy3- 601 core forward primer | IDT | 5’-/Cy3/CTGGAGAATCCCGGTGCCG-3’ | |
| Sequence-based reagent | Cy5- 601–9 forward primer | TriLink Biotechnologies | 5’-{Cyanine5-C6-NH}GCGGCC GCCCTGGAGAATCC-3’ | |
| Sequence-based reagent | Bio- 601 + 78 reverse primer | IDT | 5’-/5BioTeg/GGTACCCGG GGA TCCTCTAGAG-3’ | |
| Sequence-based reagent | 601–120F C149cy5 | Iba | 5’-GGCACGTGTCAGATATATACATCCTGTG5ATGTATTGAACA-3’ 5 = cy5 C6-Amino-2'deoxycytidine | |
| Sequence-based reagent | SNF2h K298E forward primer | IDT | 5’-GAGAAGTCTGTGTTCGAAAAATTTAATTGGAG-3’ | |
| Sequence-based reagent | SNF2h K298E reverse primer | IDT | 5’-CTCCAATTAAATTTTTCGAAC ACAGACTTCTC-3’ | |
| Sequence-based reagent | SNF2h K440A forward primer | IDT | 5’-CTCAACTCAGCAGGCG CGATGGACAAAATGAGG-3' | |
| Sequence-based reagent | SNF2h K440A reverse primer | IDT | 5’-CCTCATTTTGTCCATCGCGCCTGCTGAGTTGAG-3' | |
| Sequence-based reagent | SNF2h D442A forward primer | IDT | 5’-CTCAGCAGGCAAGATGGCGAAAATGAGGTTATTGAAC-3' | |
| Sequence-based reagent | SNF2h D442A reverse primer | IDT | 5’-GTTCAATAACCTCATTTTCGCCATCTTGCCTGCTGAG-3' | |
| Sequence-based reagent | SNF2h K443A forward primer | IDT | 5’-CAGCAGGCAAGATGGACGCGATGAGGTTATTGAACATC-3' | |
| Sequence-based reagent | SNF2h K443A reverse primer | IDT | 5’-GATGTTCAATAACCTCATCGCGTCCATCTTGCCTGCTG-3' | |
| Sequence-based reagent | SNF2h N448A forward primer | IDT | 5’-GACAAAATGAGGTTATTGGCGATCCTAATGCAGTTGAG-3' | |
| Sequence-based reagent | SNF2h N448A Reverse primer | IDT | 5’-CTCAACTGCATTAGGATCGCCAATAACCTCATTTTGTC-3' | |
| Sequence-based reagent | SNF2h W581A forward primer | IDT | 5’-GTAATTTTGTATGATTCTGATGCGAATCCCCAAGTAGATCTTC-3' | |
| Sequence-based reagent | SNF2h W581A reverse primer | IDT | 5’-GAAGATCTACTTGGGGATTCGCATCAGAATCATACAAAATTAC-3' | |
| Peptide, recombinant protein (Homo sapiens) | SNF2h | (Leonard and Narlikar, 2015) | ||
| Peptide, recombinant protein (Xenopus laevis) | Histone H2A | (Luger et al., 1997) | ||
| Peptide, recombinant protein (Xenopus laevis) | Histone H2B | (Luger et al., 1997) | ||
| Peptide, recombinant protein (Xenopus laevis) | Histone H3 | (Luger et al., 1997) | ||
| Peptide, recombinant protein (Xenopus laevis) | Histone H4 | (Luger et al., 1997) | ||
| Peptide, recombinant protein (Xenopus laevis) | Histone H3 33C | (Rowe and Narlikar, 2010) | ||
| Peptide, recombinant protein (Escherichia virus T4) | T4 DNA Ligase | New England Biolabs | Cat #: M0202L | |
| Peptide, recombinant protein (Bos taurus) | Catalase | Sigma | Cat #: E3289 | |
| Peptide, recombinant protein (Aspergillus niger) | Glucose oxidase | Sigma | Cat #: G2133 | |
| Peptide, recombinant protein (Oryctolagus cuniculus) | Lactate dehydrogenase | Sigma | Cat #: 427217 | |
| Peptide, recombinant protein (Oryctolagus cuniculus) | Pyruvate Kinase | Sigma | Cat #: 10128155001 | |
| Chemical compound, drug | ATP | GE | Cat #: 27-2056-01 | |
| Chemical compound, drug | γ-32P-ATP | Perkin Elmer | Cat #: Blu002Z250uC | |
| Chemical compound, drug | ADP | Millipore sigma | Cat #: 117105 | |
| Chemical compound, drug | Cy3-maleimide | Lumiprobe | Cat #: 21080 | |
| Chemical compound, drug | Cy5-maleimide | Lumiprobe | Cat #: 43080 | |
| Chemical compound, drug | dNTPs | Allstar Scientific | Cat #: 471-5DN | |
| Chemical compound, drug | N-(2-aminoethyl)−3-aminopropyltrimethoxysilane | United Chemicals | Cat #: A0700 | |
| Chemical compound, drug | mPEG-SVA | Laysan Bio | ||
| Chemical compound, drug | biotin-PEG-SVA | Laysan Bio | ||
| Chemical compound, drug | acetylated BSA | Promega | Cat #: R3691 | |
| Chemical compound, drug | Neutravidin | Life Technologies | A2666 | |
| Chemical compound, drug | Trolox | Sigma | Cat #: 238813 | |
| Chemical compound, drug | 10XTBE | Bio-Rad | Cat #: 161–0770 | |
| Chemical compound, drug | Acrylamide/Bis-acrylamide | Bio-rad | Cat #: 161–0146 | |
| Chemical compound, drug | HEPES | Fisher | Cat #: BP310 | |
| Chemical compound, drug | Tris Base | Thermo Fisher | Cat #: BP1525 | |
| Chemical compound, drug | NaCl | RPI | Cat #: S23020 | |
| Chemical compound, drug | KCl | Sigma | Cat #: P3911 | |
| Chemical compound, drug | MgCl2 | RPI | Cat #: M24000 | |
| Chemical compound, drug | Glycerol | Sigma | Cat #: G7893 | |
| Chemical compound, drug | NP40 (IGEPAL) | Sigma | Cat #: I8896 | |
| Chemical compound, drug | 2-Mercaptoethanol | Sigma | Cat #: M3148 | |
| Chemical compound, drug | Glucose | RPI | Cat#: G32045 | |
| Chemical compound, drug | NADH | Millipore Sigma | Cat#: 481913 | |
| Chemical compound, drug | Phosphoenol Pyruvate | Thermo Fisher | Cat#: NC9842221 | |
| Software, algorithm | Prism 6 | Graphpad | ||
| Software, algorithm | Traces | https://github.com/stephlj/Traces | ||
| Software, algorithm | pyhsmm | https://github.com/mattjj/pyhsmm | ||
| Software, algorithm | Slopey | https://github.com/stephlj/slopey | ||
| Software, algorithm | PyEM | https://github.com/asarnow/pyem | ||
| Software, algorithm | Gautomatch | http://www.mrc-lmb.cam.ac.uk/kzhang/ | ||
| Software, algorithm | SerialEM | (Mastronarde, 2005) | ||
| Software, algorithm | RELION 3.0 | (Zivanov et al., 2018) | ||
| Software, algorithm | Motioncor2 | (Zheng et al., 2017) | ||
| Software, algorithm | GCTF | (Zhang, 2016) | ||
| Software, algorithm | UCSFImage4 | (Li et al., 2015) | ||
| Software, algorithm | EMAN2 | (Tang et al., 2007) | ||
| Software, algorithm | CryoSPARC | (Punjani et al., 2017) | ||
| Software, algorithm | Diffmap.exe | http://grigoriefflab.janelia.org/diffmap | ||
| Software, algorithm | Coot | (Emsley et al., 2010) | ||
| Software, algorithm | Phenix | (Adams et al., 2010) | ||
| Software, algorithm | ImageJ | https://imagej.nih.gov/ij/ | ||
| Other | Superdex 200 increase10/300 GL | GE | Cat. #: 29091596 | |
| Other | HiTrap QXL column | GE | Cat. #: 17-5159-01 | |
| Other | Superdex 200 HiLoad 26/600 | GE | Cat. #: 28989336 | |
| Other | TALON metal affinity resin | Clontech | Cat. # 635503 |
Summary table for data collection and refinement on TVIPS 816 scintillator-based camera
https://doi.org/10.7554/eLife.46057.026| Dataset | Single-bound SNF2h-nucleosome complex at SHL + 2 | Doubly-bound SNF2h-nucleosome at SHL ± 2 |
|---|---|---|
| Microscope | TF20 (FEI) | TF20 (FEI) |
| Voltage (kV) | 200 | 200 |
| Camera | TemF816 8k × 8 k CMOS (TVIPS) | TemF816 8k × 8 k CMOS (TVIPS) |
| Magnification | 62.000 | 62.000 |
| Pixel size (Å) | 1.2 | 1.2 |
| Defocus range (µm) | −1.8: −2.9 | −1.8: −2.9 |
| Number of images | 766 | 766 |
| Total electron dose (e-/Å2) | 25 | 25 |
| Number of frames | - | - |
| Initial number of particles | 450322 | 450322 |
| Particles selected after 2D cleanup | 379540 | 379540 |
| Particles in final reconstruction | 32233 | 57060 |
| Final resolution (Å) | 8.4 | 8.4 |
Summary table for data collection and refinement on Gatan K2-Summit direct electron detector camera.
https://doi.org/10.7554/eLife.46057.027| Dataset | Single-bound SNF2h-nucleosome complex: SHL-2/SHL + 2 | Single-bound SNF2h-nucleosome complex, SHL-2 | Core nucleosome |
|---|---|---|---|
| Microscope | TF30 (FEI) | TF30 (FEI) | TF30 (FEI) |
| Voltage (kV) | 300 | 300 | 300 |
| Camera | K2 summit (Gatan) | K2 summit (Gatan) | K2 summit (Gatan) |
| Magnification | 31.000 | 31.000 | 31.000 |
| Pixel size (Å) | 1.22 | 1.22 | 1.22 |
| Defocus range (µm) | −1.5: −3.0 | −1.5: −3.0 | −1.5: −3.0 |
| Number of images | 720 | 720 | 720 |
| Total electron dose (e-/Å2) | 42 | 42 | 42 |
| Number of frames | 30 | 30 | 30 |
| Initial number of particles | 120533 | 333430 | 24993 |
| Particles selected after 2D cleanup | 95879 | no cleanup | 19363 |
| Particles in final reconstruction | 27513/6241 | 43165 | 12130 |
| Final resolution (Å) | 3.9/6.9 | 3.4 | 7.4 |
Additional files
-
Supplementary file 1
Key Resources Table.
- https://doi.org/10.7554/eLife.46057.029
-
Transparent reporting form
- https://doi.org/10.7554/eLife.46057.030





