Amidst multiple binding orientations on fork DNA, Saccharolobus MCM helicase proceeds N-first for unwinding
Figures
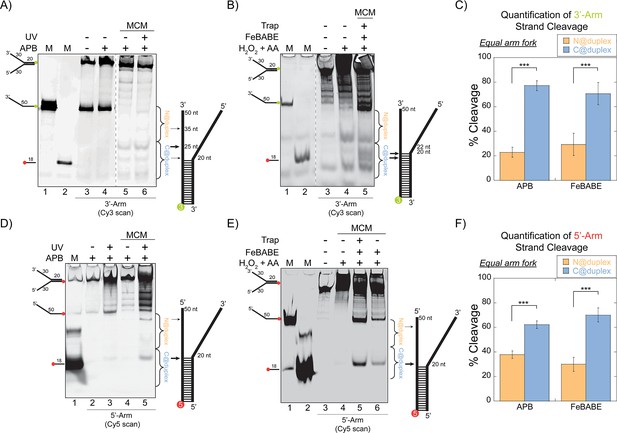
SsoMCM orientation mapping onto equal arm fork DNA substrates.
(A) APB or (B) FeBABE orientation mapping of the 3’- encircled strand labelled at the 5’- duplex end with Cy3 on an equal arm fork DNA substrate with a 20 base duplex (DNA164/165-3). SsoMCM was labelled with APB or FeBABE at C682 (within the C-term WH motif) specifically. DNA cleavage was induced and enhanced with UV light [for APB: (A), lane 6] or H2O2 and ascorbic acid (AA) [for FeBABE; (B), lane 5). Arrow thickness indicates the relative amount and position of DNA cleavage. (C) Quantification of the relative amount of DNA cleavage for bases 20-35 or 36-50 from the 5’- end indicate the relative orientation for placing the N-term (N@duplex, orange) or C-term (C@duplex, blue), respectively, closer to the duplex junction for either APB or FeBABE mapping. Similarly, (D) APB or (E) FeBABE orientation mapping of the 5’- excluded strand labelled at the 3’- duplex end with Cy5 (164-5/165). (F) Quantification of the relative amount of DNA cleavage for bases 20-35 or 36-50 from the 5’- end indicate the relative orientation for N@duplex or C@duplex, respectively, closer to the duplex junction for either APB or FeBABE mapping. DNA markers (M) indicate 18 and 50 bases and fork DNA. Error bars represent standard error from 3-5 independent experiments. The products were run on a 20% native PAGE gel. p-values are defined as *< 0.05, **< 0.01 ***<0.001.
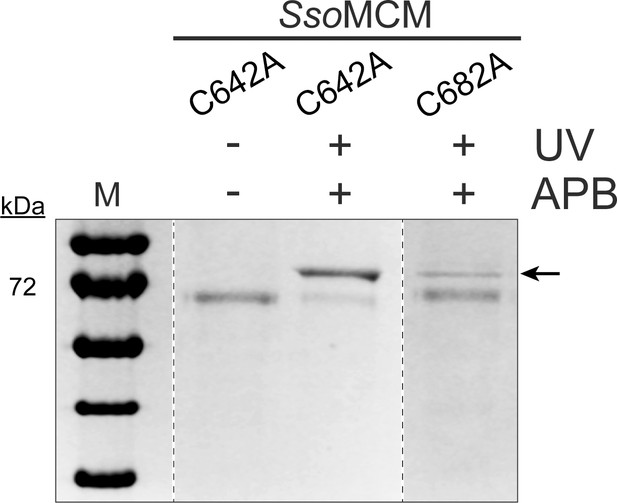
Crosslinking MCM mutants to DNA.
The UV crosslinked protein-DNA product of two SsoMCM mutants and the uncrosslinked SsoMCM C642A were run on a 10% SDS- PAGE gel. The shifted protein-DNA product is indicated by a black arrow (←). In the presence of UV and APB, SsoMCM C642A shows a higher signal for the crosslinked protein-DNA product compared to C682A mutant. Rec protein ladder is used as the standard protein marker (M).
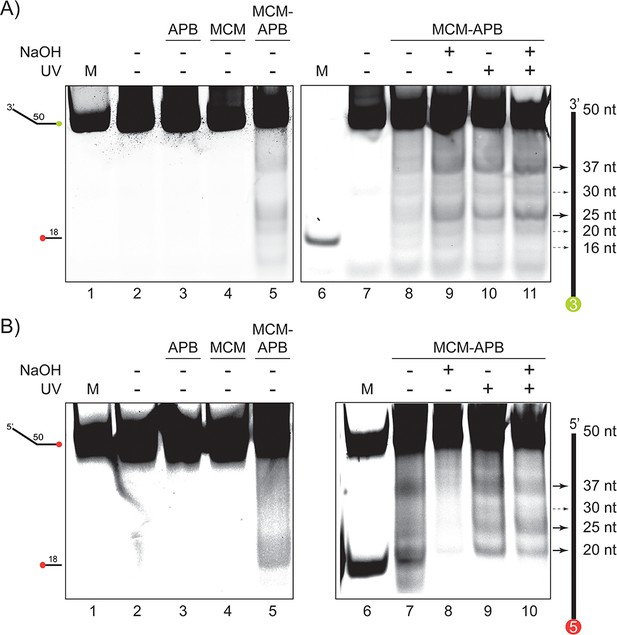
Validation of MCM-APB cleavage on ssDNA.
(A) APB cleavage of a 5’-Cy3 labelled 50 nt ssDNA. No cutting in the absence of APB or MCM (lanes 2–4). MCM-APB can cleave DNA in the absence or presence of UV or NaOH (lanes 5, 8–11). (B) APB cleavage of a 3’-Cy5 labelled 50 nt ssDNA. No cutting in the absence of APB or MCM (lanes 2–4). MCM-APB can cleave DNA in the absence or presence of UV or NaOH (lanes 5, 7–10). Markers (M) at 50 or 18 nts are indicated.
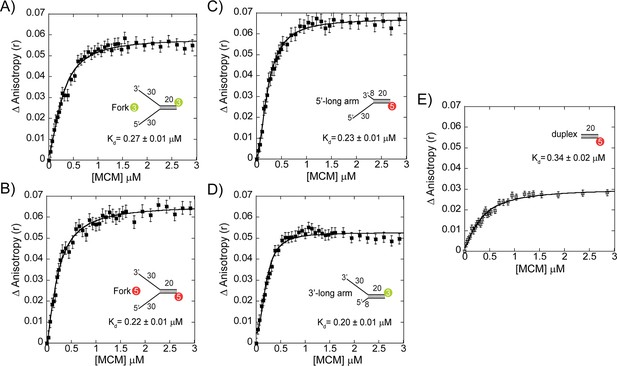
DNA binding by fluorescence anisotropy.
DNA binding was determined by fluorescence anisotropy for each of the DNA substrates. The binding constants (Kd) for equal arm fork DNA substrates (5’-Cy3 labelled or 3’-Cy3 labelled), 3’- long arm (5’-Cy3 labelled), 5’- long arm (3’-Cy5 labelled), and duplex DNA (5’-Cy5 labelled) are given in the plots. Error bars represent standard error from three independent experiments and data was fit to Equation 4.
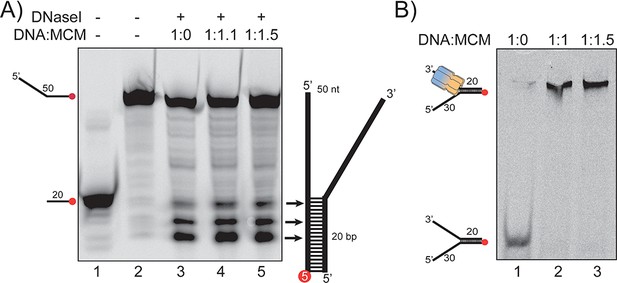
DNaseI footprinting and EMSA.
(A) DNaseI digestion of fork DNA in the absence (lane 3) and presence of SsoMCM (lanes 4–5) at different ratios. Fork DNA is labelled at the 3’ position on the duplex end with Cy5. Markers at 20 nt (lane 1) and 50 nt (lane 2) are indicated. Samples were run on 20% denaturing PAGE. (B) EMSA showing the complete formation of the DNA-protein complexes at the same concentration ratios used in (A).
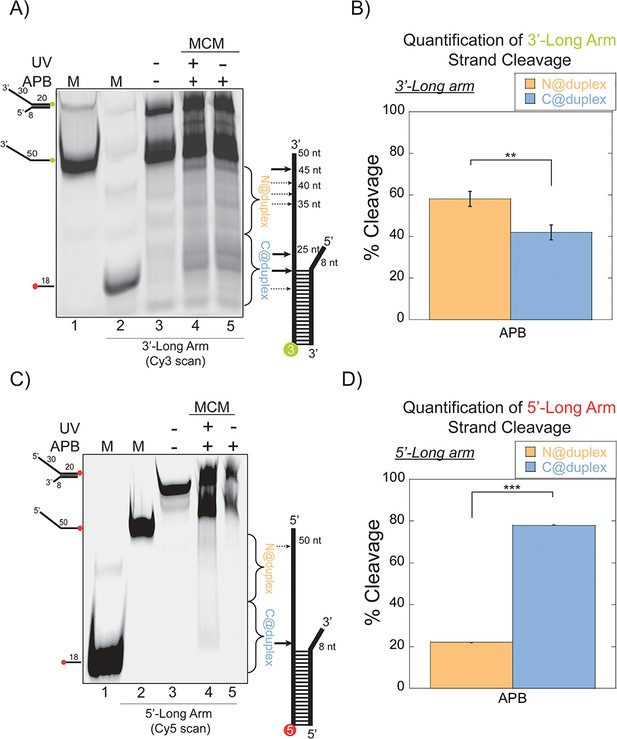
SsoMCM orientation mapping onto 3’-(DNA171/165-3) or 5’-(DNA172/164-5) long arm fork DNA substrates.
(A) APB orientation mapping of the 3’-encircled strand labelled at the 5’ duplex end with Cy3 on a 3’-long arm fork DNA substrate with a 20 base duplex. SsoMCM was labelled with APB at C682 (within the C-term WH motif) specifically. (B) Quantification of the relative amount of DNA cleavage for bases 20–35 or 36–50 from the 5’-end indicate the relative orientation for placing the N@duplex (orange) or C@duplex (blue), respectively closer to the duplex junction. Similarly, (C) APB orientation mapping of the 5’-excluded strand labelled at the 3’-duplex end with Cy5 on a 5’-long arm fork DNA substrate with a 20 base duplex. (D) Quantification of the relative amount of DNA cleavage for bases 20–35 or 36–50 from the 3’-end indicate the relative orientation for N@duplex or C@duplex, respectively, closer to the duplex junction. DNA cleavage was induced and enhanced with UV light [(A), lane 4, (C), lane 4]. Arrow thickness indicates the relative amount and position of DNA cleavage. DNA markers (M) indicate 18 and 50 bases and fork DNA. Error bars represent standard error from 3 to 5 independent experiments. The products were run on a 20% native PAGE gel. p-values are defined as *<0.05, **<0.01 ***<0.001. shorter 8 nt 3’-arm (Figure 2C–D). Here, the footprinting orientations were reversed, with a > 3:1 preference for C@duplex (Figure 2D). Therefore, on these long arm fork DNA substrates, SsoMCM can bind either the 3’- or 5’-arm in both orientations, but the preferred 3’−5’ polarity is CTD-NTD.
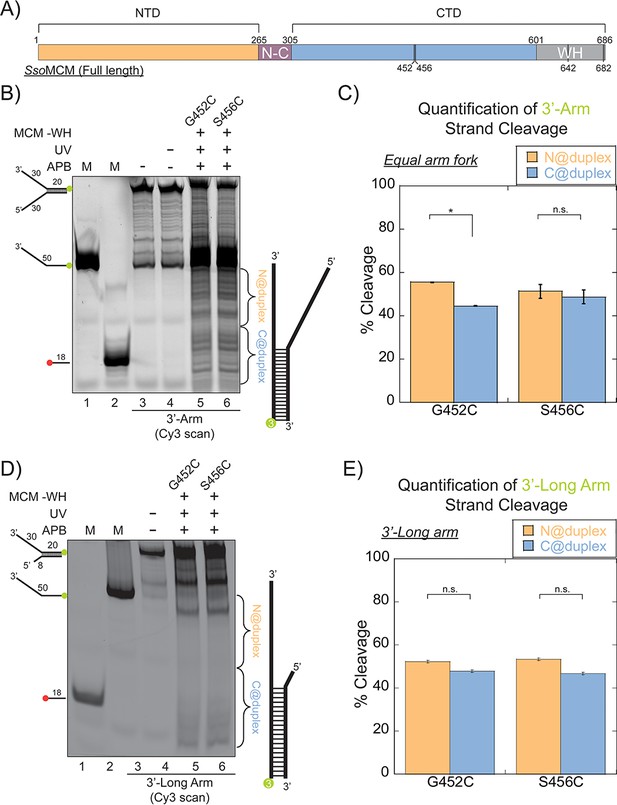
SsoMCM (-WH) orientation mapping onto fork DNA substrates.
(A) Schematic of full length SsoMCM highlighting the NTD (orange), CTD (blue), and the WH domain (grey) and cysteine sites of conjugation used. (B) Orientation mapping of SsoMCM labelled at C452 or C456 with APB for the 3’-encircled strand labelled at the 5’-duplex end with Cy3 on a forked DNA substrate with a 20 base duplex. DNA cleavage was enhanced with UV light and the products run on a 20% native PAGE gel. The brackets indicate the regions quantified (bases 20-35 or 36-50) for (C) the relative amount and position of DNA cleavage and correspond with the relative orientation of the SsoMCM hexamer; N@duplex (orange) or C@duplex (blue). (D) 3’-encircled strand labelled at the 5’-duplex end with Cy3 on a 3’-long arm fork DNA substrate (E) the relative amount and position of DNA cleavage and correspond with the relative orientation of the SsoMCM hexamer. DNA markers (M) indicate 18 and 50 bases and fork DNA. Error bars represent standard error from 3-5 independent experiments. p-values are defined as *< 0.05, **< 0.01 ***<0.001. n.s. is not significant.
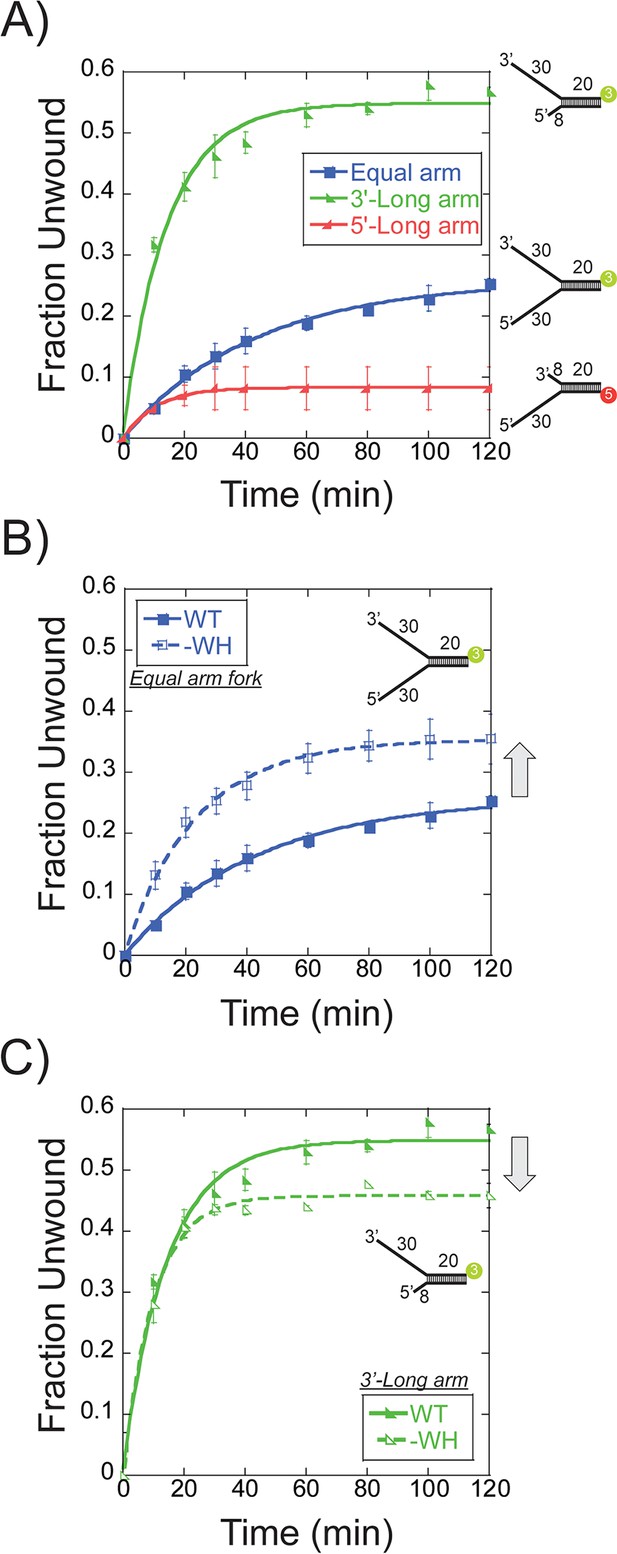
Single turnover DNA unwinding.
(A) DNA unwinding of equal arm (in blue boxes -■-, 5’-Cy3 labelled at the duplex), 3’-long arm (in green lower left triangle -◣-, 5’-Cy3 labelled at the duplex), 5’-long arm (in red lower right triangle -SsoMCM WT. Experiments were simultaneously initiated with ATP and an unlabelled ssDNA trap oligo identical to the labelled strand as described in the Materials and methods. (B) DNA unwinding of equal arm fork DNA substrate (5’-Cy3 labelled at duplex) with SsoMCM WT (in closed boxes -■-) and –WH (in open boxes -□-). There is an increase in total DNA unwound with –WH compared to WT (grey arrow). (C) DNA unwinding of 3’-long arm fork DNA substrate (5’-Cy3 labelled at duplex) with SsoMCM WT (in lower left closed triangle -◣-) and –WH (in lower left open triangle -◺ -). There is a decrease in the total DNA unwound with –WH compared to WT (grey arrow). Error bars represent standard error from 3 to 5 independent experiments and data was fit to Equation 3.
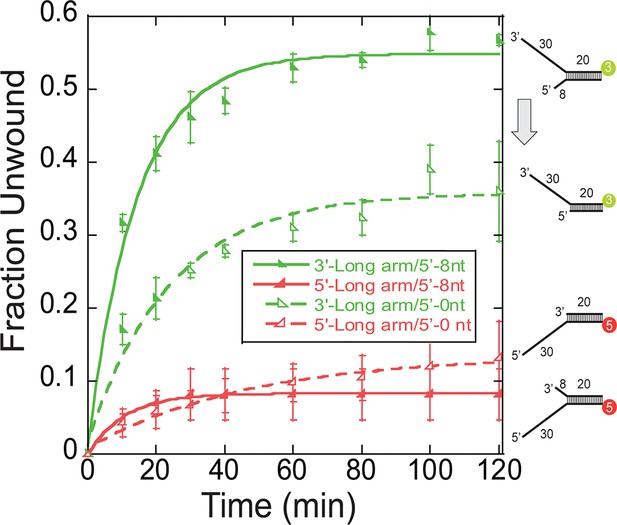
Single turnover DNA Unwinding.
DNA unwinding of 3’-long arm/5’-arm (n), (n = 0, in green open lower left triangle -◣-, n = 8 in green closed lower left triangle -◣-,5’-Cy3 labelled at the duplex), 5’-long arm/3’-arm (n), (n = 0, in red open lower right triangle -◢-, n = 8 in red open lower right triangle-◢-, 3’-Cy5 labelled at the duplex) fork DNA substrates with SsoMCM WT. Experiments were simultaneously initiated with ATP and an unlabelled ssDNA trap oligo identical to the labelled strand as described in the Materials and methods. There is a decrease in total DNA unwound with n = 0 compared to n = 8 in 3’-long arm/5’-arm(n) fork substrate (grey arrow). Error bars represent standard error from 3 to 5 independent experiments and data was fit to Equation 3.
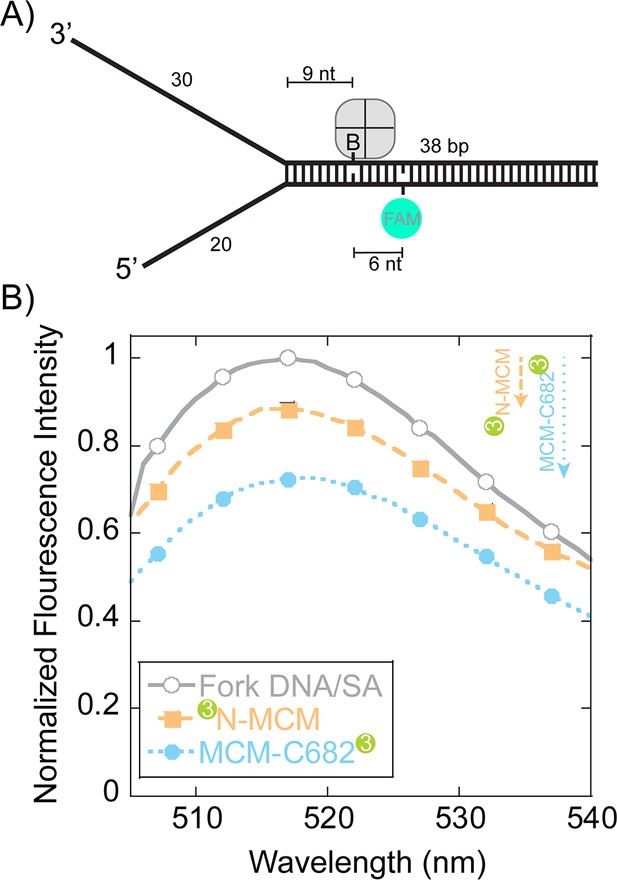
Steady-state FRET quenching of SsoMCM bound to a fork DNA.
(A) Schematic of the DNA fork substrate that includes a 30 nt 3’-arm, 20 nt 5’-arm, a biotin placed nine nt from the duplex junction and a dT-FAM placed a further six nt downstream. The Tm of the duplex was calculated to be greater than 75°C. (B) Normalized FAM quenching effects upon addition of SsoMCM labelled with Cy3 at the N-terminus (closed boxes, orange) or C682 (closed circles, blue). Background changes in fluorescence upon addition of unlabelled SsoMCM were subtracted prior to normalization.
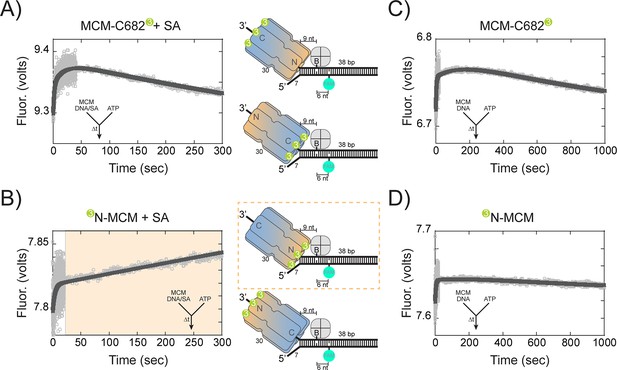
Presteady-state stopped-flow measure of SsoMCM translocation/unwinding.
SsoMCM (167 nM hexamer) labelled at (A, C) C682 or (B, D) the N-terminus was preassembled onto 3’-long arm fork DNA (125 nM) with a 30 base 3’-arm and a 7 base 5’-arm in the (A, B) presence or (C, D) absence of streptavidin (375 nM) as indicated. Translocation was initiated through rapid mixing of ATP (1 mM) and the change in fluorescence above 570 nm was monitored using a split time base at 57 oC. Data was fit to Equation 5. The blue shaded region in (B) highlights the N-first translocation mode (boxed).The following figure supplements are available for Figure 6:Figure supplement 1. Presteady-state loading of SsoMCM on DNA. SsoMCM (167 nM hexamer) labelled at (A) C682 or (B) the N-terminus and preincubated with 1 mM ATP was mixed vs 3’-long arm fork DNA (13 nM) with a 30 base 3-’arm and a 7 base 5’-arm blocked with streptavidin (38 nM) as indicated. (C) Experiments in the absence of ATP for SsoMCM labelled at the N-terminus with Cy3. The change in fluorescence above 570 nm was monitored using a split time base at 57 oC. Data was fit to Equation 5.
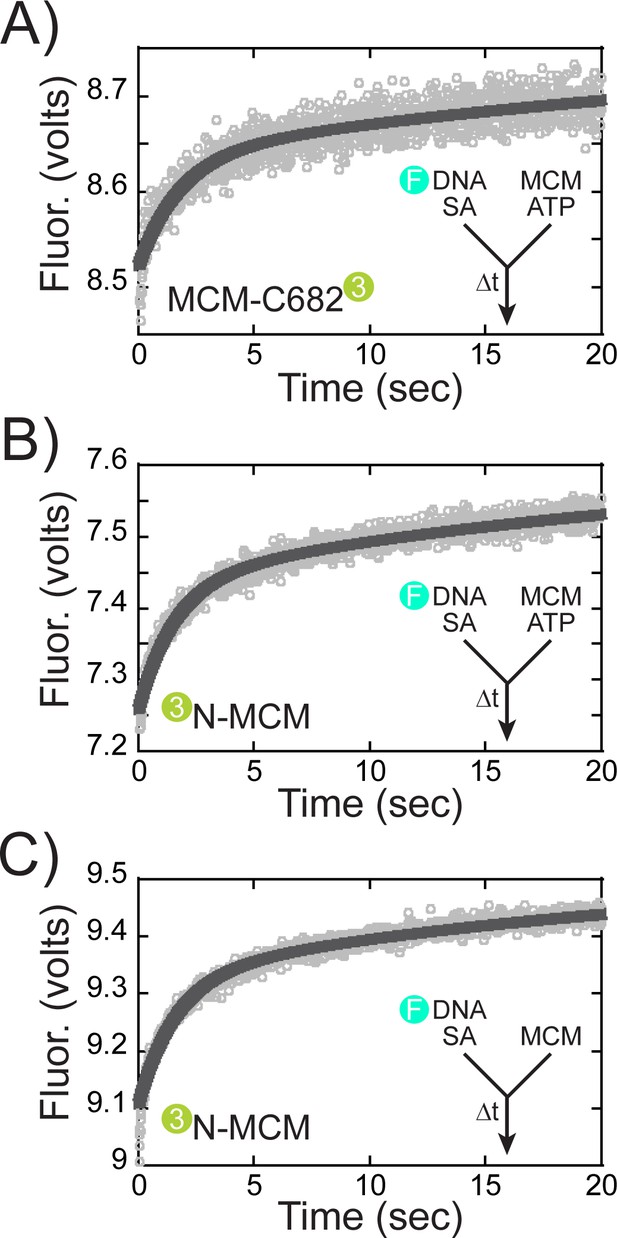
Presteady-state loading of SsoMCM on DNA.
https://doi.org/10.7554/eLife.46096.013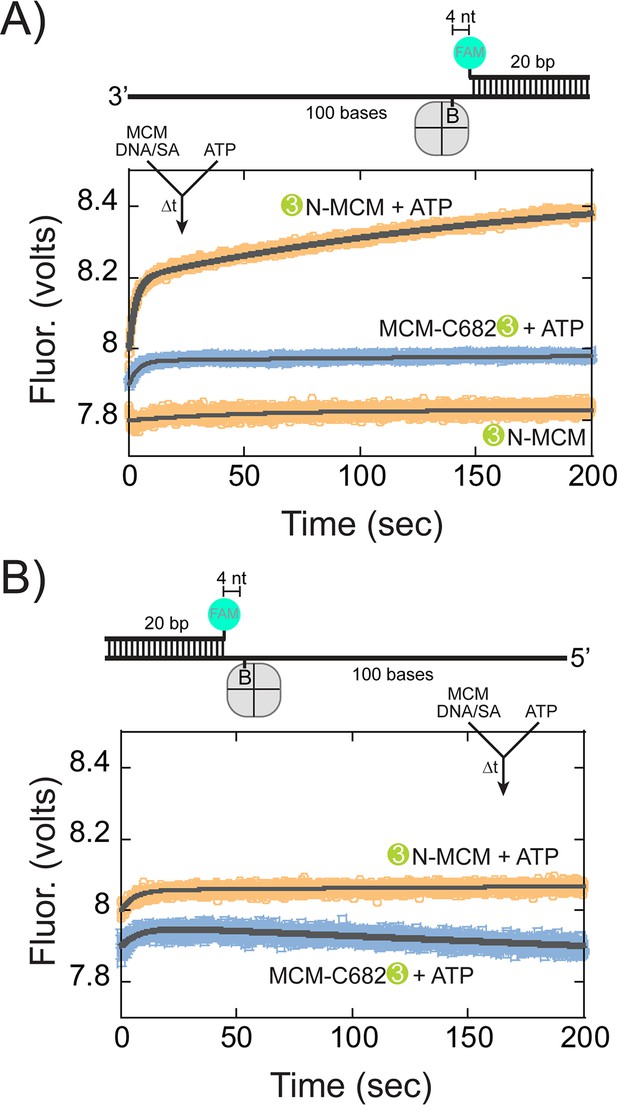
Presteady-state stopped-flow measure of SsoMCM translocation.
SsoMCM (120 nM hexamer) labelled at C682 (orange) or the N-terminus (blue) was preassembled onto (A) 3’- or (B) 5’long arm ssDNA (100 nM) with a flanking 20 bp duplex with 375 nM streptavidin. A FAM label was incorporated at the 5’ or 3’ end of the duplex, respectively, and four bases from a biotin in the long ssDNA. Translocation was initiated through rapid mixing of ATP (1 mM) and the change in fluorescence above 570 nm was monitored over time at 57°C. Data was fit to Equation 5.
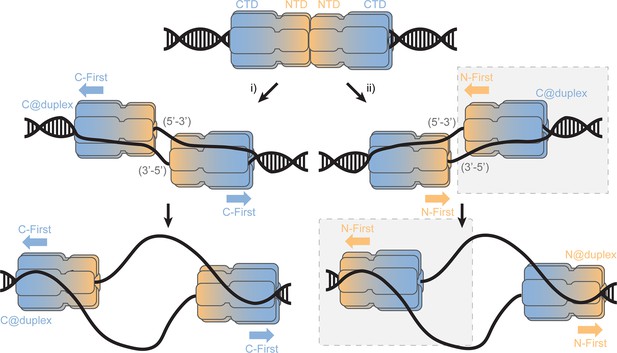
SsoMCM loading at origins.
Model for loading double hexamer MCM at an origin of replication and the two pathways (i or ii) for encircling the 5’-3’ or 3’-5’ strands placing the CTD at the duplex (C@duplex). Translocation from (i) would proceed C-first separating hexamers, while translocation from (ii) would proceed N-first bypassing each hexamer. The shaded (grey) box identifies the conformations and states consistent in this report.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Plasmid construct (E. coli) | pET30a-SsoMCM (C642A) | McGeoch et al. (2005) | ||
| Plasmid construct (E. coli) | pET30a-SsoMCM (C682A) | McGeoch et al. (2005) | ||
| Plasmid construct (E. coli) | pET30a-SsoMCM 1–612 (G452C) | This paper | Site- directed mutagenesis using primers listed in Materials and methods | |
| Plasmid construct (E. coli) | pET30a-SsoMCM1–612 (S456C) | This paper | Site- directed mutagenesis using primers listed in Materials and methods | |
| Expression strain | Rosetta 2 | Novagen | ||
| Chemical compound | 4-azidophenacyl bromide (APB) | Sigma-Aldrich | 57018-46-9 | |
| Chemical compound | ATP | Invitrogen | 51963-61-2 | |
| Chemical compound | 1-(p-Bromoacetamidobenzyl) ethylenediamine N, N,N (Fe-BABE) | Dojindo | 186136-50-5 | |
| Chemical compound | DNaseI | New England Biolabs | M0303S | |
| Chemical compound | Streptavidin | Invitrogen | 800-955-6288 | |
| Chemical compound | Cy3 succinimidyl ester | ThermoFisher | 57757-57-0 | |
| Chemical compound | Cy3 maleimide | ThermoFisher | 45-001-273 | |
| Sequence-based reagent | DNA primers and substrates | Sigma-Aldrich and IDT | Refer to Materials and methods | |
| Software, algorithm | Kaleidagraph | www.synergy.com | V4.5 |
Additional files
-
Supplementary file 1
Table and listing of all DNA sequences and templates used.
- https://doi.org/10.7554/eLife.46096.016
-
Transparent reporting form
- https://doi.org/10.7554/eLife.46096.017





