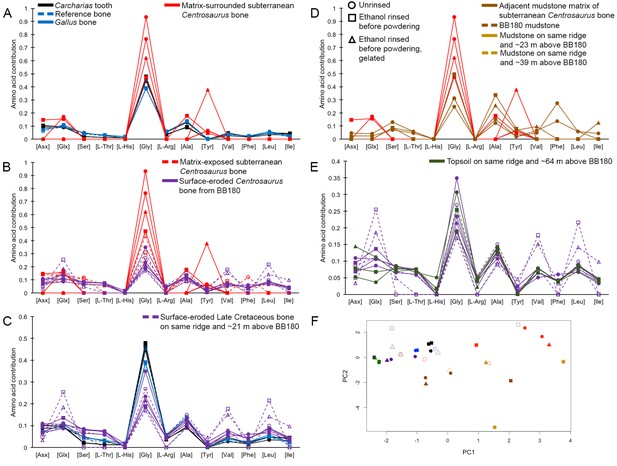
Cretaceous dinosaur bone contains recent organic material and provides an environment conducive to microbial communities
Figures
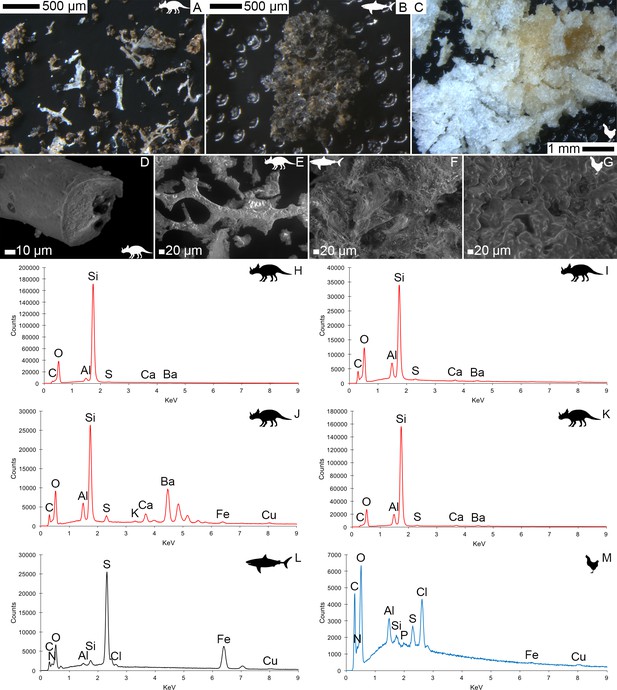
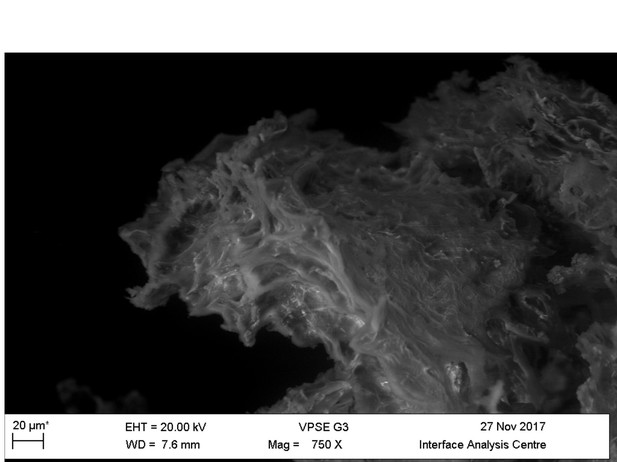
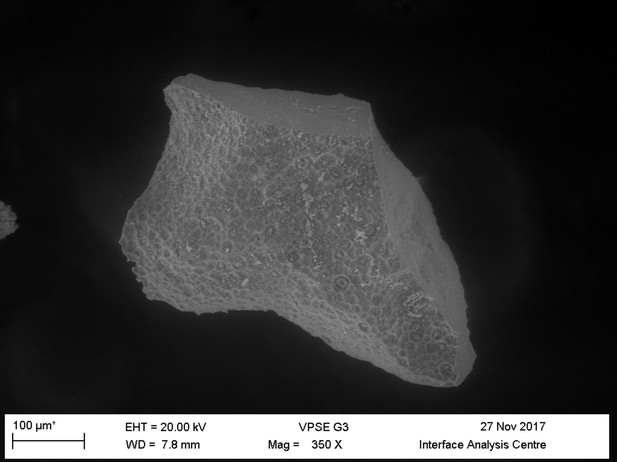
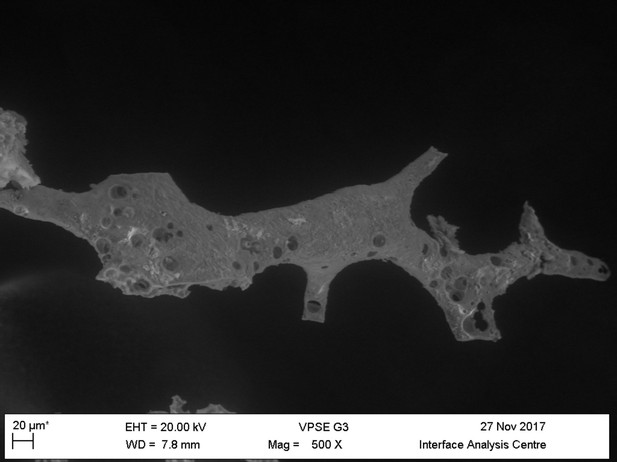
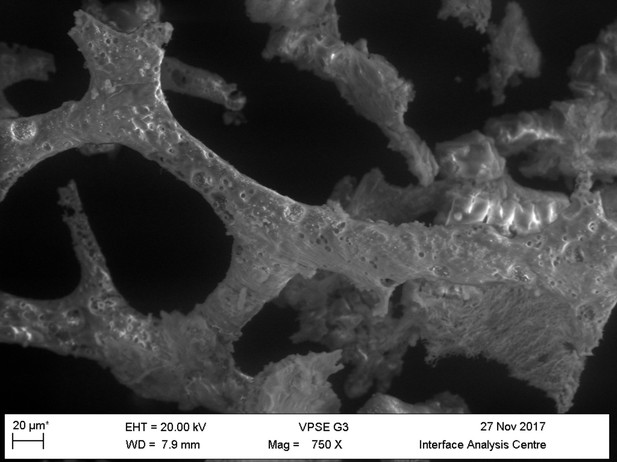
Light microscopy (A–C) and VPSEM (D–G) images and EDS spectra (H–M) of HCl demineralized, freeze-dried samples.
(A–C) Samples rested on carbon tape upon SEM stubs and the pitting was a result of prior VPSEM and EDS analysis. (A) Centrosaurus vessels and associated minerals. (B, F, L) Carcharias tooth. (C, G, M) Gallus. (D) Infilled Centrosaurus vessel. (E) Centrosaurus vessel, fibrous material along the center of the vessel, and associated reddish minerals around the vessel. (H) Centrosaurus vessel exterior from D. (I) Centrosaurus vessel infilling from D. (J) Associated reddish mineral in Centrosaurus. (K) Centrosaurus fibrous material from E. Centrosaurus samples are matrix-surrounded subterranean bone.
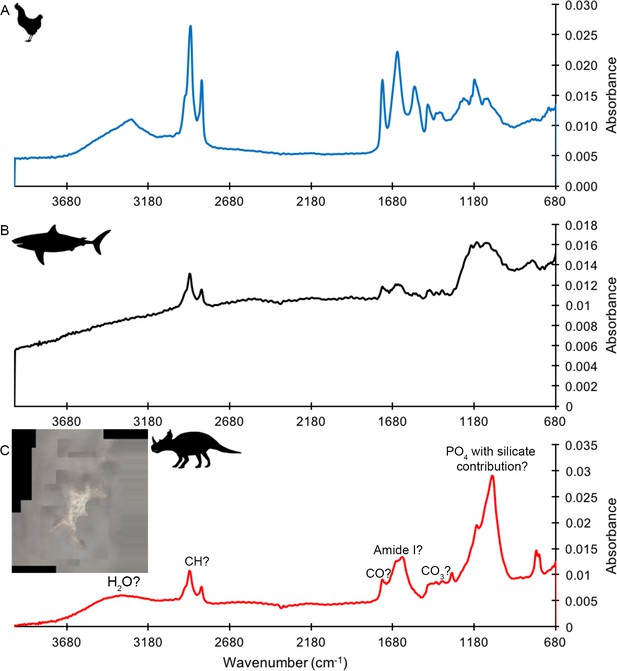
ATR FTIR spectra of HCl demineralized, freeze-dried samples.
(A) Gallus. (B) Carcharias tooth. (C) Matrix-surrounded subterranean Centrosaurus bone vessel with inset showing a composite image of the vessel that was analyzed.
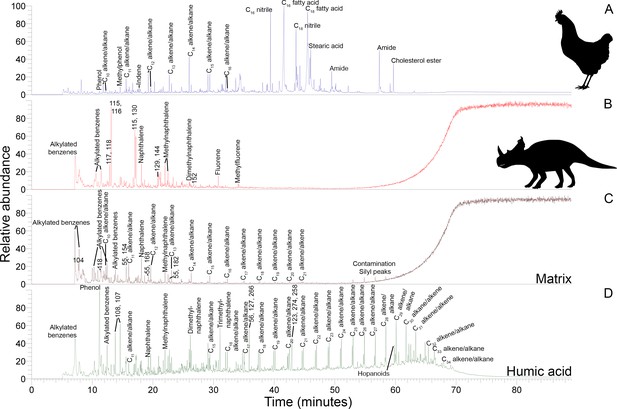
Py-GC-MS total ion chromatograms of samples ethanol rinsed before powdering.
Some of the major pyrolysis products are labeled with the compound name or prominent m/z peaks. (A) Gallus bone. (B) Matrix-surrounded subterranean Centrosaurus bone. (C) Adjacent mudstone matrix of subterranean Centrosaurus bone in B. (D) Humic acid (technical grade) powder with a series of branched and cyclic alkanes, several aromatic ions, and several hopanoid (m/z = 191, 189, 367) and steroid (m/z = 217, 129, 257) ions.
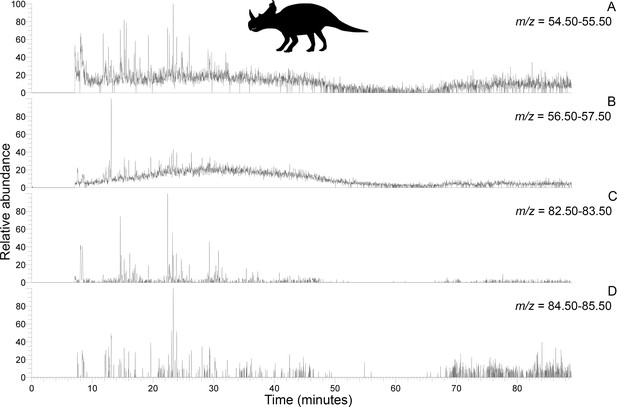
Py-GC-MS chromatograms searching for ion m/z ranges typical of n-alkanes and n-alkenes from kerogen in the matrix-surrounded subterranean Centrosaurus bone ethanol rinsed before powdering.
Potential doublets indicative of n-alkanes/n-alkenes are weakly apparent at best. A, m/z = 55. B, m/z = 57. C, m/z = 83. D, m/z = 85.
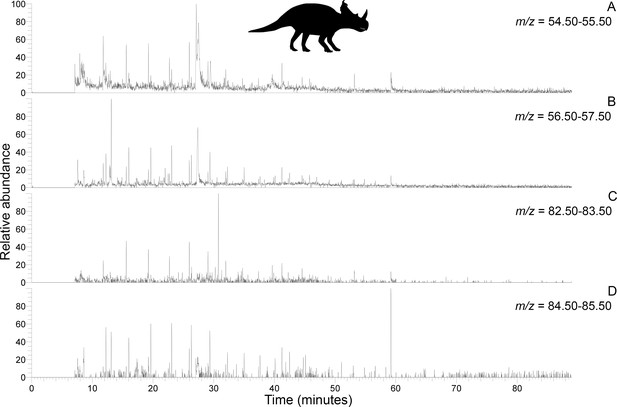
Py-GC-MS chromatograms searching for ion m/z ranges typical of n-alkanes and n-alkenes from kerogen in the uncovered subterranean Centrosaurus bone ethanol rinsed before powdering.
Doublets indicative of n-alkanes/n-alkenes are relatively more abundant than in Figure 4. A, m/z = 55. B, m/z = 57. C, m/z = 83. D, m/z = 85..
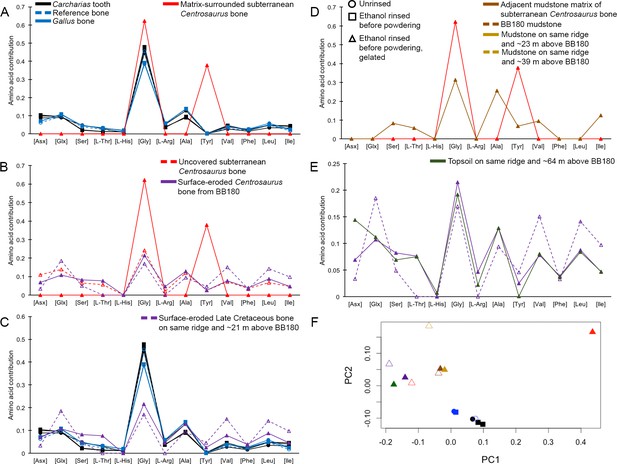
THAA compositional profiles of the KOH-treated samples based on amino acid percentages.
Lines connecting points are added to aid visualization. (A) Late Cretaceous subterranean bone compared to non-aseptically collected Pleistocene-Holocene teeth (with a repeated measurement for the ethanol rinsed sample) and modern bone. (B) Late Cretaceous subterranean bone compared to surface-eroded Late Cretaceous bone from the same outcrop. (C) Surface-eroded Late Cretaceous bone compared to Pleistocene-Holocene teeth and modern bone. (D) Late Cretaceous subterranean bone aseptically collected compared to the adjacent mudstone matrix. (E) Surface-eroded Late Cretaceous bone compared to topsoil at higher elevation (i.e., prairie level) on the same ridge. (F) PCA on non-normalized amino acid percentages (i.e. percentages that do not require further normalization) (see A–E legends). PC1 and PC2 describe 55.04% and 22.66% of the data variation, respectively. See Appendix 1 (Appendix 1—figure 21; Appendix 1—table 9) for PCA summary. Color and symbol coding is constant throughout.
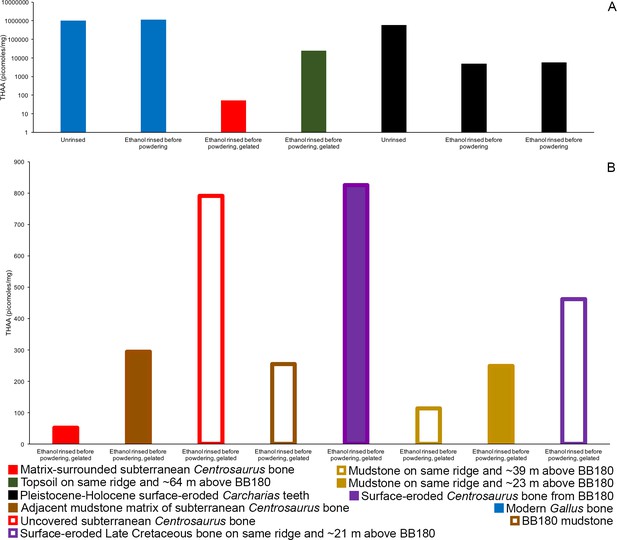
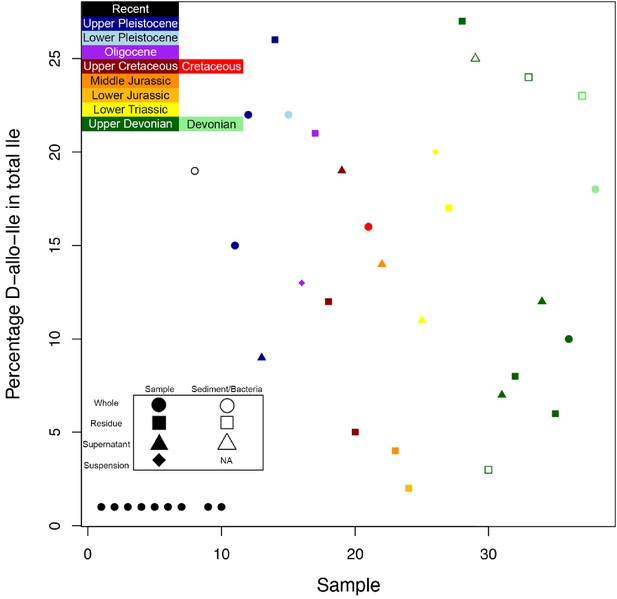
THAA concentrations (summed total of all amino acids measured) of the KOH-treated samples.
(A) Logarithmic scale comparison of modern bone, matrix-surrounded subterranean Late Cretaceous bone, Pleistocene-Holocene surface-eroded teeth (with a repeated measurement for the ethanol rinsed sample), and topsoil on same ridge and ~64 m above BB180. (B) Comparison between fossil Late Cretaceous bone and mudstone.
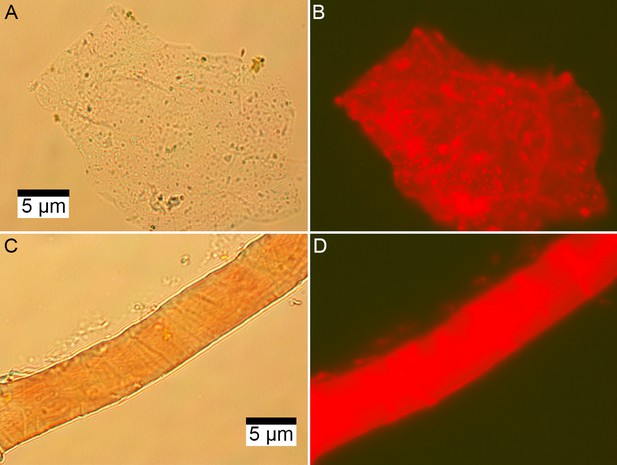
Microscopic images of EDTA demineralized, PI stained matrix-surrounded subterranean Centrosaurus bone.
(A–B) Fibrous material. (C–D) Vessel. (A, C) Transmission light. (B, D) Fluorescence.
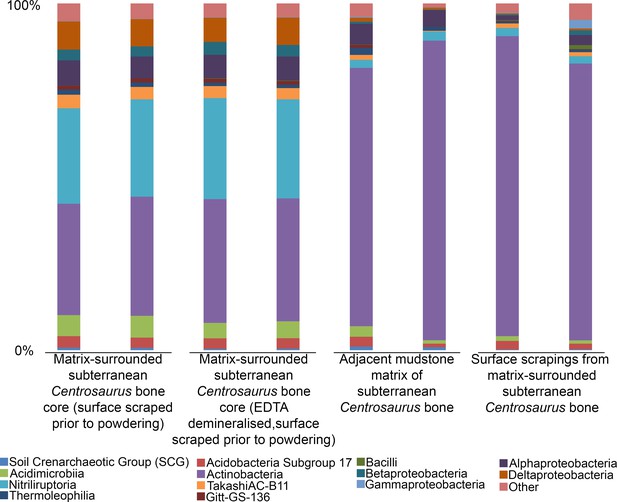
Comparison of microbial community (class level) from matrix-surrounded subterranean Centrosaurus bone and adjacent mudstone matrix.
There are two replicates per sample. Classes with <1% representation in all replicates and samples are combined into an ‘other’ category.
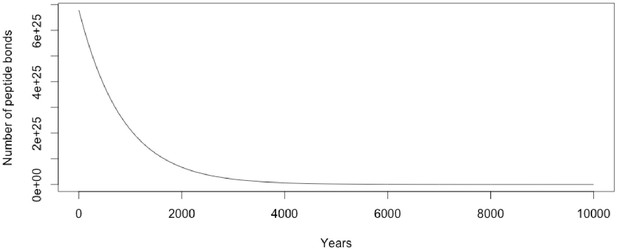
Decay curve of average human proteome based on a half life of 600 years as described by the equation in the text.
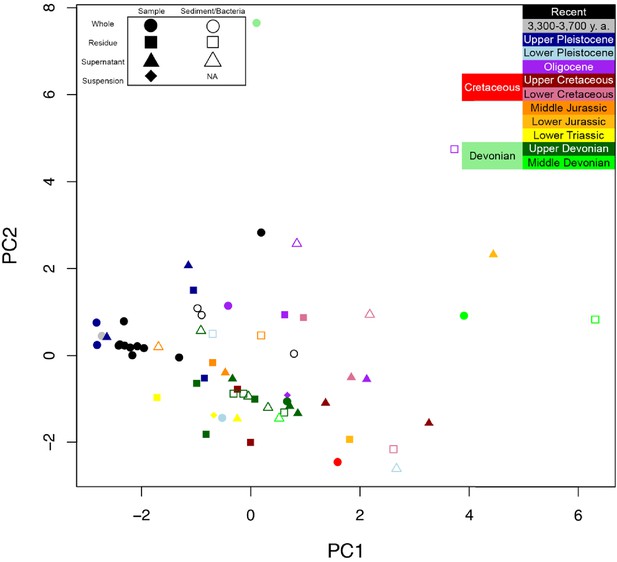
PCA of data from Armstrong et al. (1983).
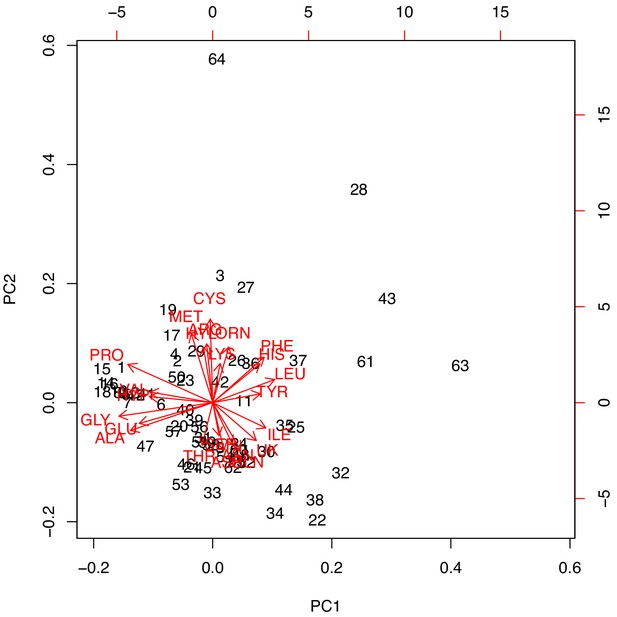
PCA biplot of data from Armstrong et al. (1983).
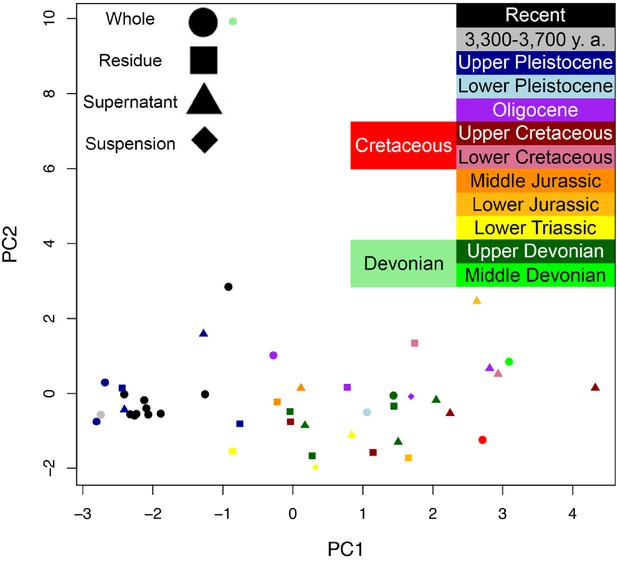
PCA on reduced Armstrong et al. (1983).
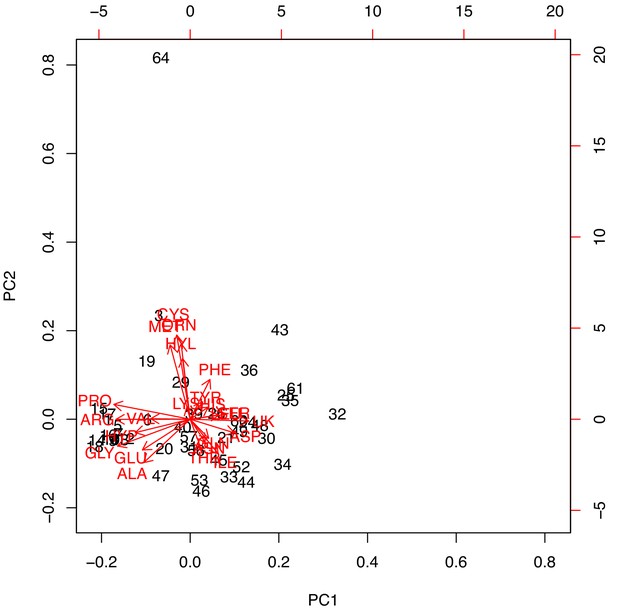
PCA biplot on reduced Armstrong et al. (1983).
Amino acid epimerisation data in Armstrong et al. (1983).
Amino acid concentration data in Armstrong et al. (1983).

Ridge on which BB180 is located. View looking east at mouth of Jackson Coulee, Dinosaur Provincial Park, AB.

Foil placed on top of sediment and matrix-surrounded Centrosaurus tibia portion prior to flipping with an awl.
Uncovered distal end of tibia is visible to the right of foil.

Centrosaurus tibia after matrix-surrounded sample and uncovered distal end were collected.

Aseptic collection of fossil samples in BB180 on July 8, 2016.
Only skin of the face, wrists, and shins was exposed above the fossils. Shins remained uncovered to act as a thermal window for health and safety reasons to avoid overheating and to reduce the likelihood of contamination by lowering body temperature to reduce perspiration that might fall onto the samples. The body was positioned downhill of the bones at all times to compensate. Photograph by Kentaro Chiba.
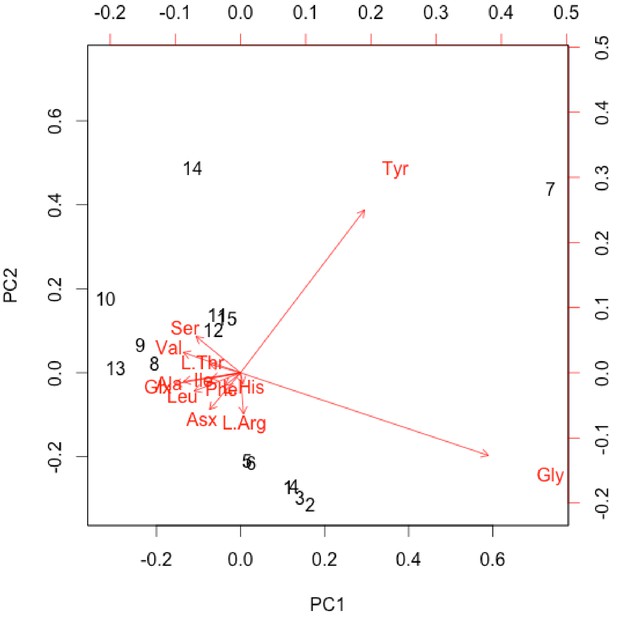
Non-normalised PCA biplot of THAA composition of only the sufficiently treated samples associated with the plot in Figure 6F.
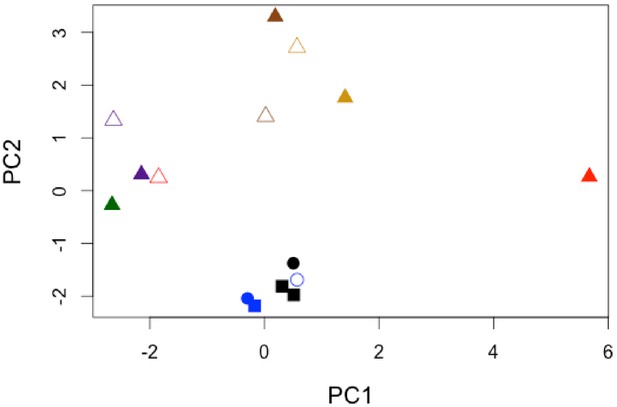
Normalised PCA of THAA composition of only the sufficiently treated samples.
prcomp() function in R scale set to ‘TRUE’. Color and shape coding identical to that in Appendix 1—figure 24.
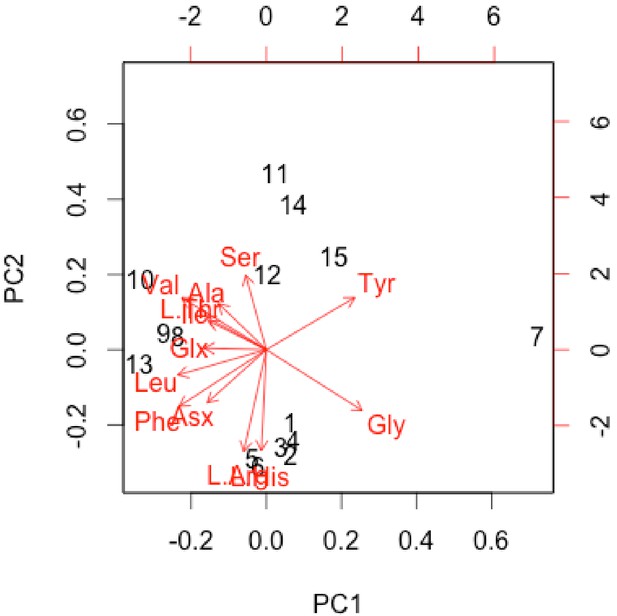
Normalised PCA biplot of THAA composition of only the sufficiently treated samples associated with Appendix 1—figure 22.
THAA compositional profiles of the samples based on amino acid percentages.
(A) Late Cretaceous subterranean bone (red) compared to non-aseptically collected Pleistocene-Holocene teeth (black) and modern bone (blue). (B) Late Cretaceous subterranean bone (red) compared to surface-eroded Late Cretaceous bone from the same outcrop (purple). (C) Surface-eroded Late Cretaceous bone (purple) compared to Pleistocene-Holocene teeth (black) and modern bone (blue). (D) Late Cretaceous subterranean bone aseptically collected (red) compared to the adjacent mudstone matrix (brown). (E) Surface-eroded Late Cretaceous bone (purple) compared to topsoil at higher elevation (i.e., prairie level) on the same ridge (green). (F) PCA on normalised amino acid percentages (see A–E legends). See Appendix 1 (Appendix 1—figure 25; Appendix 1—table 11) for PCA summary. Color and symbol coding is constant throughout.
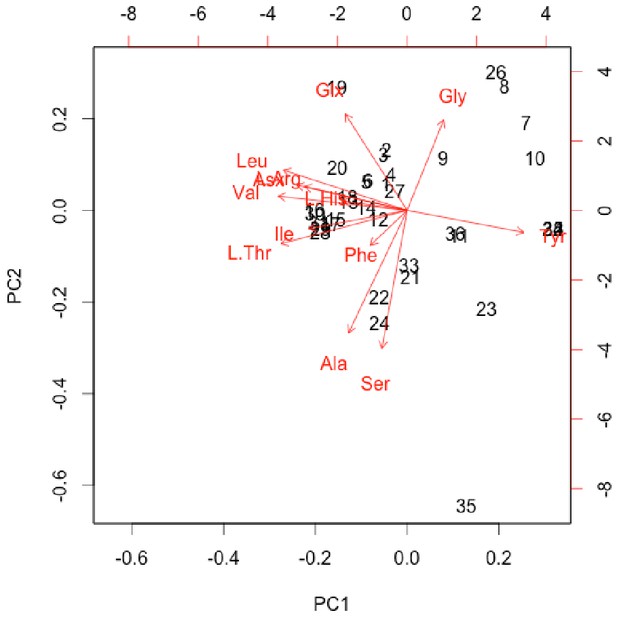
Normalised PCA biplot of THAA composition associated with the plot in Appendix 1—figure 24F.
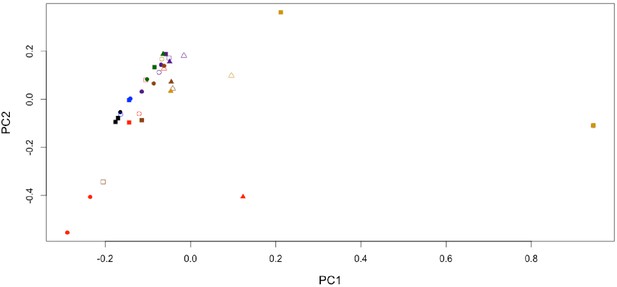
Non-normalised PCA of THAA composition.
prcomp() function in R scale set to ‘FALSE’. Color and shape coding identical to that in Appendix 1—figure 24.
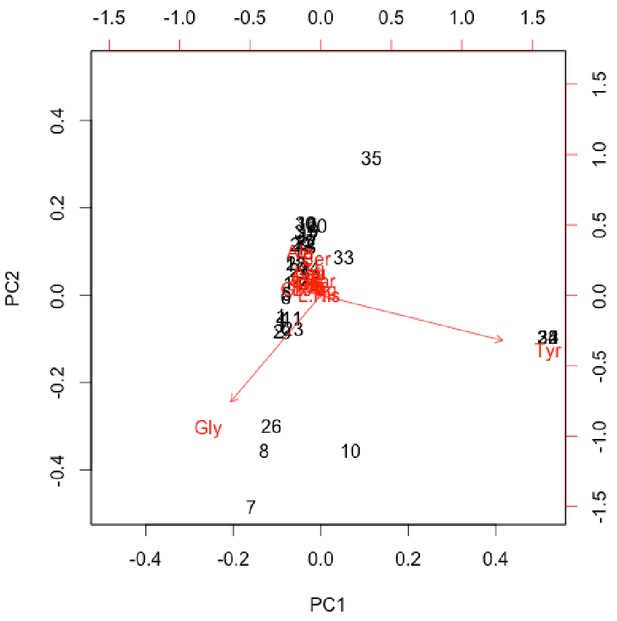
Non-normalised PCA biplot of THAA composition associated with Appendix 1—figure 26.
prcomp() function in R scale set to ‘FALSE’.
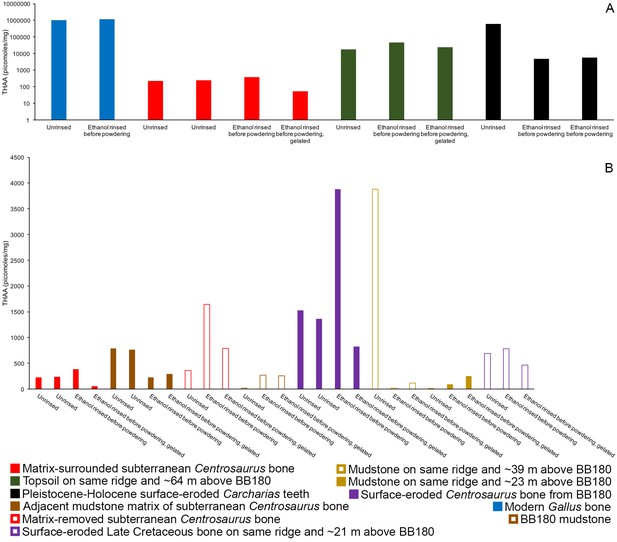
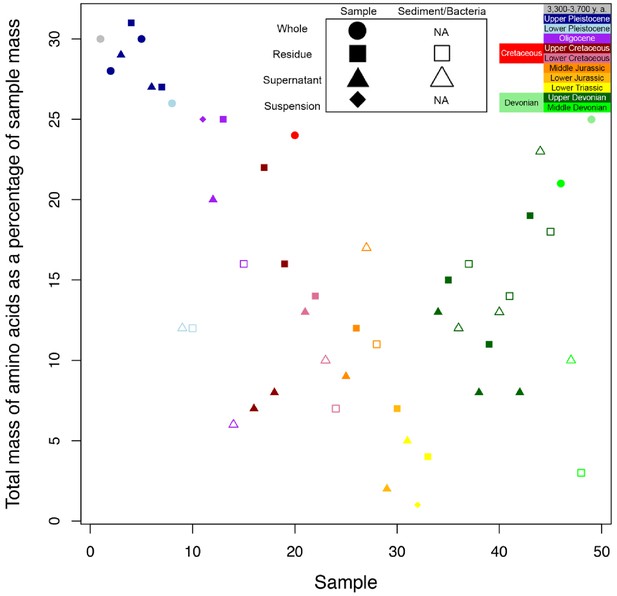
THAA concentrations (summed total of all amino acids measured) of the samples.
(A) Logarithmic scale comparison of modern Gallus bone (blue), matrix-surrounded subterranean Centrosaurus bone (red), Pleistocene-Holocene surface-eroded shark teeth (black, with a repeated measurement for the ethanol rinsed sample), and topsoil on same ridge and ~64 m above BB180 (green). (B) Comparison between fossil Late Cretaceous bone and mudstone. Matrix-surrounded subterranean Centrosaurus bone (solid red), adjacent mudstone matrix of subterranean Centrosaurus bone (solid brown), uncovered subterranean Centrosaurus bone (open red), BB180 mudstone (open brown), surface-eroded Centrosaurus bone from BB180 (solid purple), mudstone on same ridge and ~39 m above BB180 (open tan), mudstone on same ridge and ~23 m above BB180 (solid tan), and surface-eroded Late Cretaceous bone on same ridge and ~21 m above BB180 (open purple). Gelated replicates likely provide the most accurate measurements given the peak reduction present in the non-gelated replicates.
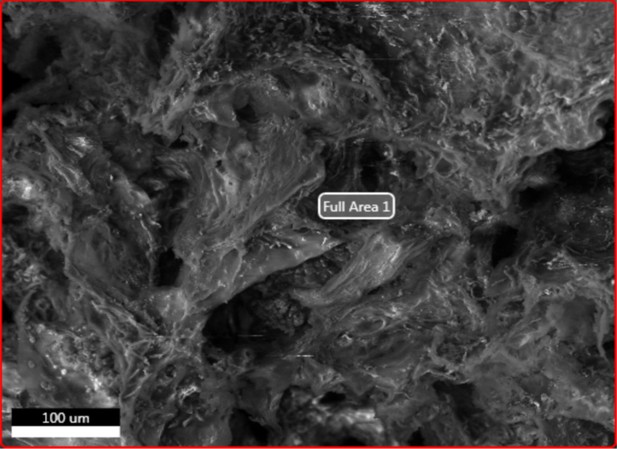
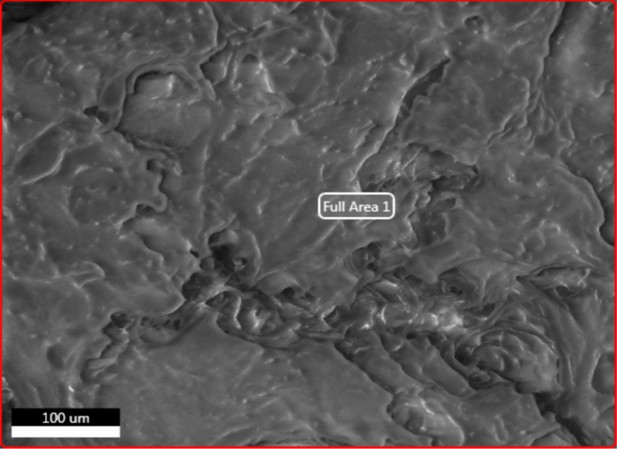
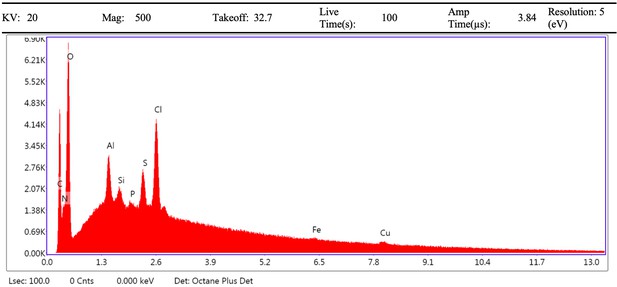
Electron image of demineralised Pleistocene-Holocene shark tooth full area 1 EDS analysis.

Electron image of mineral grain from demineralised Late Cretaceous Centrosaurus bone area A.
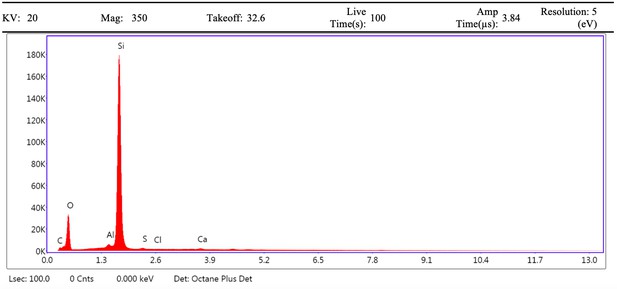
EDS spectrum of mineral grain from demineralised Late Cretaceous Centrosaurus bone area A.
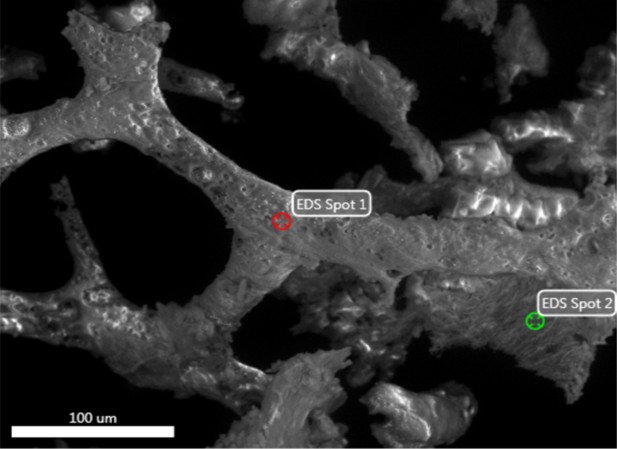
Electron image of vessel (spot 1) and fibrous mass (spot 2) from demineralised Late Cretaceous Centrosaurus bone area B.
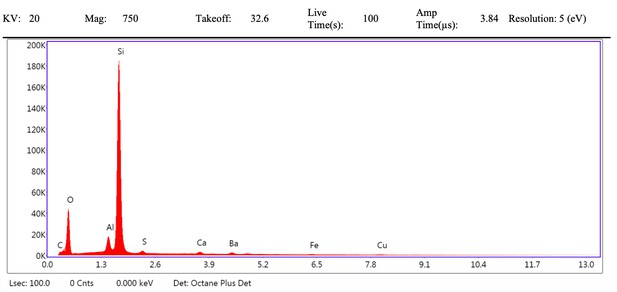
EDS spectrum of vessel (spot 1) from demineralised Late Cretaceous Centrosaurus bone area B.
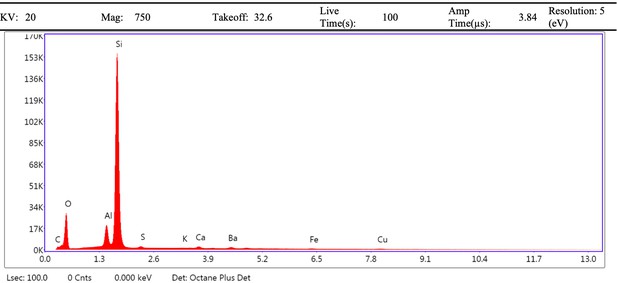
EDS spectrum of fibrous mass (spot 2) from demineralised Late Cretaceous.
Centrosaurus bone area B.
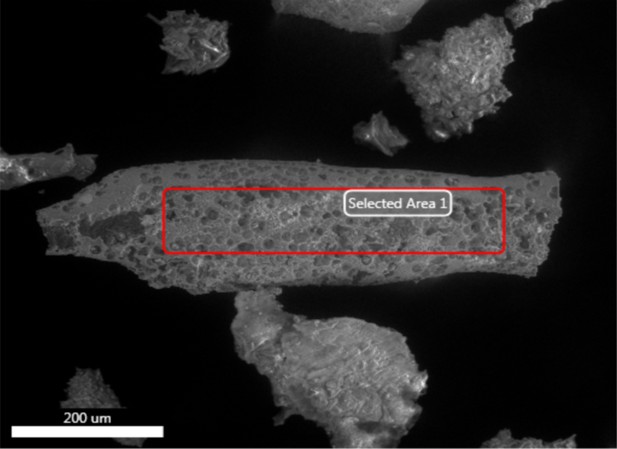
Electron image of another vessel from demineralised Late Cretaceous Centrosaurus bone area C.
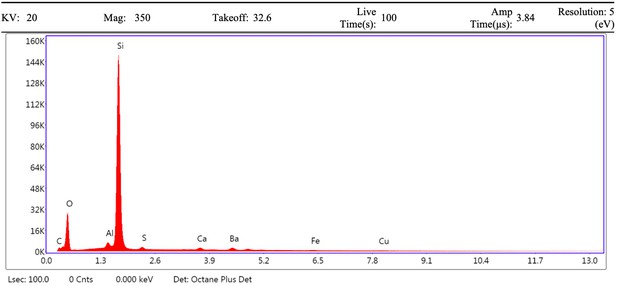
EDS spectrum of vessel from demineralised Late Cretaceous Centrosaurus bone area C.
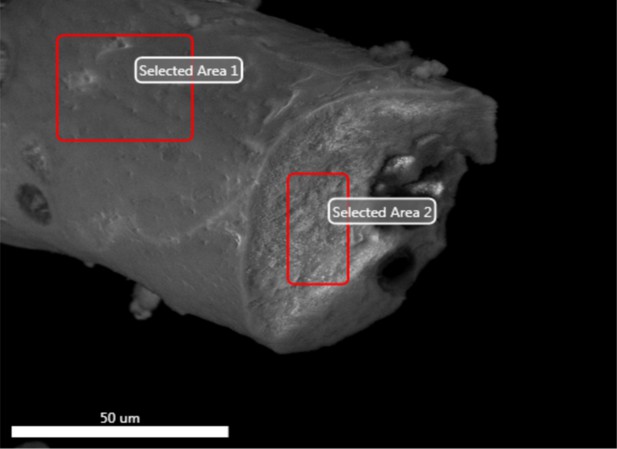
Electron image of a different vessel exterior (region 1) and interior (region 2) from demineralised Late Cretaceous Centrosaurus bone area D.
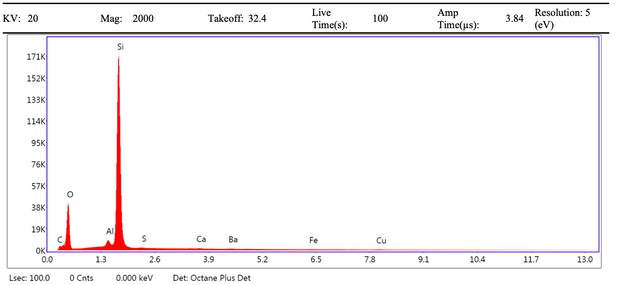
EDS spectrum of vessel exterior (region 1) from demineralised Late Cretaceous Centrosaurus bone area D.
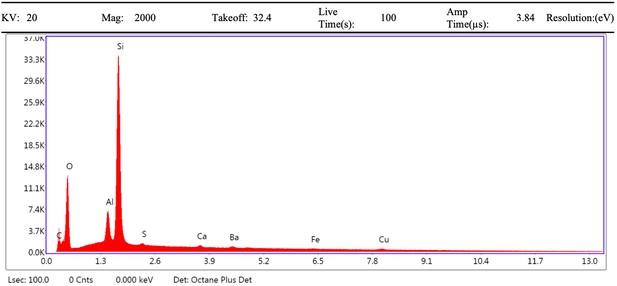
EDS spectrum of vessel interior (region 2) from demineralised Late Cretaceous Centrosaurus bone area D.
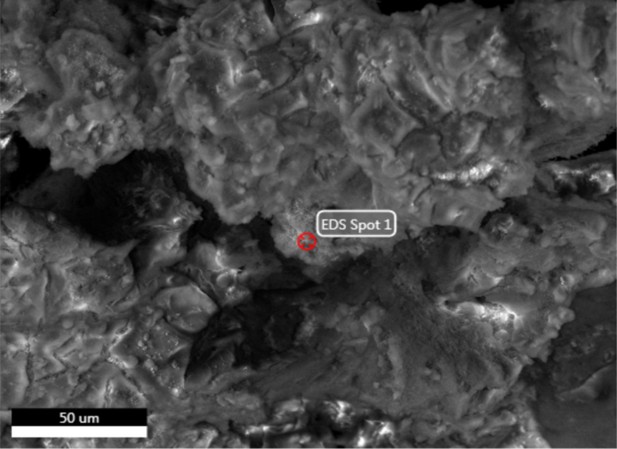
Electron image of mineral grain from demineralised Late Cretaceous Centrosaurus bone area E with spot one analysis shown.
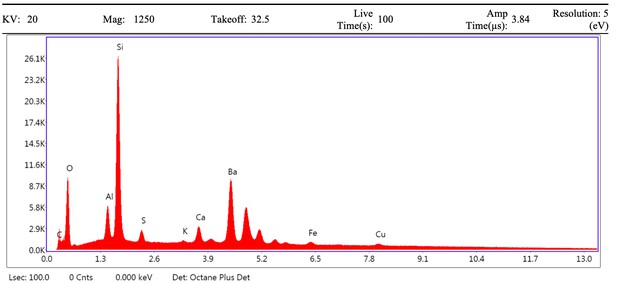
EDS spectrum of mineral grain from demineralised Late Cretaceous Centrosaurus bone area E, spot 1.
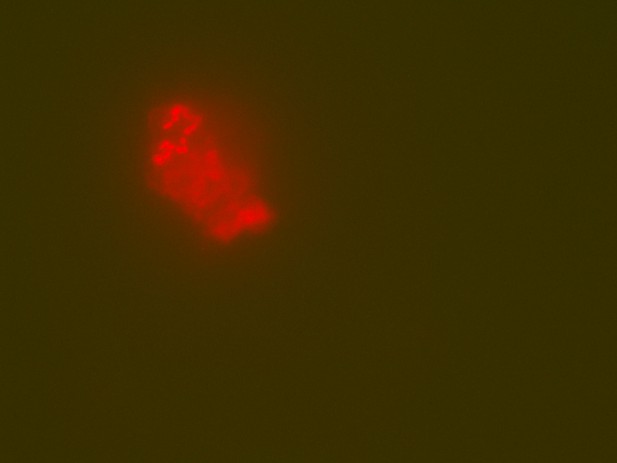
EDTA demineralised Late Cretaceous surface eroded fossil bone at 691 m elevation in Dinosaur Provincial Park showing possible cell clusters as might be expected in a biofilm (image 1).
Sample ID as in Appendix 1—table 2: 16B.

EDTA demineralised Late Cretaceous surface eroded fossil bone at 691 m elevation in Dinosaur Provincial Park showing possible cell clusters as might be expected in a biofilm (image 2).
Sample ID as in Appendix 1—table 2: 16B.

EDTA demineralised Late Cretaceous surface eroded fossil bone at 691 m elevation in Dinosaur Provincial Park showing possible cell clusters as might be expected in a biofilm (image 3).
Sample ID as in Appendix 1—table 2: 16B.

EDTA demineralised Late Cretaceous surface eroded fossil bone at 691 m elevation in Dinosaur Provincial Park showing possible cell clusters as might be expected in a biofilm (image 4).
Sample ID as in Appendix 1—table 2: 16B.
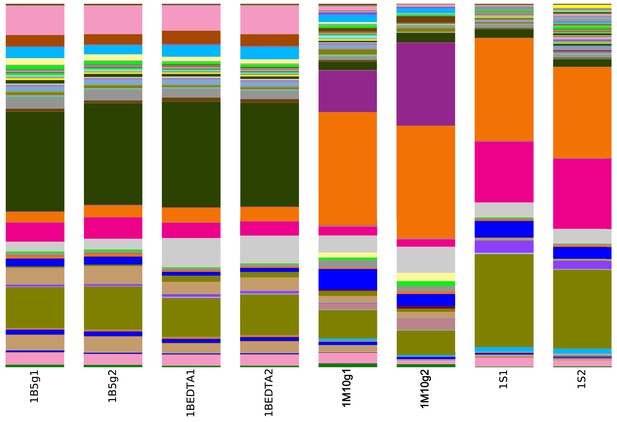
16S rRNA amplicon sequence diversity at the species level of matrix-surrounded subterranean Late Cretaceous Centrosaurus bone and adjacent mudstone matrix.
There are two replicates per sample. Sample ID’s are as in Appendix 1—table 2: B = matrix surrounded subterranean Centrosaurus bone core (surface scraped prior to powdering; 5g indicates that this sample was analysed using the DNeasy PowerMax Soil kit), BEDTA = matrix surrounded subterranean Centrosaurus bone core (EDTA demineralised, surface scraped prior to powdering), M = adjacent mudstone matrix of subterranean Centrosaurus bone (10g indicates that the samples were analysed using the DNeasy PowerMax Soil kit), S = Surface scrapings from matrix-surrounded subterranean Centrosaurus bone. The dark green bands are sequences phylogenetically close to Euzebya. See Source data 1 for a full listing of taxa. Both the bone core data and the EDTA demineralized bone core data are technique replicates deriving from single DNA extractions of each sample category, while both the mudstone matrix and surface scrapings data columns are from two separate DNA extractions of each sample category (Appendix 1—table 25).
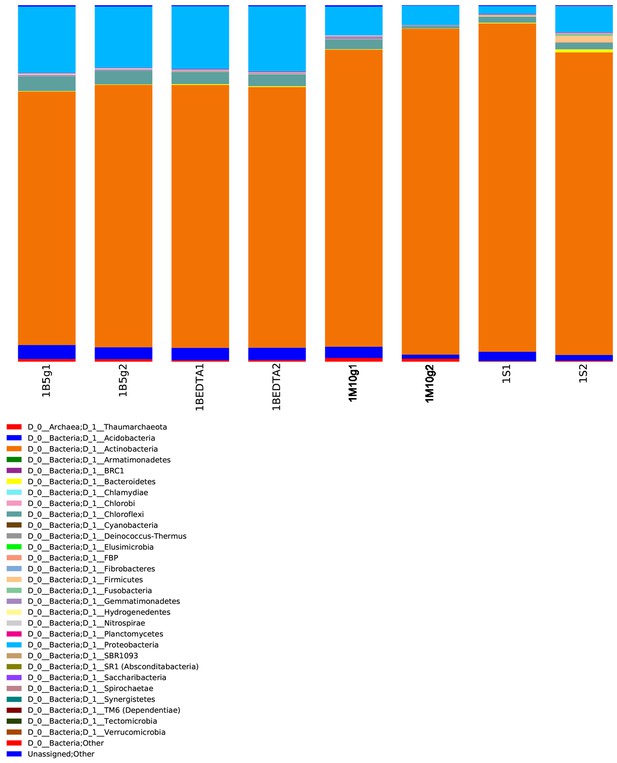
Comparison of microbial community (phylum level) from matrix-surrounded subterranean.
Centrosaurus bone, bone scrapings, and adjacent mudstone matrix. There are two replicates per sample. Sample ID’s are as in Appendix 1—table 2: B = matrix surrounded subterranean Centrosaurus bone core (surface scraped prior to powdering; 5g indicates that this sample was analysed using the DNeasy PowerMax Soil kit), BEDTA = matrix surrounded subterranean Centrosaurus bone core (EDTA demineralised, surface scraped prior to powdering), M = adjacent mudstone matrix of subterranean Centrosaurus bone (10g indicates that the samples were analysed using the DNeasy PowerMax Soil kit), S = Surface scrapings from matrix-surrounded subterranean Centrosaurus bone. Both the bone core data and the EDTA demineralized bone core data are technique replicates deriving from single DNA extractions of each sample category, while both the mudstone matrix and surface scrapings data columns are from two separate DNA extractions of each sample category (Appendix 1—table 25).
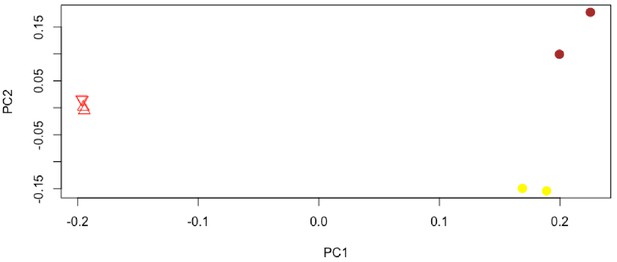
PCA of species-level 16S rRNA amplicon sequence data (percentages without additional normalization).
Red triangles with one vertex upward are from the bone core, red triangles with one vertex downward are the EDTA demineralized bone core, yellow circles are the bone surface scrapings, and brown circles are the mudstone. PC1 and PC2 account for 75.87% and 21.65% of the variation in the data, respectively.
Tables
Comparison of Late Cretaceous, Pleistocene-Holocene, and modern amino acid racemization values of the KOH-treated samples.
NA indicates that amino acid concentration was below detection limit.
| Sample treatment | Asx D/L | Glx D/L | Ser D/L | Ala D/L | Val D/L | |
|---|---|---|---|---|---|---|
| Matrix-surrounded subterranean Centrosaurus bone | ||||||
| Ethanol rinsed before powdering, gelated | NA | NA | NA | NA | NA | |
| Subterranean Centrosaurus bone uncovered from matrix before collection | ||||||
| Ethanol rinsed before powdering, gelated | 0.21 | 0.55 | 0 | 0.21 | 0 | |
| Adjacent mudstone matrix of subterranean Centrosaurus bone | ||||||
| Ethanol rinsed before powdering, gelated | NA | NA | 0 | 0.30 | 0 | |
| Surface-eroded Centrosaurus bone from BB180 | ||||||
| Ethanol rinsed before powdering, gelated | 0 | 0 | 0 | 0 | 0 | |
| Surface-eroded Late Cretaceous bone on same ridge and ~ 21 m above BB180 | ||||||
| Ethanol rinsed before powdering, gelated | 0 | 0.95 | 0 | 0.32 | 0.90 | |
| Topsoil on same ridge and ~ 64 m above BB180 | ||||||
| Ethanol rinsed before powdering, gelated | 0.14 | 0.14 | 0.05 | 0.09 | 0.04 | |
| Pleistocene-Holocene surface-eroded Carcharias teeth | ||||||
| Unrinsed | 0.21 | 0.04 | 0.09 | 0.03 | 0.01 | |
| Ethanol rinsed before powdering | 0.51 | 0.15 | 0.30 | 0.16 | 0.11 | |
| Ethanol rinsed before powdering | 0.53 | 0.15 | 0.30 | 0.17 | 0.11 | |
| Modern Gallus bone | ||||||
| Unrinsed | 0.05 | 0.03 | 0 | 0.02 | 0 | |
| Ethanol rinsed before powdering | 0.06 | 0.03 | 0 | 0.02 | 0 | |
Carbon data from Late Cretaceous fossil bone, mudstone, topsoil, and younger bone.
| Sample | % mass after HCl demineralization | C % (organic fraction) | F14C (organic fraction) |
|---|---|---|---|
| Matrix-surrounded subterranean Centrosaurus bone core (surface scraped prior to powdering) | 53.98 | 2.777 | 0.0149 |
| Adjacent mudstone matrix of subterranean Centrosaurus bone | 82.27 | 1.32 | 0.0573 |
| Topsoil on same ridge and ~ 64 m above BB180 | 91.63 | 2 | 0.766 |
| Mudstone on same ridge and ~ 23 m above BB180 | 90.38 | 0.89 | 0.0628 |
| Surface-eroded Late Cretaceous bone core on same ridge and ~ 21 m above BB180 (surface scraped prior to powdering) | 43.4 | 1.63 | 0.0422 |
| Yarnton bovine right femur (82–71 ka, Cook et al., 2012) | 16.73 | 44.9 | 0.0056* |
-
*This sample was used for blank correction in the AMS analyses, therefore this value is not blank-subtracted.
DNA concentrations in mudstone matrix and bone quantified with Qubit fluorometry.
| Sample | Average DNA concentration (ng/μL) | Total DNA (ng) | DNA per 1 g of bone or mudstone (ng/g) |
|---|---|---|---|
| Matrix-surrounded subterranean Centrosaurus bone core (surface scraped prior to powdering) | 0.79 | 3965 | 793 |
| Adjacent mudstone matrix of subterranean Centrosaurus bone | 0.03 | 164 | 16.4 |
| Laboratory blank | Below detection (<0.01*) |
-
*Note: the detection limit corresponds to the actual concentration of DNA in the assay tube (0.0005 ng/µL) after 200 times dilution of the original sample according to the manufacturer’s protocol.
Average human body composition and hydrolysis calculations.
Source is Janaway et al., 2009.
| Substance | % of body mass | Average adult body mass (kg) | Mass of substance (kg) | Mass of substance (Da) | Typical amino acid (Da) | Number of amino acids | Maximum number of peptide bonds | Number of bonds surviving |
|---|---|---|---|---|---|---|---|---|
| Protein | 20 | 62 | 12.4 | 7.46728E + 27 | 110 | 6.78844E + 25 | 6.78844E + 25 | −1.25963E + 27 |
| H2O (Da) | Number of H2O molecules | % H2O molecules used up in hydrolysis | ||||||
| Water | 64 | 62 | 39.68 | 2.38953E + 28 | 18 | 1.32752E + 27 | 5.113636364 |
Sample ID key and descriptions.
| Sample bag # | Originally sent to | HPLC ID | Py-GC/MS ID | DNA extraction, fluorescence microscopy, 16S rRNA amplicon ID | Location | Type | Details |
|---|---|---|---|---|---|---|---|
| 1 (TMP 2016.016.0007) | Princeton | NA | NA | 1B, 1S, 1F | Dinosaur Provincial Park, Alberta, Canada | Bone | Centrosaurus rib aseptically collected within immediately surrounding sediment. One end of the rib was first exposed. Aseptic protocol was then implemented to expose more of the rib. A rib section was isolated with the surrounding sediment kept in situ. Foil was placed on top of this section. The bone was sawed on its ends and then flipped by prying underneath with an awl. Mudstone tended to fracture during flipping. More foil was added after flipping to encase the whole sample. |
| 1 (TMP 2016.016.0007) | Princeton | NA | NA | 1M | Dinosaur Provincial Park, Alberta, Canada | Mudstone matrix | Mudstone matrix surrounding and collected with the matrix-surrounded Centrosaurus rib sample. Had tendency to fracture when manipulated. |
| 2 (TMP 2016.016.0007) | Princeton | NA | NA | 2B, 2S | Dinosaur Provincial Park, Alberta, Canada | Bone | Uncovered Centrosaurus rib section immediately adjacent to matrix-surrounded section of sample bag #1. |
| 6 (TMP 2016.016.0013) | Princeton | NA | NA | 6B, 6S | Dinosaur Provincial Park, Alberta, Canada | Bone | Surface bone eroded out of BB180, either excavated in past years and left or naturally eroded. About eight steps away from quarry cliff-face. 667 m elevation. |
| 8 (TMP 2016.016.0014) | Princeton | NA | NA | 8M | Dinosaur Provincial Park, Alberta, Canada | Mudstone sediment | Sediment from BB180 at bone producing layer. Sampled ~ 30 cm away from the sampled rib and tibia (sample bags #1–4). 670 m elevation. |
| 10 (TMP 2016.016.0015) | Princeton | NA | NA | 10, 10T | Dinosaur Provincial Park, Alberta, Canada | Topsoil | Topsoil from same ridge as BB180. 734 m elevation. |
| 11 (TMP 2016.016.0016) | Princeton | NA | NA | 11M | Dinosaur Provincial Park, Alberta, Canada | Mudstone sediment | Sediment on same ridge as BB180 from 709 m elevation. Outcrop was dug into by several cm before sampling. |
| 13 (TMP 2016.016.0017) | Princeton | NA | NA | 13, 13M | Dinosaur Provincial Park, Alberta, Canada | Mudstone sediment | Sediment on same ridge as BB180 from 693 m elevation. Outcrop was dug into by several cm before sampling. |
| 16 (TMP 2016.016.0018) | Princeton | NA | NA | 16B, 16S | Dinosaur Provincial Park, Alberta, Canada | Bone | Surface bone eroded out of same ridge as BB180 but at 691 m elevation. Unknown taxon. Near sample bags #13–14. |
| NA | Bristol | 1 | 1 | NA | Sainsbury’s Bristol, UK | Bone | Chicken bone purchased from grocery store with meat removed. |
| 3 (TMP 2016.016.0008) | Bristol | 2 | 2 | NA | Dinosaur Provincial Park, Alberta, Canada | Bone | Centrosaurus tibia aseptically collected within immediately surrounding sediment. One end of the tibia was first exposed. Aseptic protocol was then implemented to expose more of the tibia. A tibia section was isolated with the surrounding sediment kept in situ. Foil was placed on top of this section. The bone was sawed on its ends and then flipped by prying underneath with an awl. Mudstone tended to fracture during flipping. More foil was added after flipping to encase the whole sample. |
| 3 (TMP 2016.016.0008) | Bristol | 3 | 3 | NA | Dinosaur Provincial Park, Alberta, Canada | Mudstone matrix | Mudstone matrix surrounding and collected with the matrix-surrounded Centrosaurus tibia. Had tendency to fracture when manipulated. |
| 4 (TMP 2016.016.0008) | Bristol | 4 | 4 | NA | Dinosaur Provincial Park, Alberta, Canada | Bone | Uncovered Centrosaurus tibia region immediately adjacent to matrix-surrounded section of sample bag #3. |
| 5 (TMP 2016.016.0013) | Bristol | 6 | 6 | NA | Dinosaur Provincial Park, Alberta, Canada | Bone | Surface bone eroded out of BB180, either excavated in past years and left or naturally eroded. About eight steps away from quarry cliff-face. 667 m elevation. |
| 7 (TMP 2016.016.0014) | Bristol | 5 | 5 | NA | Dinosaur Provincial Park, Alberta, Canada | Mudstone sediment | Sediment from BB180 at bone producing layer. Sampled ~ 30 cm away from the sampled rib and tibia (sample bags #1–4). 670 m elevation. |
| 9 (TMP 2016.016.0015) | Bristol | 7 | 7 | NA | Dinosaur Provincial Park, Alberta, Canada | Topsoil | Topsoil from same ridge as BB180. 734 m elevation. |
| 12 (TMP 2016.016.0016) | Bristol | 8 | 8 | NA | Dinosaur Provincial Park, Alberta, Canada | Mudstone sediment | Sediment on same ridge as BB180 from 709 m elevation. Outcrop was dug into by several cm before sampling. |
| 14 (TMP 2016.016.0017) | Bristol | 9 | 9 | NA | Dinosaur Provincial Park, Alberta, Canada | Sediment | Sediment on same ridge as BB180 from 693 m elevation. Outcrop was dug into by several cm before sampling. |
| 15 (TMP 2016.016.0018) | Bristol | 10 | 10 | NA | Dinosaur Provincial Park, Alberta, Canada | Bone | Surface bone eroded out of same ridge as BB180 but at 691 m elevation. Unknown taxon. Near sample bags #13–14. |
Pilot samples of fossil Mesozoic bone THAA composition.
Concentrations in picomoles/mg.
| Taxon | Details | Location | Approximate age | Notes | [Asx] | [Glx] | [Ser] | [L-Thr] | [L-His] | [Gly] | [L-Arg] | [Ala] | [Tyr] | [Val] | [Phe] | [Leu] | [Ile] | [Total] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hadrosaur? | unrinsed, exterior, Dinosaur Park Fm | Dinosaur Provincial Park, Alberta, Canada | Late Cretaceous | signal suppression | 81 | 126 | 153 | 0 | 81.61 | 433 | 24 | 119 | 1.0 | 30 | 65 | 61 | 0 | 1174 |
| hadrosaur? | unrinsed, exterior, Dinosaur Park Fm | Dinosaur Provincial Park, Alberta, Canada | Late Cretaceous | signal suppression | 140 | 175 | 230 | 67 | 0.00 | 568 | 0 | 108 | 2.0 | 52 | 76 | 0 | 0 | 1417 |
| hadrosaur? | unrinsed, interior, Dinosaur Park Fm | Dinosaur Provincial Park, Alberta, Canada | Late Cretaceous | signal suppression | 57 | 97 | 135 | 28 | 0.00 | 432 | 30 | 73 | 3.0 | 26 | 38 | 0 | 0 | 919 |
| hadrosaur? | unrinsed, interior, Dinosaur Park Fm | Dinosaur Provincial Park, Alberta, Canada | Late Cretaceous | signal suppression | 58 | 102 | 123 | 31 | 0.00 | 354 | 30 | 84 | 4.0 | 30 | 32 | 31 | 23 | 902 |
| hadrosaur? | gelated, unrinsed, interior, Dinosaur Park Fm | Dinosaur Provincial Park, Alberta, Canada | Late Cretaceous | weaker signal suppression | 46 | 84 | 104 | 25 | 0.00 | 356 | 25 | 72 | 12.0 | 23 | 39 | 27 | 18 | 831 |
| sauropod? | unrinsed,, high in siderite, Morrison Fm | near Grass Range, Montana, USA | Late Jurassic | strong signal suppression | 0 | 127 | 0 | 0 | 0.00 | 482 | 0 | 149 | 13.0 | 0 | 0 | 0 | 0 | 772 |
| hadrosaur? | ethanol rinsed, exterior, Dinosaur Park Fm | Dinosaur Provincial Park, Alberta, Canada | Late Cretaceous | strong signal suppression | 84 | 119 | 164 | 0 | 0.00 | 465 | 95 | 0 | 6.0 | 0 | 59 | 0 | 0 | 991 |
| hadrosaur? | ethanol rinsed, exterior, Dinosaur Park Fm | Dinosaur Provincial Park, Alberta, Canada | Late Cretaceous | strong signal suppression | 68 | 105 | 145 | 39 | 0.00 | 348 | 0 | 76 | 7.0 | 0 | 0 | 0 | 0 | 789 |
| hadrosaur? | gelated. ethanol rinsed, exterior, Dinosaur Park Fm | Dinosaur Provincial Park, Alberta, Canada | Late Cretaceous | weaker signal suppression | 56 | 93 | 132 | 35 | 0.00 | 294 | 0 | 62 | 14.0 | 37 | 40 | 32 | 21 | 816 |
| hadrosaur? | ethanol rinsed, interior, Dinosaur Park Fm | Dinosaur Provincial Park, Alberta, Canada | Late Cretaceous | signal suppression | 60 | 116 | 174 | 30 | 0.00 | 315 | 0 | 64 | 8.0 | 28 | 38 | 0 | 0 | 832 |
| hadrosaur? | ethanol rinsed, interior, Dinosaur Park Fm | Dinosaur Provincial Park, Alberta, Canada | Late Cretaceous | signal suppression | 57 | 104 | 164 | 38 | 73.67 | 286 | 0 | 56 | 9.0 | 28 | 33 | 30 | 16 | 895 |
| hadrosaur? | gelated, ethanol rinsed, interior, Dinosaur Park Fm | Dinosaur Provincial Park, Alberta, Canada | Late Cretaceous | weaker signal suppression | 50 | 98 | 149 | 32 | 42.43 | 280 | 13 | 58 | 15.0 | 26 | 39 | 30 | 18 | 848 |
| sauropod? | gelated, ethanol rinsed, high in siderite, Morrison Fm | near Grass Range, Montana, USA | Late Jurassic | still strong signal suppression | 0 | 98 | 241 | 129 | 0.00 | 997 | 0 | 0 | 16.0 | 0 | 0 | 0 | 0 | 1481 |
| hadrosaur? | unrinsed, exterior, Dinosaur Park Fm | Dinosaur Provincial Park, Alberta, Canada | Late Cretaceous | no signal suppression | 9 | 3 | 7 | 1 | 0.00 | 21 | 1 | 12 | 12.0 | 6 | 4 | 1 | 5 | 82 |
| hadrosaur? | unrinsed, exterior, Dinosaur Park Fm | Dinosaur Provincial Park, Alberta, Canada | Late Cretaceous | no signal suppression | 8 | 4 | 40 | 8 | 5.98 | 37 | 1 | 15 | 13.0 | 6 | 5 | 4 | 4 | 152 |
| Alioramus | loaned from American Museum of Natural History | Mongolia | Late Cretaceous | poor amino acid profile | 0 | 0 | 0 | 0 | 0.00 | 0 | 0 | 0 | 17.0 | 0 | 0 | 0 | 0 | 17 |
| Alioramus | ethanol rinsed, loaned from American Museum of Natural History | Mongolia | Late Cretaceous | decent amino acid profile | 29 | 40 | 89 | 0 | 0.00 | 98 | 0 | 61 | 19.2 | 0 | 0 | 0 | 0 | 336 |
| Alioramus | gelated, ethanol rinsed, loaned from American Museum of Natural History | Mongolia | Late Cretaceous | poor amino acid profile | 0 | 0 | 0 | 0 | 0.00 | 0 | 0 | 0 | 21.4 | 0 | 0 | 0 | 0 | 21 |
Pilot samples of fossil Mesozoic bone THAA D/L values.
| Taxon | Details | Location | Approximate age | Notes | Asx D/L | Glx D/L | Ser D/L | Arg D/L | Ala D/L | Val D/L | Phe D/L | Leu D/L | Ile D/L | Tyr D/L |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hadrosaur? | unrinsed, exterior, Dinosaur Park Fm | Dinosaur Provincial Park, Alberta, Canada | Late Cretaceous | signal suppression | 0.185 | 0.121 | 0.000 | 0.000 | 0.552 | 0.000 | 0.000 | 1.071 | NA | NA |
| hadrosaur? | unrinsed, exterior, Dinosaur Park Fm | Dinosaur Provincial Park, Alberta, Canada | Late Cretaceous | signal suppression | 0.185 | 0.000 | 0.000 | NA | 0.000 | 0.000 | 0.000 | NA | NA | NA |
| hadrosaur? | unrinsed, interior, Dinosaur Park Fm | Dinosaur Provincial Park, Alberta, Canada | Late Cretaceous | signal suppression | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | NA | NA | 0.000 |
| hadrosaur? | unrinsed, interior, Dinosaur Park Fm | Dinosaur Provincial Park, Alberta, Canada | Late Cretaceous | signal suppression | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | NA |
| hadrosaur? | gelated, unrinsed, interior, Dinosaur Park Fm | Dinosaur Provincial Park, Alberta, Canada | Late Cretaceous | weaker signal suppression | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | NA |
| sauropod? | unrinsed, high in siderite, Morrison Fm | near Grass Range, Montana, USA | Late Jurassic | strong signal suppression | NA | 0.000 | NA | NA | 0.000 | NA | NA | NA | NA | NA |
| hadrosaur? | ethanol rinsed, exterior, Dinosaur Park Fm | Dinosaur Provincial Park, Alberta, Canada | Late Cretaceous | strong signal suppression | 0.000 | 0.000 | 0.000 | 0.000 | NA | NA | 0.000 | NA | NA | 0.000 |
| hadrosaur? | ethanol rinsed, exterior, Dinosaur Park Fm | Dinosaur Provincial Park, Alberta, Canada | Late Cretaceous | strong signal suppression | 0.000 | 0.000 | 0.000 | NA | 0.000 | NA | NA | NA | NA | NA |
| hadrosaur? | gelated, ethanol rinsed, exterior, Dinosaur Park Fm | Dinosaur Provincial Park, Alberta, Canada | Late Cretaceous | weaker signal suppression | 0.000 | 0.000 | 0.000 | NA | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | NA |
| hadrosaur? | ethanol rinsed, interior, Dinosaur Park Fm | Dinosaur Provincial Park, Alberta, Canada | Late Cretaceous | signal suppression | 0.000 | 0.000 | 0.000 | NA | 0.000 | 0.000 | 0.000 | NA | NA | 0.000 |
| hadrosaur? | ethanol rinsed, interior, Dinosaur Park Fm | Dinosaur Provincial Park, Alberta, Canada | Late Cretaceous | signal suppression | 0.000 | 0.000 | 0.000 | NA | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | NA |
| hadrosaur? | gelated, ethanol rinsed, interior, Dinosaur Park Fm | Dinosaur Provincial Park, Alberta, Canada | Late Cretaceous | weaker signal suppression | 0.000 | 0.000 | 0.000 | 1.842 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | NA |
| sauropod? | gelated, ethanol rinsed, high in siderite, Morrison Fm | near Grass Range, Montana, USA | Late Jurassic | still strong signal suppression | NA | 0.000 | 0.000 | NA | NA | NA | NA | NA | NA | NA |
| hadrosaur? | unrinsed, exterior, Dinosaur Park Fm | Dinosaur Provincial Park, Alberta, Canada | Late Cretaceous | no signal suppression | 0.000 | 0.426 | 0.895 | 0.000 | 0.000 | 0.000 | 0.428 | 0.000 | 0.000 | 0.000 |
| hadrosaur? | unrinsed, exterior, Dinosaur Park Fm | Dinosaur Provincial Park, Alberta, Canada | Late Cretaceous | no signal suppression | 0.084 | 0.163 | 0.014 | 1.524 | 0.089 | 0.108 | 0.245 | 0.000 | 0.000 | 0.134 |
| Alioramus | loaned from American Museum of Natural History | Mongolia | Late Cretaceous | poor amino acid profile | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Alioramus | ethanol rinsed, loaned from American Museum of Natural History | Mongolia | Late Cretaceous | decent amino acid profile | 0.000 | 0.000 | 0.000 | NA | 0.000 | NA | NA | NA | NA | NA |
| Alioramus | gelated, ethanol rinsed, loaned from American Museum of Natural History | Mongolia | Late Cretaceous | poor amino acid profile | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
Shark teeth THAA composition.
Concentrations in picomoles/mg.
| Species | Details | Location | Approximate age | Notes | [Asx] | [Glx] | [Ser] | [L-Thr] | [L-His] | [Gly] | [L-Arg] | [Ala] | [Tyr] | [Val] | [Phe] | [Leu] | [Ile] | [Total] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carcharias taurus | unrinsed, unnamed Pleistocene-Holocene sediments | Ponte Vedra Beach, Florida, USA | Quaternary | high concentration | 52656 | 53337 | 29149 | 17064 | 4643.25 | 266160 | 31884 | 76481 | 5.0 | 16577 | 9366 | 20462 | 19071 | 596855 |
| Carcharias taurus | ethanol rinsed, unnamed Pleistocene-Holocene sediments | Ponte Vedra Beach, Florida, USA | Quaternary | weak signal suppression | 478 | 471 | 104 | 65 | 58.31 | 2309 | 188 | 430 | 10.0 | 194 | 93 | 229 | 195 | 4824 |
| Carcharias taurus | ethanol rinsed, unnamed Pleistocene-Holocene sediments | Ponte Vedra Beach, Florida, USA | Quaternary | weak signal suppression | 588 | 534 | 115 | 73 | 61.79 | 2617 | 203 | 537 | 11.0 | 261 | 120 | 272 | 255 | 5647 |
Shark teeth THAA D/L values.
| Species | Details | Location | Approximate age | Notes | Asx D/L | Glx D/L | Ser D/L | Arg D/L | Ala D/L | Val D/L | Phe D/L | Leu D/L | Ile D/L | Tyr D/L |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carcharias taurus | unrinsed, unnamed Pleistocene- Holocene sediments | Ponte Vedra Beach, Florida, USA | Quaternary | high concentration | 0.209 | 0.039 | 0.092 | 0.047 | 0.027 | 0.011 | 0.028 | 0.026 | 0.165 | 0.114 |
| Carcharias taurus | ethanol rinsed, unnamed Pleistocene- Holocene sediments | Ponte Vedra Beach, Florida, USA | Quaternary | weak signal suppression | 0.512 | 0.153 | 0.301 | 0.236 | 0.155 | 0.114 | 0.000 | 0.092 | 0.107 | 0.000 |
| Carcharias taurus | ethanol rinsed, unnamed Pleistocene- Holocene sediments | Ponte Vedra Beach, Florida, USA | Quaternary | weak signal suppression | 0.527 | 0.154 | 0.295 | 0.358 | 0.166 | 0.112 | 0.158 | 0.094 | 0.106 | NA |
Aseptically collected samples and chicken control THAA composition.
See Appendix 1—table 2 for Sample ID’s. e = ethanol rinsed before powdering. d = gelated. Concentrations in picomoles/mg.
| Sample ID | [Asx] | [Glx] | [Ser] | [L-Thr] | [L-His] | [Gly] | [L-Arg] | [Ala] | [Tyr] | [Val] | [Phe] | [Leu] | [Ile] | [Total] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 71588 | 105116 | 45564 | 33327 | 17233.81 | 385402 | 58057 | 136671 | 14.7 | 38380 | 26397 | 57808 | 24829 | 1000385 |
| 1e | 81260 | 124003 | 51060 | 36547 | 22168.25 | 445966 | 66261 | 157216 | 17.4 | 42057 | 28714 | 56845 | 27166 | 1139278 |
| 2 | 0 | 0 | 0 | 0 | 0.00 | 209 | 0 | 0 | 14.8 | 0 | 0 | 0 | 0 | 224 |
| 2 | 0 | 41 | 0 | 0 | 0.00 | 182 | 0 | 0 | 15.0 | 0 | 0 | 0 | 0 | 239 |
| 2e | 56 | 60 | 0 | 0 | 0.00 | 181 | 0 | 68 | 17.6 | 0 | 0 | 0 | 0 | 383 |
| 2ed | 0 | 0 | 0 | 0 | 0.00 | 33 | 0 | 0 | 19.8 | 0 | 0 | 0 | 0 | 52 |
| 3 | 18 | 20 | 56 | 35 | 0.00 | 245 | 0 | 97 | 15.2 | 38 | 216 | 46 | 0 | 787 |
| 3 | 35 | 32 | 99 | 45 | 0.00 | 190 | 0 | 117 | 15.4 | 49 | 105 | 45 | 33 | 765 |
| 3e | 0 | 0 | 20 | 0 | 0.00 | 111 | 0 | 76 | 17.8 | 0 | 0 | 0 | 0 | 224 |
| 3ed | 0 | 0 | 24 | 17 | 0.00 | 92 | 0 | 75 | 20.0 | 28 | 0 | 0 | 37 | 294 |
| 4 | 29 | 32 | 43 | 0 | 0.00 | 158 | 0 | 41 | 15.6 | 0 | 43 | 0 | 0 | 360 |
| 4e | 234 | 227 | 177 | 108 | 0.00 | 509 | 0 | 220 | 18.0 | 148 | 0 | 0 | 0 | 1642 |
| 4ed | 87 | 110 | 51 | 45 | 0.00 | 191 | 17 | 95 | 20.2 | 57 | 29 | 53 | 36 | 791 |
| 5 | 0 | 0 | 0 | 0 | 0.00 | 0 | 0 | 0 | 15.8 | 0 | 0 | 0 | 0 | 16 |
| 5e | 0 | 64 | 0 | 0 | 0.00 | 184 | 0 | 0 | 18.1 | 0 | 0 | 0 | 0 | 266 |
| 5ed | 15 | 40 | 16 | 15 | 0.00 | 79 | 0 | 33 | 20.3 | 16 | 0 | 19 | 0 | 255 |
| 6 | 104 | 131 | 100 | 91 | 0.00 | 533 | 66 | 164 | 15.9 | 77 | 92 | 102 | 50 | 1526 |
| 6 | 148 | 144 | 116 | 97 | 0.00 | 318 | 52 | 163 | 16.1 | 89 | 59 | 104 | 55 | 1361 |
| 6e | 367 | 528 | 305 | 284 | 66.15 | 723 | 177 | 503 | 18.3 | 253 | 160 | 332 | 162 | 3878 |
| 6ed | 57 | 89 | 68 | 63 | 0.00 | 177 | 38 | 106 | 20.5 | 64 | 32 | 72 | 38 | 825 |
| 7 | 932 | 643 | 1516 | 1157 | 904.34 | 5438 | 898 | 2528 | 16.3 | 1153 | 675 | 1226 | 651 | 17737 |
| 7e | 3541 | 3068 | 3467 | 3424 | 799.16 | 11568 | 2024 | 6377 | 18.5 | 3508 | 1808 | 3972 | 2061 | 45636 |
| 7ed | 3505 | 2717 | 1674 | 1818 | 178.64 | 4659 | 541 | 3125 | 20.7 | 1947 | 900 | 2035 | 1148 | 24268 |
| 8 | 393 | 306 | 262 | 282 | 0.00 | 834 | 142 | 516 | 16.5 | 323 | 175 | 415 | 217 | 3880 |
| 8e | 0 | 0 | 0 | 0 | 0.00 | 0 | 0 | 0 | 18.7 | 0 | 0 | 0 | 0 | 19 |
| 8ed | 9 | 11 | 15 | 7 | 0.00 | 24 | 0 | 17 | 20.9 | 9 | 0 | 0 | 0 | 114 |
| 9 | 0 | 0 | 0 | 0 | 0.00 | 0 | 0 | 0 | 16.7 | 0 | 0 | 0 | 0 | 17 |
| 9e | 0 | 0 | 36 | 0 | 0.00 | 0 | 0 | 34 | 18.9 | 0 | 0 | 0 | 0 | 89 |
| 9ed | 18 | 48 | 45 | 0 | 0.00 | 82 | 0 | 35 | 21.1 | 0 | 0 | 0 | 0 | 249 |
| 10 | 59 | 77 | 47 | 40 | 0.00 | 185 | 38 | 104 | 16.9 | 54 | 0 | 68 | 0 | 688 |
| 10e | 39 | 199 | 0 | 0 | 0.00 | 148 | 0 | 70 | 19.1 | 139 | 0 | 169 | 0 | 782 |
| 10ed | 15 | 86 | 23 | 0 | 0.00 | 78 | 0 | 43 | 21.3 | 69 | 15 | 65 | 45 | 462 |
Aseptically collected samples and chicken control THAA D/L values.
See Appendix 1—table 2 for Sample ID’s. e = ethanol rinsed before powdering. d = gelated.
| Sample ID | Asx D/L | Glx D/L | Ser D/L | Arg D/L | Ala D/L | Val D/L | Phe D/L | Leu D/L | Ile D/L | Tyr D/L |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.053 | 0.027 | 0.000 | 0.096 | 0.015 | 0.000 | 0.019 | 0.022 | 0.000 | 0.000 |
| 1e | 0.055 | 0.029 | 0.000 | 0.081 | 0.016 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 2 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 2 | NA | 0.000 | NA | NA | NA | NA | NA | NA | NA | NA |
| 2e | 0.000 | 0.000 | NA | NA | 0.000 | NA | NA | NA | NA | NA |
| 2ed | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 3 | 0.000 | 0.000 | 0.000 | NA | 0.000 | 0.000 | 0.000 | 0.000 | NA | NA |
| 3 | 0.000 | 0.000 | 0.000 | NA | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | NA |
| 3e | NA | NA | 0.000 | NA | 0.338 | NA | NA | NA | NA | NA |
| 3ed | NA | NA | 0.000 | NA | 0.299 | 0.000 | NA | NA | 0.000 | NA |
| 4 | 0.000 | 0.000 | 0.000 | NA | 0.000 | NA | 0.000 | NA | NA | NA |
| 4e | 0.000 | 0.000 | 0.000 | NA | 0.000 | 0.000 | NA | NA | NA | NA |
| 4ed | 0.214 | 0.550 | 0.000 | 0.989 | 0.207 | 0.000 | 0.000 | 0.000 | 0.000 | NA |
| 5 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 5e | NA | 0.000 | NA | NA | NA | NA | NA | NA | NA | NA |
| 5ed | 0.000 | 0.727 | 0.000 | NA | 0.000 | 0.000 | NA | 0.000 | NA | NA |
| 6 | 0.000 | 0.000 | 0.000 | 1.996 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | NA |
| 6 | 0.110 | 0.000 | 0.000 | 1.645 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | NA |
| 6e | 0.082 | 0.084 | 0.000 | 0.889 | 0.078 | 0.000 | 0.000 | 0.000 | 0.000 | NA |
| 6ed | 0.000 | 0.000 | 0.000 | 1.338 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | NA |
| 7 | 0.418 | 0.378 | 0.020 | 0.887 | 0.071 | 0.000 | 0.000 | 0.056 | 0.000 | 0.000 |
| 7e | 0.130 | 0.123 | 0.038 | 0.856 | 0.080 | 0.038 | 0.048 | 0.070 | 0.000 | 0.000 |
| 7ed | 0.135 | 0.139 | 0.045 | 0.639 | 0.089 | 0.040 | 0.049 | 0.063 | 0.000 | NA |
| 8 | 0.125 | 0.000 | 0.000 | 1.148 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 8e | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 8ed | 0.000 | 0.000 | 0.000 | NA | 0.000 | 0.000 | NA | NA | NA | NA |
| 9 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 9e | NA | NA | 0.000 | NA | 0.000 | NA | NA | NA | NA | NA |
| 9ed | 0.000 | 1.055 | 0.000 | NA | 0.000 | NA | NA | NA | NA | NA |
| 10 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | NA | 0.000 | NA | NA |
| 10e | 0.000 | 0.538 | NA | NA | 0.000 | 0.750 | NA | 0.675 | NA | NA |
| 10ed | 0.000 | 0.951 | 0.000 | NA | 0.323 | 0.901 | 0.000 | 0.751 | 0.677 | NA |
Summary of variable loadings for the non-normalised PCA of THAA composition of only the sufficiently treated samples associated with Figure 6F.
Proportion of variance for each principal component in parentheses.
| PC1 (55.04%) | PC2 (22.66%) | |
|---|---|---|
| Asx | −0.100509897 | −0.18614726 |
| Glx | −0.214376490 | −0.05665852 |
| Ser | −0.144286512 | 0.18384010 |
| L. Thr | −0.098805431 | 0.04356834 |
| L. His | 0.007992613 | −0.05584073 |
| Gly | 0.805294439 | −0.41882274 |
| L. Arg | 0.009652324 | −0.20786057 |
| Alo | −0.185328976 | −0.04346374 |
| Tyr | 0.401827150 | 0.82375170 |
| Val | −0.186019066 | 0.10343138 |
| Phe | −0.048918108 | −0.06420351 |
| Leu | −0.150726139 | −0.09272073 |
| Ile | −0.095795908 | −0.02887370 |
Summary of variable loadings for the normalised PCA of THAA composition of only the sufficiently treated samples associated with Appendix 1—figure 22.
Proportion of variance for each principal component in parentheses.
| PC1 (31.64%) | PC2 (25.42%) | |
|---|---|---|
| Asx | −0.24729982 | −0.247493801 |
| Glx | −0.25925017 | 0.007025074 |
| Ser | −0.08672307 | 0.347552010 |
| L. Thr | −0.25282104 | 0.153203703 |
| L. His | −0.02073369 | −0.470955522 |
| Gly | 0.40229882 | −0.284333925 |
| L. Arg | −0.09443713 | −0.475273301 |
| Alo | −0.20019839 | 0.215195078 |
| Tyr | 0.37012033 | 0.243369947 |
| Val | −0.35372396 | 0.243119598 |
| Phe | −0.36466490 | −0.264010157 |
| Leu | −0.36975651 | −0.118102708 |
| Ile | −0.23897848 | 0.132700762 |
Summary of variable loadings for the normalised PCA of THAA composition associated with the plot in Appendix 1—figure 24F.
Proportion of variance for each principal component in parentheses.
| PC1 (30.01%) | PC2 (15.97%) | |
|---|---|---|
| Asx | −0.3356269 | 0.10557119 |
| Glx | −0.18736532 | 0.40091324 |
| Ser | −0.07675319 | −0.57396909 |
| L. Thr | −0.38235418 | −0.13868623 |
| L. His | −0.19702491 | 0.04051298 |
| Gly | 0.11068434 | 0.37647168 |
| L. Arg | −0.31147285 | 0.09964743 |
| Alo | −0.17744465 | −0.50906983 |
| Tyr | 0.35387327 | −0.09153454 |
| Val | −0.39069929 | 0.05903456 |
| Phe | −0.11192988 | −0.14530964 |
| Leu | −0.37568445 | 0.16792075 |
| Ile | −0.29841571 | −0.07628272 |
Summary of variable loadings for the non-normalised PCA of THAA composition associated with Appendix 1—figure 26.
Proportion of variance for each principal component in parentheses.
| PC1 (62.09%) | PC2 (24.49%) | |
|---|---|---|
| Asx | −0.061334126 | 0.101860246 |
| Glx | −0.101689512 | 0.042538223 |
| Ser | −0.017584775 | 0.227710833 |
| L. Thr | −0.026788291 | 0.091854076 |
| L. His | −0.006334389 | 0.006290775 |
| Gly | −0.438716137 | −0.826790362 |
| L. Arg | −0.024360271 | 0.030568893 |
| Alo | −0.079762251 | 0.26975096 |
| Tyr | 0.883822618 | −0.350122096 |
| Val | −0.035750618 | 0.142845467 |
| Phe | −0.029804014 | 0.062677703 |
| Leu | −0.040620928 | 0.139913899 |
| Ile | −0.021077307 | 0.060901383 |
Comparison of Late Cretaceous, Pleistocene-Holocene, and modern amino acid racemisation values.
NA indicates that amino acid concentration was below detection limit.
| Sample treatment | Asx D/L | Glx D/L | Ser D/L | Ala D/L | Val D/L | |
|---|---|---|---|---|---|---|
| Matrix-surrounded subterranean Centrosaurus bone | ||||||
| Unrinsed | NA | NA | NA | NA | NA | |
| Unrinsed | NA | 0 | NA | NA | NA | |
| Ethanol rinsed before powdering | 0 | 0 | NA | 0 | NA | |
| Ethanol rinsed before powdering, gelated | NA | NA | NA | NA | NA | |
| Subterranean Centrosaurus bone uncovered from matrix before collection | ||||||
| Unrinsed | 0 | 0 | 0 | 0 | NA | |
| Ethanol rinsed before powdering | 0 | 0 | 0 | 0 | 0 | |
| Ethanol rinsed before powdering, gelated | 0.214 | 0.550 | 0 | 0.207 | 0 | |
| Adjacent mudstone matrix of subterranean Centrosaurus bone | ||||||
| Unrinsed | 0 | 0 | 0 | 0 | 0 | |
| Unrinsed | 0 | 0 | 0 | 0 | 0 | |
| Ethanol rinsed before powdering | NA | NA | 0 | 0.338 | NA | |
| Ethanol rinsed before powdering, gelated | NA | NA | 0 | 0.299 | 0 | |
| Surface-eroded Centrosaurus bone from BB180 | ||||||
| Unrinsed | 0 | 0 | 0 | 0 | 0 | |
| Unrinsed | 0.110 | 0 | 0 | 0 | 0 | |
| Ethanol rinsed before powdering | 0.082 | 0.084 | 0 | 0.078 | 0 | |
| Ethanol rinsed before powdering, gelated | 0 | 0 | 0 | 0 | 0 | |
| Surface-eroded Late Cretaceous bone on same ridge and ~ 21 m above BB180 | ||||||
| Unrinsed | 0 | 0 | 0 | 0 | 0 | |
| Ethanol rinsed before powdering | 0 | 0.538 | NA | 0 | 0.750 | |
| Ethanol rinsed before powdering, gelated | 0 | 0.951 | 0 | 0.323 | 0.901 | |
| Topsoil on same ridge and ~ 64 m above BB180 | ||||||
| Unrinsed | 0.418 | 0.378 | 0.020 | 0.071 | 0 | |
| Ethanol rinsed before powdering | 0.130 | 0.123 | 0.038 | 0.080 | 0.038 | |
| Ethanol rinsed before powdering, gelated | 0.135 | 0.139 | 0.045 | 0.089 | 0.040 | |
| Pleistocene-Holocene surface-eroded Carcharias teeth | ||||||
| Unrinsed | 0.209 | 0.039 | 0.092 | 0.027 | 0.011 | |
| Ethanol rinsed before powdering | 0.512 | 0.153 | 0.301 | 0.155 | 0.114 | |
| Ethanol rinsed before powdering | 0.527 | 0.154 | 0.295 | 0.166 | 0.112 | |
| Modern Gallus bone | ||||||
| Unrinsed | 0.053 | 0.027 | 0 | 0.015 | 0 | |
| Ethanol rinsed before powdering | 0.055 | 0.029 | 0 | 0.016 | 0 | |
EDS eZAF Smart Quant results of demineralised modern chicken bone full area 1.
| Element | Weight % | Atomic % | Net int. | Error % | Kratio | Z | R | A | F |
|---|---|---|---|---|---|---|---|---|---|
| C K | 21.88 | 27.49 | 191.17 | 8.98 | 0.0666 | 1.0413 | 0.9803 | 0.2921 | 1.0000 |
| N K | 24.85 | 26.76 | 138.36 | 10.57 | 0.0451 | 1.0169 | 0.9906 | 0.1786 | 1.0000 |
| O K | 44.17 | 41.65 | 471.57 | 9.96 | 0.0679 | 0.9957 | 0.9999 | 0.1543 | 1.0000 |
| AlK | 1.77 | 0.99 | 134.20 | 7.25 | 0.0103 | 0.8846 | 1.0363 | 0.6545 | 1.0066 |
| SiK | 0.61 | 0.33 | 54.63 | 9.51 | 0.0043 | 0.9038 | 1.0422 | 0.7658 | 1.0100 |
| P K | 0.21 | 0.10 | 16.93 | 23.70 | 0.0016 | 0.8679 | 1.0477 | 0.8576 | 1.0160 |
| S K | 1.53 | 0.72 | 132.12 | 4.25 | 0.0127 | 0.8848 | 1.0529 | 0.9237 | 1.0199 |
| ClK | 4.10 | 1.74 | 322.09 | 2.47 | 0.0334 | 0.8416 | 1.0578 | 0.9553 | 1.0138 |
| FeK | 0.24 | 0.07 | 7.47 | 55.87 | 0.0022 | 0.7593 | 1.0862 | 1.0212 | 1.1884 |
| CuK | 0.63 | 0.15 | 12.85 | 30.91 | 0.0061 | 0.7264 | 1.0858 | 1.0175 | 1.2983 |
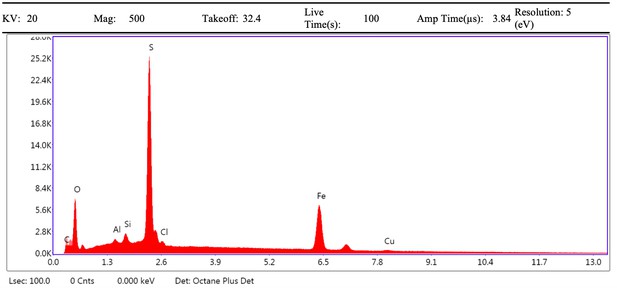
EDS eZAF Smart Quant results of demineralised Pleistocene-Holocene shark tooth full area 1.
| Element | Weight % | Atomic % | Net int. | Error % | Kratio | Z | R | A | F |
|---|---|---|---|---|---|---|---|---|---|
| C K | 25.65 | 42.51 | 150.16 | 10.58 | 0.0401 | 1.1074 | 0.9367 | 0.1411 | 1.0000 |
| O K | 27.79 | 34.58 | 542.95 | 9.50 | 0.0597 | 1.0620 | 0.9592 | 0.2024 | 1.0000 |
| AlK | 0.39 | 0.29 | 35.97 | 12.82 | 0.0021 | 0.9479 | 1.0018 | 0.5587 | 1.0102 |
| SiK | 0.88 | 0.62 | 99.39 | 7.89 | 0.0060 | 0.9691 | 1.0088 | 0.6901 | 1.0169 |
| S K | 21.63 | 13.43 | 2464.07 | 2.58 | 0.1807 | 0.9499 | 1.0217 | 0.8695 | 1.0112 |
| ClK | 0.76 | 0.43 | 68.59 | 10.21 | 0.0054 | 0.9039 | 1.0277 | 0.7737 | 1.0166 |
| FeK | 22.45 | 8.00 | 847.32 | 2.19 | 0.1927 | 0.8189 | 1.0658 | 1.0039 | 1.0442 |
| CuK | 0.44 | 0.14 | 10.24 | 41.09 | 0.0037 | 0.7847 | 1.0692 | 0.9849 | 1.0840 |
EDS eZAF Smart Quant results of mineral grain from demineralised Late Cretaceous Centrosaurus bone area A.
| Element | Weight % | Atomic % | Net int. | Error % | Kratio | Z | R | A | F |
|---|---|---|---|---|---|---|---|---|---|
| C K | 8.68 | 14.27 | 101.93 | 99.99 | 0.0107 | 1.0874 | 0.9545 | 0.1133 | 1.0000 |
| O K | 40.70 | 50.27 | 2568.99 | 8.50 | 0.1113 | 1.0415 | 0.9759 | 0.2625 | 1.0000 |
| AlK | 1.06 | 0.78 | 326.72 | 5.09 | 0.0075 | 0.9274 | 1.0163 | 0.7418 | 1.0360 |
| SiK | 48.46 | 34.09 | 16392.38 | 2.65 | 0.3872 | 0.9479 | 1.0229 | 0.8410 | 1.0025 |
| S K | 0.40 | 0.25 | 80.67 | 9.76 | 0.0023 | 0.9284 | 1.0350 | 0.6279 | 1.0056 |
| ClK | 0.01 | 0.01 | 2.31 | 60.70 | 0.0001 | 0.8832 | 1.0405 | 0.7259 | 1.0084 |
| CaK | 0.69 | 0.34 | 125.27 | 8.44 | 0.0057 | 0.8950 | 1.0552 | 0.9103 | 1.0202 |
EDS eZAF Smart Quant results of vessel (spot 1) from demineralised Late Cretaceous Centrosaurus bone area B.
| Element | Weight % | Atomic % | Net int. | Error % | Kratio | Z | R | A | F |
|---|---|---|---|---|---|---|---|---|---|
| C K | 5.61 | 9.66 | 81.05 | 99.99 | 0.0071 | 1.1005 | 0.9464 | 0.1152 | 1.0000 |
| O K | 40.95 | 52.94 | 3343.32 | 8.29 | 0.1212 | 1.0546 | 0.9683 | 0.2807 | 1.0000 |
| AlK | 3.75 | 2.88 | 1306.45 | 4.48 | 0.0253 | 0.9400 | 1.0097 | 0.6987 | 1.0256 |
| SiK | 44.51 | 32.78 | 16780.97 | 3.33 | 0.3320 | 0.9609 | 1.0165 | 0.7739 | 1.0031 |
| S K | 0.87 | 0.56 | 212.94 | 7.26 | 0.0052 | 0.9414 | 1.0290 | 0.6248 | 1.0067 |
| CaK | 0.96 | 0.50 | 211.92 | 6.87 | 0.0081 | 0.9079 | 1.0501 | 0.9059 | 1.0253 |
| BaL | 2.46 | 0.37 | 177.23 | 8.15 | 0.0184 | 0.6537 | 1.2526 | 1.0822 | 1.0599 |
| FeK | 0.40 | 0.15 | 46.67 | 13.64 | 0.0035 | 0.8100 | 1.0708 | 0.9944 | 1.0873 |
| CuK | 0.49 | 0.16 | 36.93 | 17.79 | 0.0044 | 0.7756 | 1.0733 | 1.0044 | 1.1609 |
EDS eZAF Smart Quant results of fibrous mass (spot 2) from demineralised Late Cretaceous Centrosaurus bone area B.
| Element | Weight % | Atomic % | Net int. | Error % | Kratio | Z | R | A | F |
|---|---|---|---|---|---|---|---|---|---|
| C K | 5.94 | 10.38 | 62.73 | 99.99 | 0.0070 | 1.1021 | 0.9464 | 0.1076 | 1.0000 |
| O K | 36.53 | 47.94 | 2200.10 | 8.49 | 0.1021 | 1.0562 | 0.9684 | 0.2646 | 1.0000 |
| AlK | 5.47 | 4.26 | 1553.47 | 4.17 | 0.0385 | 0.9413 | 1.0098 | 0.7276 | 1.0272 |
| SiK | 48.16 | 36.01 | 14261.52 | 3.30 | 0.3611 | 0.9622 | 1.0166 | 0.7771 | 1.0027 |
| S K | 0.57 | 0.37 | 105.79 | 8.71 | 0.0033 | 0.9426 | 1.0291 | 0.6074 | 1.0061 |
| K K | 0.06 | 0.03 | 12.05 | 55.96 | 0.0005 | 0.8927 | 1.0453 | 0.8536 | 1.0188 |
| CaK | 0.79 | 0.42 | 135.54 | 6.59 | 0.0066 | 0.9091 | 1.0501 | 0.8985 | 1.0229 |
| BaL | 1.43 | 0.22 | 80.41 | 12.01 | 0.0107 | 0.6546 | 1.2527 | 1.0784 | 1.0639 |
| FeK | 0.34 | 0.13 | 31.14 | 16.61 | 0.0030 | 0.8109 | 1.0708 | 0.9951 | 1.0943 |
| CuK | 0.71 | 0.24 | 42.62 | 16.37 | 0.0065 | 0.7765 | 1.0733 | 1.0049 | 1.1673 |
EDS eZAF Smart Quant results of vessel from demineralised Late Cretaceous Centrosaurus bone area C.
| Element | Weight % | Atomic % | Net int. | Error % | Kratio | Z | R | A | F |
|---|---|---|---|---|---|---|---|---|---|
| C K | 7.78 | 13.45 | 85.90 | 99.99 | 0.0100 | 1.1029 | 0.9436 | 0.1160 | 1.0000 |
| O K | 37.94 | 49.22 | 2221.65 | 8.49 | 0.1064 | 1.0572 | 0.9657 | 0.2653 | 1.0000 |
| AlK | 1.56 | 1.20 | 407.62 | 5.19 | 0.0104 | 0.9426 | 1.0074 | 0.6911 | 1.0273 |
| SiK | 46.30 | 34.22 | 13551.97 | 3.16 | 0.3543 | 0.9636 | 1.0143 | 0.7913 | 1.0032 |
| S K | 0.84 | 0.54 | 154.40 | 7.87 | 0.0049 | 0.9442 | 1.0269 | 0.6217 | 1.0071 |
| CaK | 1.01 | 0.52 | 169.40 | 6.80 | 0.0086 | 0.9108 | 1.0482 | 0.9041 | 1.0273 |
| BaL | 3.73 | 0.56 | 203.06 | 7.22 | 0.0279 | 0.6559 | 1.2507 | 1.0806 | 1.0545 |
| FeK | 0.41 | 0.15 | 35.85 | 17.08 | 0.0036 | 0.8128 | 1.0694 | 0.9912 | 1.0797 |
| CuK | 0.43 | 0.14 | 24.38 | 21.06 | 0.0038 | 0.7784 | 1.0721 | 1.0027 | 1.1492 |
EDS eZAF Smart Quant results of vessel exterior (region 1) from demineralised Late Cretaceous Centrosaurus bone area D.
| Element | Weight % | Atomic % | Net int. | Error % | Kratio | Z | R | A | F |
|---|---|---|---|---|---|---|---|---|---|
| C K | 10.99 | 17.49 | 159.50 | 99.99 | 0.0148 | 1.0832 | 0.9565 | 0.1245 | 1.0000 |
| O K | 43.55 | 52.05 | 3194.43 | 8.39 | 0.1224 | 1.0374 | 0.9778 | 0.2708 | 1.0000 |
| AlK | 1.62 | 1.15 | 541.46 | 4.85 | 0.0110 | 0.9237 | 1.0178 | 0.7166 | 1.0296 |
| SiK | 42.37 | 28.85 | 15672.00 | 2.92 | 0.3265 | 0.9440 | 1.0244 | 0.8142 | 1.0026 |
| S K | 0.16 | 0.10 | 38.75 | 15.68 | 0.0010 | 0.9246 | 1.0364 | 0.6496 | 1.0060 |
| CaK | 0.31 | 0.15 | 65.74 | 9.96 | 0.0026 | 0.8914 | 1.0564 | 0.9211 | 1.0232 |
| BaL | 0.53 | 0.07 | 36.74 | 27.53 | 0.0040 | 0.6416 | 1.2593 | 1.0954 | 1.0750 |
| FeK | 0.17 | 0.06 | 19.37 | 24.43 | 0.0015 | 0.7947 | 1.0756 | 1.0031 | 1.1088 |
| CuK | 0.29 | 0.09 | 21.53 | 21.86 | 0.0027 | 0.7607 | 1.0773 | 1.0090 | 1.2037 |
EDS eZAF Smart Quant results of vessel interior (region 2) from demineralised Late Cretaceous Centrosaurus bone area D.
| Element | Weight % | Atomic % | Net int. | Error % | Kratio | Z | R | A | F |
|---|---|---|---|---|---|---|---|---|---|
| C K | 25.09 | 35.59 | 183.44 | 99.99 | 0.0467 | 1.0660 | 0.9650 | 0.1747 | 1.0000 |
| O K | 43.49 | 46.31 | 999.60 | 8.89 | 0.1049 | 1.0204 | 0.9857 | 0.2365 | 1.0000 |
| AlK | 4.18 | 2.64 | 475.03 | 4.96 | 0.0265 | 0.9080 | 1.0246 | 0.6878 | 1.0165 |
| SiK | 24.25 | 14.71 | 3016.29 | 3.57 | 0.1723 | 0.9279 | 1.0309 | 0.7630 | 1.0034 |
| S K | 0.17 | 0.09 | 16.84 | 23.85 | 0.0012 | 0.9087 | 1.0424 | 0.7368 | 1.0077 |
| CaK | 0.43 | 0.18 | 33.69 | 13.24 | 0.0037 | 0.8760 | 1.0616 | 0.9572 | 1.0302 |
| BaL | 1.18 | 0.15 | 29.89 | 27.69 | 0.0090 | 0.6305 | 1.2647 | 1.1172 | 1.0818 |
| FeK | 0.19 | 0.06 | 7.87 | 55.09 | 0.0017 | 0.7809 | 1.0796 | 1.0094 | 1.1220 |
| CuK | 1.01 | 0.27 | 26.63 | 17.38 | 0.0091 | 0.7473 | 1.0805 | 1.0120 | 1.1953 |
EDS eZAF Smart Quant results of mineral grain from demineralised Late Cretaceous Centrosaurus bone area E, spot 1.
| Element | Weight % | Atomic % | Net int. | Error % | Kratio | Z | R | A | F |
|---|---|---|---|---|---|---|---|---|---|
| C K | 8.12 | 19.85 | 76.35 | 99.99 | 0.0181 | 1.2324 | 0.8623 | 0.1813 | 1.0000 |
| O K | 21.32 | 39.16 | 732.39 | 8.55 | 0.0718 | 1.1857 | 0.8869 | 0.2838 | 1.0000 |
| AlK | 4.94 | 5.38 | 431.02 | 7.60 | 0.0225 | 1.0649 | 0.9356 | 0.4237 | 1.0087 |
| SiK | 21.74 | 22.74 | 2311.26 | 6.07 | 0.1233 | 1.0899 | 0.9440 | 0.5169 | 1.0068 |
| S K | 1.45 | 1.33 | 146.92 | 8.19 | 0.0096 | 1.0705 | 0.9597 | 0.6086 | 1.0162 |
| K K | 0.27 | 0.20 | 27.82 | 18.41 | 0.0024 | 1.0174 | 0.9810 | 0.8363 | 1.0548 |
| CaK | 2.62 | 1.92 | 249.09 | 4.58 | 0.0257 | 1.0373 | 0.9875 | 0.8814 | 1.0698 |
| BaL | 36.01 | 7.70 | 1044.05 | 2.70 | 0.2929 | 0.7495 | 1.1854 | 1.0526 | 1.0312 |
| FeK | 1.34 | 0.71 | 59.05 | 10.93 | 0.0120 | 0.9325 | 1.0202 | 0.9145 | 1.0442 |
| CuK | 2.19 | 1.01 | 63.31 | 7.26 | 0.0203 | 0.8976 | 1.0306 | 0.9591 | 1.0793 |
Non-demineralised Dinosaur Provincial Park samples.
| Sample | Mass analysed (mg) | C % |
|---|---|---|
| Matrix-surrounded subterranean bone (not scrapped) | 3.219 | 2.3 |
| Adjacent mudstone matrix of subterranean bone | 2.112 | 1.11 |
| BB180 surface bone (not scrapped) | 3.184 | 4.19 |
| Topsoil | 2.235 | 1.26 |
| Mudstone 693 m elevation | 3.413 | 1.15 |
Pilot tests on demineralised Dinosaur Provincial Park samples (0.5 M HCl).
| Sample | Mass (mg) bulk powder before demineralisation | Mass (mg) after demineralisation | % mass surviving demineralisation | Mass (mg) analysed | C % | % change in [C] from bulk powder |
|---|---|---|---|---|---|---|
| Matrix-surrounded subterranean bone (not scrapped) | 299.7 | 7.536 | 2.514514515 | 4.035 | 13.48 | 486.0869565 |
| Adjacent mudstone matrix of subterranean bone | 308.5 | 253.8 | 82.26904376 | 3.499 | 1.28 | 15.31531532 |
| BB180 surface bone (not scrapped) | 307.1 | 71.99 | 23.44187561 | 10.051 | 0.13 | −96.8973747 |
| Topsoil | 307.2 | 281.5 | 91.63411458 | 6.21 | 1.43 | 13.49206349 |
| Mudstone 693 m elevation | 300.4 | 271.5 | 90.37949401 | 5.005 | 0.92 | −20 |
| Surface bone 691 m elevation (core) | 341 | 148 | 43.40175953 | 7.903 | 1.47 | NA |
Quebit fluorometer test results on Dinosaur Provincial Park samples.
Sample ID’s as in Appendix 1—table 2.
| Bag number | Type | Replicate | Sample ID | Kit | Ng of DNA / μL | Concentration | Concentrated ng of DNA / μL | ||
|---|---|---|---|---|---|---|---|---|---|
| First read | Second read | First read | Second read | ||||||
| NA | Blank | NA | Blank | Power Viral | Below detection | Below detection | |||
| 10 | Topsoil | 1 | 10 | Power Viral | 0.151 | 0.133 | |||
| 13 | Mudstone | 1 | 13 | Power Viral | Below detection | Below detection | |||
| 16 | Scrappings | 1 | 16S | Power Viral | 0.172 | 0.164 | |||
| 16 | Bone | 1 | 16B1 | Power Viral | 0.424 | 0.404 | |||
| 16 | Bone | 2 | 16B2 | Power Viral | 0.592 | 0.55 | |||
| NA | Blank | NA | Blank | Power Viral | Below detection | Below detection | |||
| 1 | 'Float' bone in matrix | 1 | 1F1 | Power Viral | 0.0926 | 0.0908 | |||
| 1 | Scrappings | 1 | 1S1 | Power Viral | 0.128 | 0.127 | |||
| 1 | Scrappings | 2 | 1S2 | Power Viral | 0.0238 | 0.0236 | |||
| 1 | Bone | 1 | 1B1 | Power Viral | 0.0382 | 0.0376 | |||
| 1 | Bone | 2 | 1B2 | Power Viral | 0.0544 | 0.0546 | |||
| 1 | Mudstone | 1 | 1M1 | Power Viral | Below detection | Below detection | |||
| 1 | Mudstone | 2 | 1M2 | Power Viral | Below detection | Below detection | |||
| NA | Blank | NA | Blank | Dneasy PowerMax Soil | Below detection | Below detection | x 25 | Below detection | Below detection |
| 1 | Bone | 3 | 1B5g | Dneasy PowerMax Soil | 0.798 | 0.788 | x 25 | 11.1 | 10.5 |
| 1 | Mudstone | 3 | 1M10g1 | Dneasy PowerMax Soil | 0.0334 | 0.0322 | x 25 | 0.626 | 0.612 |
| 1 | Mudstone | 4 | 1M10g2 | Dneasy PowerMax Soil | 0.0624 | 0.0596 | x 25 | 0.586 | 0.586 |
| 8 | Mudstone | 1 | 8M1 | Dneasy PowerMax Soil | 0.096 | 0.0924 | x 25 | 1.64 | 1.58 |
| 8 | Mudstone | 2 | 8M2 | Dneasy PowerMax Soil | 0.144 | 0.141 | x 25 | 1.6 | 1.55 |
| 11 | Mudstone | 1 | 11M1 | Dneasy PowerMax Soil | Below detection | Below detection | x 25 | 0.0306 | 0.0292 |
| 11 | Mudstone | 2 | 11M2 | Dneasy PowerMax Soil | Below detection | Below detection | x 25 | 0.0218 | 0.021 |
| 13 | Mudstone | 2 | 13M1 | Dneasy PowerMax Soil | Below detection | Below detection | x 25 | 0.123 | 0.119 |
| 13 | Mudstone | 3 | 13M2 | Dneasy PowerMax Soil | 0.0166 | 0.016 | x 25 | 0.141 | 0.138 |
| 2 | Scrappings | 1 | 2S1 | Power Viral | 0.167 | 0.168 | |||
| 2 | Scrappings | 2 | 2S2 | Power Viral | 0.113 | 0.109 | |||
| 2 | Bone | 1 | 2B1 | Power Viral | 0.134 | 0.131 | |||
| 2 | Bone | 2 | 2B2 | Power Viral | 0.118 | 0.116 | |||
| 6 | Scrappings | 1 | 6S1 | Power Viral | Below detection | Below detection | |||
| 6 | Scrappings | 2 | 6S2 | Power Viral | 0.0114 | Below detection | |||
| 6 | Bone | 1 | 6B1 | Power Viral | Below detection | Below detection | |||
| 6 | Bone | 2 | 6B2 | Power Viral | 0.0102 | Below detection | |||
| 1 | Float | 2 | 1F2 | Power Viral | 0.029 | 0.0278 | |||
| 10 | Topsoil | 2 | 10T2 | Power Viral | 1.04 | 1.02 | |||
| 6 | Scrappings | 3 | 6S3 | Power Viral | x 2 | 0.208 | 0.206 | ||
| 6 | Bone | 3 | 6B3 | Power Viral | x 2 | 0.0422 | 0.0424 | ||
| 1 | Bone (EDTA demineralised) | 1 | 1BEDTA | Dneasy PowerMax Soil | 0.148 | 0.145 | x 20 | 3.52 | 3.44 |
| 6 | Bone (EDTA demineralised) | 1 | 6BEDTA | Dneasy PowerMax Soil | 0.0144 | 0.013 | x 20 | 0.324 | 0.318 |
Cell abundance calculations for dinosaur bone and adjacent mudstone matrix from amino acid and DNA abundance based on Lomstein et al. (2012), Onstott et al. (2014), and Magnabosco et al. (2018).
| Amino acids | |||
|---|---|---|---|
| Bone | Mudstone | ||
| picomoles/mg | 50 | 300 | picomoles/mg |
| nanomoles/g of bone | 50 | 300 | nanomoles/g of mudstone |
| g/mole | 117.4 | 113.8 | g/mole |
| grams of AA/g | 8.39E-06 | 4.88E-05 | grams of AA/g |
| g of cells/g of bone | 1.68E-05 | 9.75E-05 | g of cells/g of mudstone |
| g dry wt/cell | 4.00E-14 | 4.00E-14 | g dry wt/cell |
| cells/gram | 4.19E + 08 | 2.44E + 09 | cells/gram |
| DNA | |||
| Bone | Mudstone | ||
| ng/g | 793 | 16.4 | ng/g |
| DNA g/g of bone | 7.93E-07 | 1.64E-08 | DNA g/g of mudstone |
| DNA g/cell | 3.00E-15 | 3.00E-15 | DNA g/cell |
| cells/g of bone | 2.64E + 08 | 5.47E + 06 | cells/g of mudstone |
Pairwise F values from PERMANOVA of species-level sequence percentages.
| Mudstone | Bone surface scrapings | Bone core | EDTA demineralized bone core | |
|---|---|---|---|---|
| Mudstone | - | 17.62 | 47.05 | 46.28 |
| Bone surface scrapings | 17.62 | - | 162.8 | 168.6 |
| Bone core | 47.05 | 162.8 | - | 33.41 |
| EDTA demineralized bone core | 46.28 | 168.6 | 33.41 | - |
Additional files
-
Source data 1
Raw data for ATR FTIR, EDS, Py-GC-MS, and 16S rRNA amplicon sequencing.
- https://cdn.elifesciences.org/articles/46205/elife-46205-data1-v2.zip
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/46205/elife-46205-transrepform-v2.docx