Massive antibody discovery used to probe structure–function relationships of the essential outer membrane protein LptD
Figures
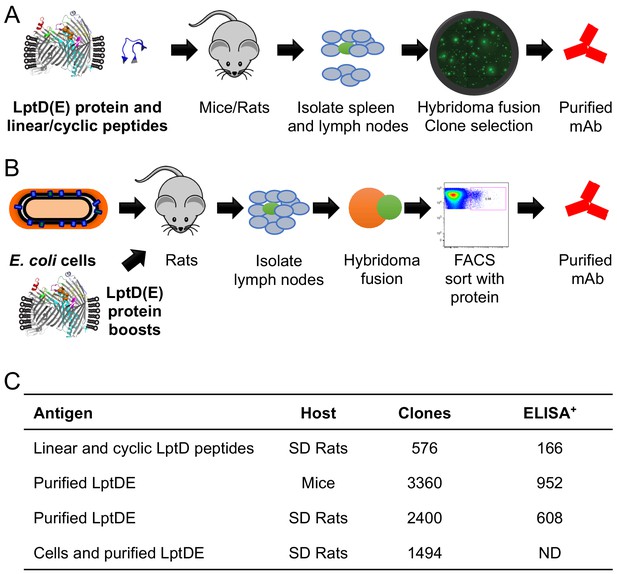
Immunization schematics and α-LptD mAb library characteristics.
(A) Cartoon of LptD immunization campaigns with purified E. coli LptDE protein, LptD cyclic peptides, and linear peptides. 10 rounds of protein or peptide injections were performed. Clones were selected from hybridoma fusions. (B) Cartoon of targeted boost-and-sort strategy in which SD rats were initially primed with E. coli K-12 bacteria, followed by two boosts with the recombinant LptDE protein reconstituted in amphipol matrix. Cell hybridoma fusions were sorted with fluorescently-labeled LptDE to enrich LptDE+ hybridomas. (C) Overview of the individual immunization campaigns. The total number of hybridoma clones isolated and binding to LptDE by ELISA were determined for each campaign. ELISA positive antibodies had a signal 3x above background. ELISA measurements were not determined (ND) for the hybridoma clones from the boost-and-sort approach.
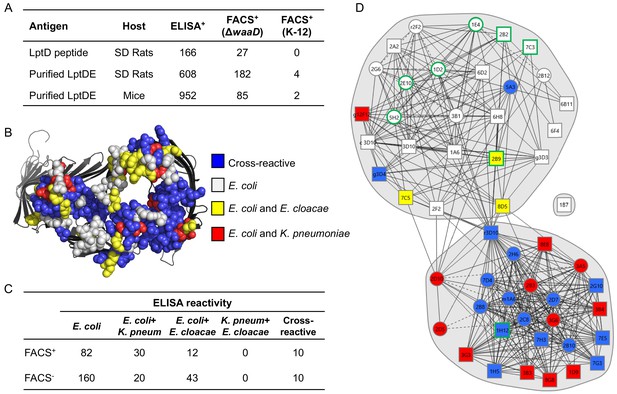
Whole cell FACS binding and epitope binning of α-LptD mAbs.
(A) LptD mAb campaign summary of ELISA+ LptDE mAbs tested for surface binding on E. coli ΔwaaD and E. coli K-12 by FACS assay. Antibodies were scored FACS+ if the MFI was 2x above an isotype control. (B) Amino acids comprising the 13 ECLs of LptD are highlighted on the structure as spheres. The spheres are color-coded based on amino acid conservation between different sequence comparisons as indicated in the key. Structure is of Shigella flexneri LptDE with LptE removed rendered in PyMol (PDB 4Q35 [Qiao et al., 2014]). (C) 134 FACS+ and 233 FACS- antibodies were characterized for cross-reactive binding to purified LptDE from two closely-related Enterobacteriaceae species: Klebsiella pneumoniae (K. pneum) and Enterobacter cloacae (E. cloacae) by ELISA. ELISA positive antibodies had a signal 3x above background. (D) 52 FACS+ α-LptD mAbs were characterized for epitope binning patterns using a high-throughput SPR-based method to determine pairwise mAb binding competition. Antibodies are color-coded based on ELISA cross-reactivity profiles as indicated in the key. Circle designates data obtained from both capture and detection antibodies. Square designates a single capture or detection data point. Characterization of all antibodies described in Figure 4—figure supplement 1 and those with green-outlined squares or circles are shown in Figure 4B.
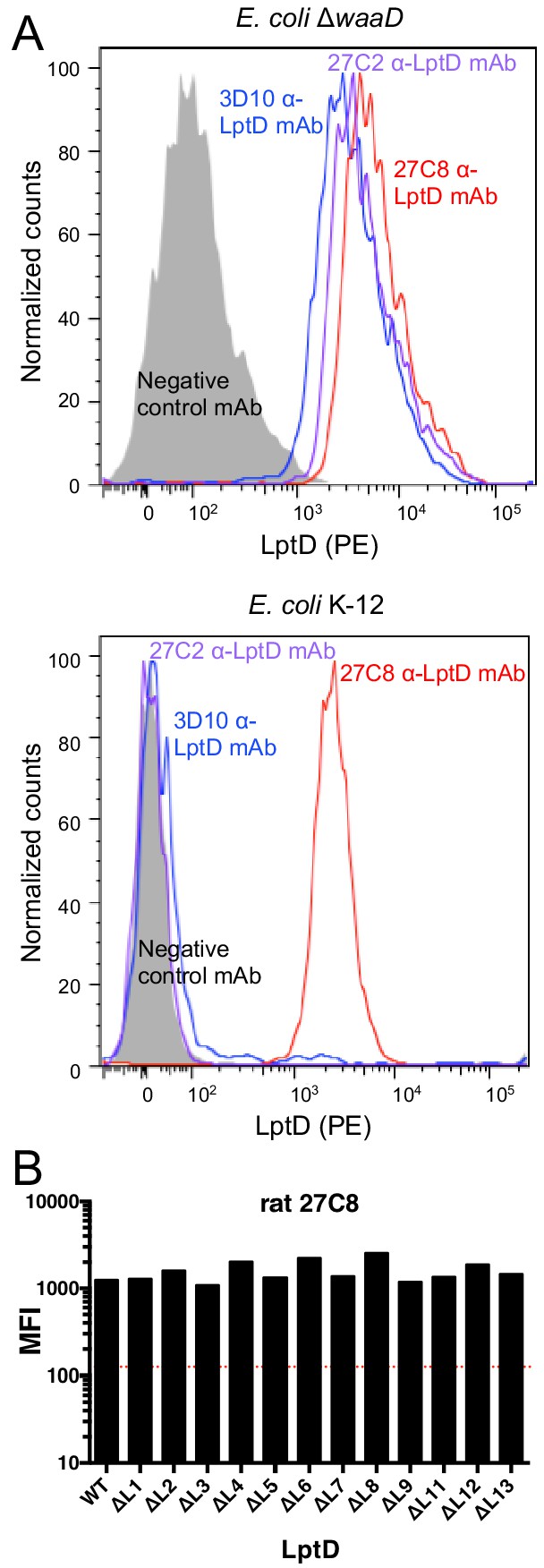
α-LptD mAb binding to E. coli K-12 and E. coli ΔwaaD strains by FACS analysis.
(A) Representative FACS traces for three ELISA+ α-LptD mAbs, 27C2 (purple), 3D10 (blue), and 27C8 (red), with E. coli ΔwaaD (top) and E. coli K-12 (bottom) cells. A non-binding isotype control mAb is shown in grey. 10,000 events were counted for each antibody and counts at each fluorescent intensity were plotted. (B) FACS analysis of a representative E. coli K-12 FACS+ α-LptD mAb (27C8) for binding to E. coli ΔwaaD strains expressing wild-type LptD and the 12 viable loop deletion strains. Loop 10 could not be assessed (see text). The mean fluorescent intensities (MFIs) are plotted. The red dashed line represents a descriptive MFI level that is 10% that of the WT control.
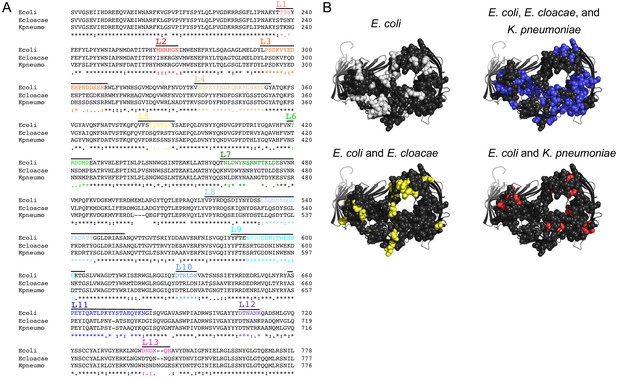
LptD ECL conservation among different Enterobacteriaceae species.
(A) Amino acid sequence alignment of LptD β-barrel region (amino acids 181–778) for E. coli (Ecoli), E. cloacae (Ecloacae), and K. pneumoniae (Kpneumo). Extent of ECLs based on available structures is indicated as lines above the sequences and the boundaries for the ECL deletions (L1–L13) are indicated by colors that match those in Figure 3A. (B) Extracellular amino acids are displayed as spheres on the LptD structure based on Shigella flexneri LptDE (PDB 4Q35 [Qiao et al., 2014]) with LptE removed. Spheres are colored based on sequence alignment if they are unique to E. coli (light grey) or identical between E. coli and E. cloacae (yellow), E. coli and K. pneumoniae (red), and all three species (blue).
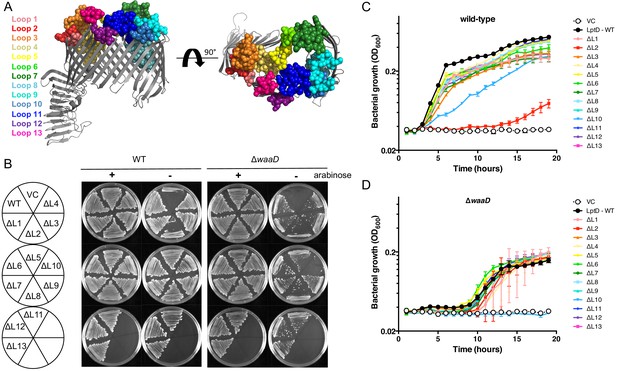
LptD ECLs are dispensable for E. coli growth.
(A) The ECL boundaries of LptD are highlighted as spheres and color-coded as indicated. Sequence boundaries are indicated Figure 2—figure supplement 2A. Structure based on S. flexneri LptDE with LptE removed rendered in PyMol (PDB 4Q35 [Qiao et al., 2014]). (B) LptD loop mutants were expressed on a plasmid, pLMG18gm, in a conditional lptD mutant strain in which the chromosomal copy of wild-type lptD is under arabinose control. Mutants were streaked onto LB gentamicin agar supplemented with or without 0.2% arabinose and imaged after overnight growth. Representative image of three biological triplicates is shown. Growth was determined in both the E. coli wild-type and E. coli ΔwaaD strain backgrounds. Vector control (VC) does not encode a copy of lptD and requires arabinose induction of the chromosomal lptD copy to grow. Growth curves of lptD loop mutants expressed in E. coli (C) K-12 and (D) ΔwaaD strain backgrounds are shown. Strains were grown from a starting inoculum of OD600 0.001 in LB gentamicin without arabinose and monitored for bacterial growth by measuring OD600 over 15 hr. Means and standard deviations from biological triplicates are plotted. Statistical analysis of growth rates for each curve are shown in Supplementary file 1.
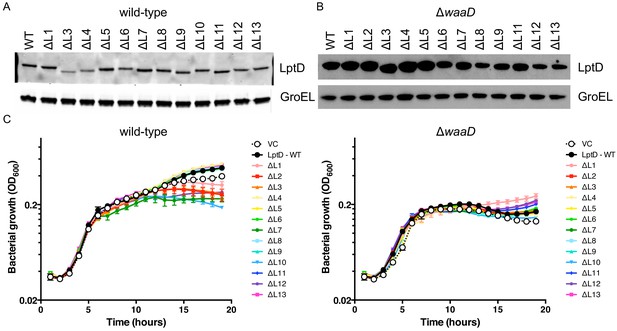
Expression of lptD loop mutants does not affect growth of E. coli expressing a wild-type copy of lptD.
Representative Western blots of (A) log-phase E. coli K-12 wild-type liquid cultures and (B) E. coli ΔwaaD cultures from overnight agar plates expressing each lptD loop mutant. Strains were grown in LB gentamicin with no arabinose. LptDΔL2 was severely growth delayed in E. coli K-12 and LptDΔL10 was not viable in E. coli ΔwaaD so these could not be evaluated for protein expression. (C) Growth curves of lptD loop mutants expressed in a E. coli K-12 (left) and E. coli ΔwaaD (right) strain background are shown. Strains were grown from a starting inoculum of OD600 0.001 in LB gentamicin supplemented with arabinose and monitored for bacterial growth by measuring OD600 over 15 hr in the presence of arabinose. Means and standard deviations for biological triplicates are plotted and comparisons of growth rates are shown in Supplementary file 1.
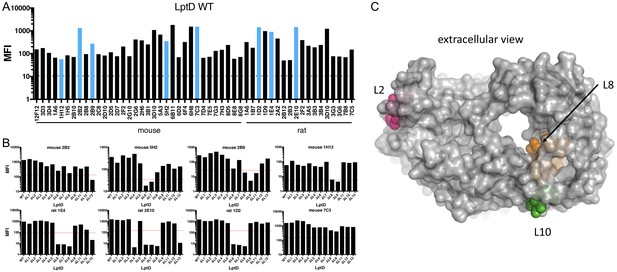
Mapping α-LptD mAb binding using LptD loop variants.
(A) 52 FACS+ LptD mAbs were screened for FACS binding to E. coli ΔwaaD expressing wild-type lptD. The mean fluorescent intensities (MFIs) are plotted. The blue bars highlight the eight representative mAbs shown below in 4B. Antibodies originating from different hosts are labeled. The dotted line is the MFI of an LptD FACS- antibody. (B) FACS analysis of 8 representative α-LptD mAbs for binding to E. coli ΔwaaD expressing wild-type LptD and the 12 viable loop deletions. Loop 10 could not be assessed (see text). The MFIs above background are plotted. The red dashed lines represent a descriptive MFI level that is 10% that of the WT control. The remaining antibodies from (A) are shown in Figure 4—figure supplement 1. (C) Top view of LptD structure from E. coli LptDE with LptE removed highlighting extracellular L2 (pink), L8 (orange), and L10 (green) rendered in PyMol (PDB 4RHB [Malojcic et al., 2015]).
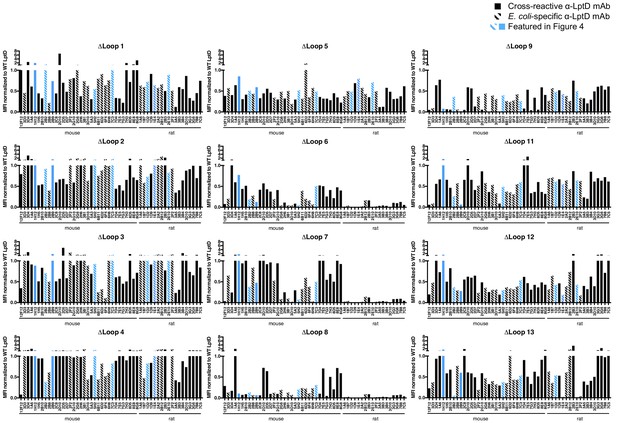
Comprehensive FACS analysis on all lptD loop mutants.
52 FACS +LptD mAbs were screened for FACS binding to E. coli ΔwaaD expressing each lptD loop mutant (except L10, see text). The mean fluorescent intensities (MFIs) above background are normalized to the MFIs of E. coli ΔwaaD expressing wild-type lptD and plotted. The antibodies are in the same order as Figure 4A and the blue bars highlight the eight representative mAbs shown in Figure 4B. The solid bars represent antibodies that are ELISA+ for at least two species and hashed bars represent antibodies that are ELISA+ only for E. coli LptDE (Figure 2D).
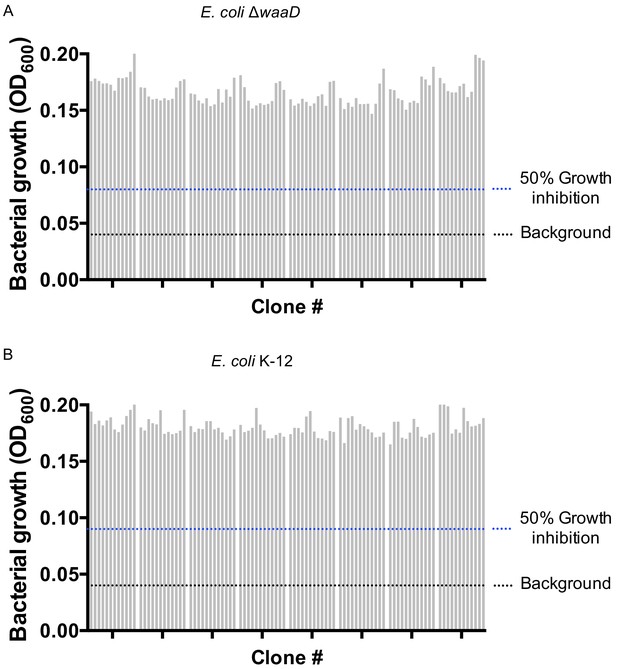
Functional analysis of α-LptD antibodies.
Bacterial growth was measured (OD600) for (A) E. coli ΔwaaD and (B) E. coli K-12 after treatment with one representative plate of α-LptDE mAbs at 10 µg/mL for 4 hr. Black dashed line indicates media only control. Blue dashed line indicates 50% growth inhibition compared to no treatment.
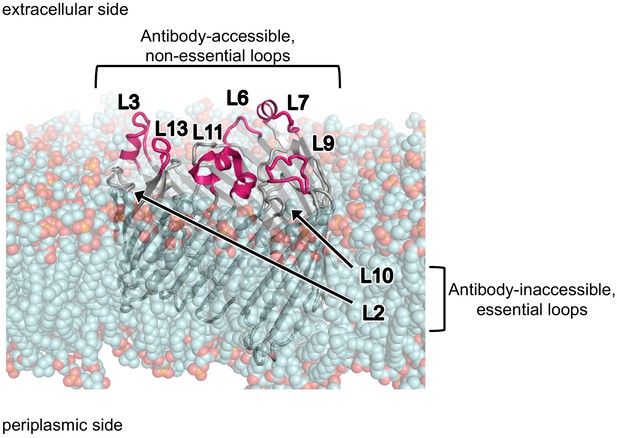
Antibody accessible ECLs of LptD.
Side view of LptD structure rendered in PyMol from E. coli LptDE with LptE removed (PDB 4RHB [Malojcic et al., 2015]). LptD was placed in a standard phosphatidylethanolamine membrane context (shown as cyan and red space fill) using MemProtMD database (Stansfeld et al., 2015). Antibody accessible, non-essential ECLs L3, L6, L7, L9, L11, and L13 are highlighted in pink and antibody inaccessible ECLs L2 and L10 are indicated.
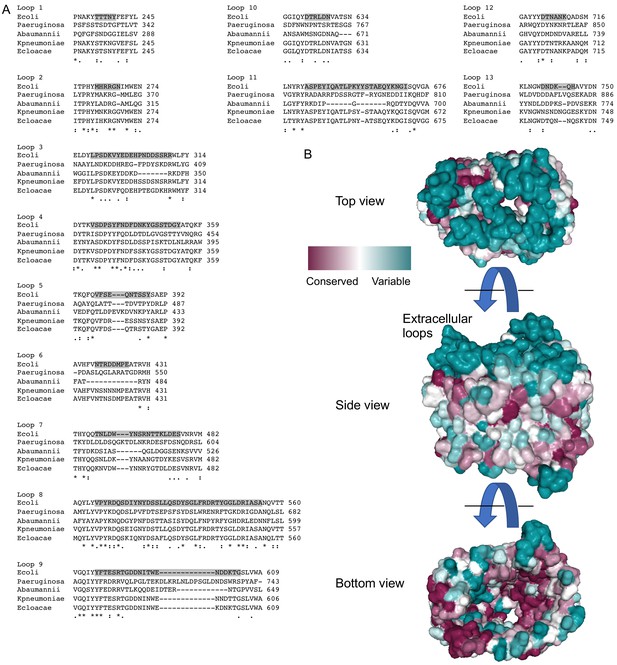
Diversity of the ECLs of LptD.
(A) Amino acid sequence alignments of each LptD ECL comparing E. coli (Ecoli), Pseudomonas aeruginosa (Paeruginosa), Acinetobacter baumannii (Abaumannii), K. pneumoniae (Kpneumoniae), and E. cloacae (Ecloacae). Boundaries for the ECLs in E. coli are highlighted. (B) Consurf analysis (consurf.tau.ac.il/2016) of LptD β-barrel with 150 sequences that are 35–95% identical to LptD from the E. coli (PDB 4RHB [Malojcic et al., 2015]) using the default settings. Deeper maroon colors are conserved and deeper cyan colors are more variable.
Tables
Removal of extracellular LptD loops sensitizes E. coli to OM-excluded antibiotics.
https://doi.org/10.7554/eLife.46258.009| MIC (µg/ml)* | ||||
|---|---|---|---|---|
| Vancomycin | Rifampicin | |||
| LptD† | WT‡ | ΔwaaD§ | WT | ΔwaaD |
| WT | 128 | 64 | 8 | 0.125 |
| ΔL1 | 128 | 64 | 4 | 0.125 |
| ΔL2 | 8 | 4 | 0.25 | 0.0625 |
| ΔL3 | 128 | 16 | 4 | 0.125 |
| ΔL4 | 16 | 1 | 0.25 | 0.0156 |
| ΔL5 | 128 | 64 | 8 | 0.125 |
| ΔL6 | 128 | 64 | 2 | 0.0625 |
| ΔL7 | 128 | 64 | 4 | 0.125 |
| ΔL8 | 16 | 4 | 0.125 | 0.0625 |
| ΔL9 | 128 | 128 | 4 | 0.125 |
| ΔL10 | 64 | NG | 2 | NG |
| ΔL11 | 128 | 64 | 8 | 0.125 |
| ΔL12 | 128 | 64 | 8 | 0.125 |
| ΔL13 | 128 | 64 | 8 | 0.125 |
-
*Minimum inhibitory concentration (MIC) is the lowest concentration of antibiotic that completely inhibits bacterial growth.
†Conditional lptD E. coli strains carry an arabinose-inducible wild-type lptD and a plasmid-encoded copy of lptD with the indicated loop deletions (as indicated in Figure 3A). Only the plasmid copy of lptD is expressed in the absence of arabinose.
-
‡The wild-type (WT) strain is a conditional E. coli K-12 with a chromosomal arabinose-inducible lptD.
§The ΔwaaD strain is a conditional E. coli ΔwaaD mutant with a chromosomal arabinose-inducible lptD.
mAbs to accessible ECLs do not inhibit essential function of LptD.
https://doi.org/10.7554/eLife.46258.013| Growth inhibitory α-LptD mAbs* | |||||
|---|---|---|---|---|---|
| Antigen | Host | Clones | K-12† | ΔwaaD‡ | ΔwaaD + Rif.§ |
| Linear and cyclic LptD peptides | SD Rats | 576 | 0 | 0 | 0 |
| Purified LptDE | Mice | 3360 | 0 | 0 | 0 |
| Purified LptDE | SD Rats | 2400 | 0 | 0 | 0 |
| Cells and purified LptDE | SD Rats | 1494 | 0 | 0 | 0 |
-
*For each antibody campaign, bacterial growth was measured (OD600) for E. coli ΔwaaD and E. coli K-12 after treatment with each antibody at 10 µg/mL for 4 hr. Growth inhibition was calculated as a percentage of growth compared to an untreated control. 50% growth inhibition was considered positive.
†WT (wild-type) is E. coli K-12.
-
‡ΔwaaD is an E. coli ΔwaaD.
§ΔwaaD + Rif. is E. coli ΔwaaD grown in the presence sub-inhibitory rifampicin
Additional files
-
Supplementary file 1
Statistical analysis of the initial growth rates for conditional lptD deletion strains complemented with lptD loop mutants.
Bacterial growth curves in Figure 3 and Figure 3—figure supplement 1 were analyzed by determining the doubling time (dt) during exponential growth phase and compared via the unpaired Student’s t test. The Bonferroni correction was applied to control for multiple comparisons.
- https://doi.org/10.7554/eLife.46258.016
-
Supplementary file 2
Strains, plasmids, and primers used in this study.
Names, descriptions, and references for all key resources (bacterial strains, plasmid constructs, and primers) as described in the text.
- https://doi.org/10.7554/eLife.46258.017
-
Supplementary file 3
Sequences of linear and cyclic peptides used for immunizations.
LptD peptides used for immunizations (Figure 1) were designed based on sequence conservation, surface exposure and loop location.
- https://doi.org/10.7554/eLife.46258.018
-
Transparent reporting form
- https://doi.org/10.7554/eLife.46258.019





