The GluTR-binding protein is the heme-binding factor for feedback control of glutamyl-tRNA reductase
Figures
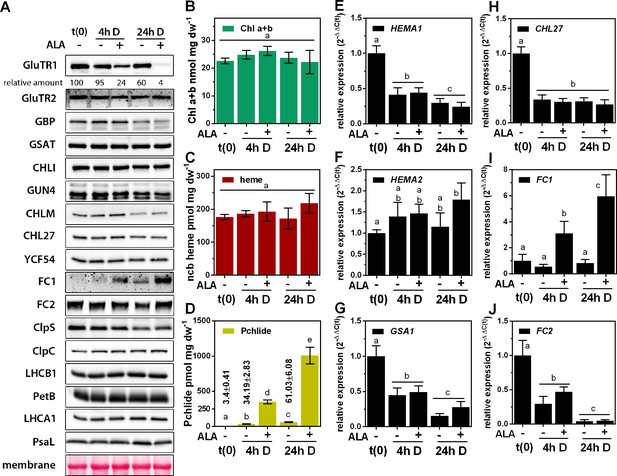
Post-translational destabilization of GluTR1 upon feeding with 5-aminolevulinic acid (ALA) in the dark.
Fourteen-day-old, light-grown Arabidopsis Col-0 wild-type plants were incubated in buffer without (-) or with 1 mM ALA (+) in darkness (D) for the indicated times. Samples were harvested prior to treatment (t0) and at 4 hr and 24 hr after the onset of treatment. (A) Western blot analysis of proteins involved in tetrapyrrole biosynthesis (GluTR1/2, GBP, CHLI, GUN4, CHLM, CHL27, YCF54, FC1, and FC2), Clp protease (ClpS and C) and components of the photosynthetic electron transfer chain (LHCA1, LHCB1, PetB, PsaL). t(0) = sample harvested from light-grown seedlings prior to ALA treatment. Signal intensities for GluTR1 relative to t(0) are shown at the top. (B – D) Levels of chlorophylls (Chl) a and b (B), non-covalently bound (ncb) heme (C) and protochlorophyllide (Pchlide) (D) found in control and ALA-treated seedlings. Data are given as means ± sd (n = 4). (E – J) qRT-PCR analysis of HEMA1 (coding for glutamyl-tRNA reductase 1), HEMA2 (GluTR2), GSA1 (glutamate-1-semialdehyde 2,1-aminomutase 1), CHL27 (a subunit of aerobic cyclase) and FC1/2 (ferrochelatase 1 and 2) mRNAs. Data are given as means ± sd (n = 4). The lowercase letters in panels (B – J) indicate statistical groups determined by Student’s t-test (p<0.05).
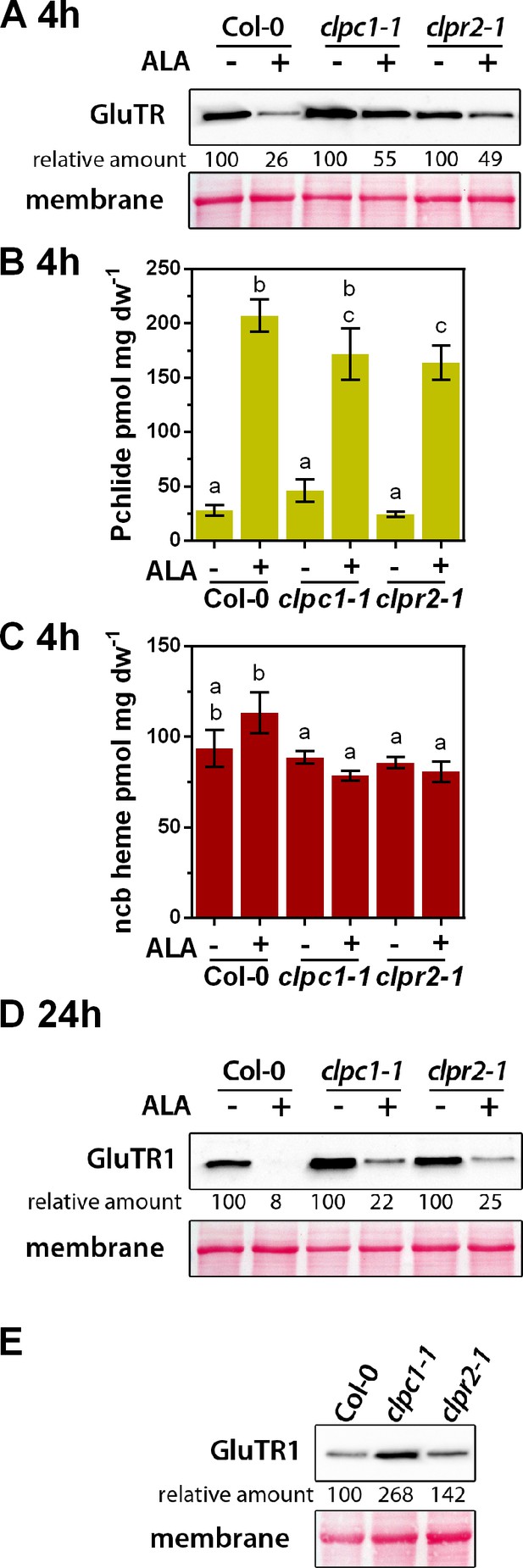
ALA-induced proteolysis of GluTR1 is attenuated in clpc1-1 and clpr2-1 mutants.
(A) Western blot analysis of 14-day-old GluTR1 in Col-0, clpc1-1 and clpr2-1 leaves incubated in buffer without (-) or with (+) 1 mM ALA for 4 hr in darkness. Signal intensities for GluTR1 relative to Col-0 without ALA treatment are shown. (B, C) Pchlide (B) and non-covalently bound (ncb) heme (C) contents of genotypes analyzed in (A) after 4 hr of treatment. Data are given as means ± sd (n = 3). Letters indicate statistical groups determined by Student’s t-test (p<0.05). (D) Western analysis of GluTR1 in Col-0, clpc1-1 and clpr2-1 incubated in buffer without (-) or with (+) 1 mM ALA for 24 hr in darkness. Signal intensities for GluTR1 in protein extracts of the genotypes treated without (- ALA = 100%) and with ALA (+ALA) are shown. (E) GluTR1 content of Col-0, clpc1-1 and clpr2-1 seedlings grown under short-day conditions. Samples were harvested 2 hr after the onset of light. Signal intensities for GluTR1 are shown relative to Col-0.
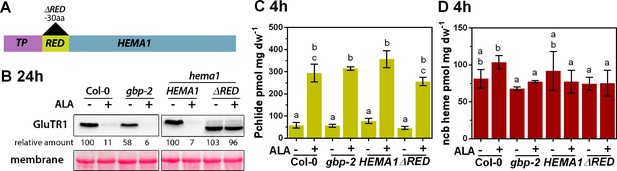
ALA-induced proteolysis depends on the N-terminal segment of GluTR1 and on GBP.
(A) Schematic presentation of the constructs used to complement a hema1 knockout mutant. In addition to the full-length coding sequence of HEMA1, a construct coding for a truncated version of GluTR1 (ΔRED), which lacks 30 amino acids (aa) of the regulatory domain, was used to complement the hema1 mutant. Transgenes were under the control of the native HEMA1 promoter. TP, transit peptide. (B) GluTR1 content of 14-day-old Col-0, gbp-2, hema1/HEMA1, and hema1/ΔRED leaves incubated in buffer without (-) or with (+) 1 mM ALA for 24 hr in darkness. Signal intensities for GluTR1 are shown relative to the untreated WT (left) or the hema1/HEMA1 line (right). (C, D) Pchlide (C) and ncb heme content (D) of genotypes analyzed in (B) after 4 hr of treatment. Data are given as mean ± sd (n = 3). Letters indicate statistical groups determined by Student’s t-test (p<0.05).
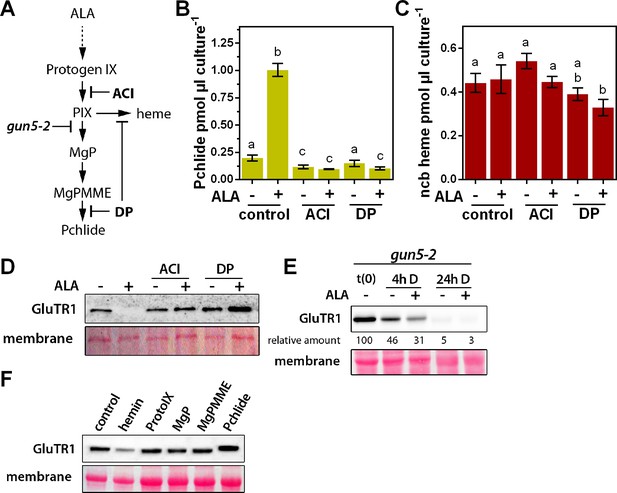
Identification of the stimulus responsible for triggering GluTR1 degradation.
A photoautotrophic Arabidopsis cell culture was used, together with mutants for the tetrapyrrole biosynthesis pathway, and isolated chloroplasts. (A) Overview of the tetrapyrrole biosynthesis pathway. Acifluorfen (ACI) prevents the formation of protoporphyrin IX (PIX) by inhibiting protoporphyrinogen oxidase. The iron chelator 2,2'-dipyridyl (DP) inhibits the reactions catalyzed by the ferrochelatases (FC) and aerobic cyclase, respectively. Knockout of GUN5 (gun5-2) prevents synthesis of Mg-protoporphyrin IX (MgP) and downstream intermediates. (B, C) Pchlide (B) and ncb heme (C) contents of photoautotrophically cultured Arabidopsis cells (PA) incubated in the presence (+) or absence of (-) 1 mM ALA. In addition to ALA, cells were treated with 100 µM ACI or 500 µM DP, respectively. PA cultures were pre-incubated with ACI or DP for 2 hr in darkness, before the buffer was exchanged for media supplemented with ACI, DP, and ALA. Treatment was performed for 16 hr in the dark. Data are given as mean ± sd (n = 4). Letters indicate statistical groups determined by Student’s t-test (p<0.05). (D) Levels of GluTR1 in cell cultures incubated in the absence (- ALA) or presence (+ALA) of added ALA and with or without ACI or DP, respectively. (E) Levels of GluTR1 in leaves of gun5-2 knockout mutants incubated without (-) or with (+) 1 mM ALA in the dark (D) for the indicated times. Signal intensities for GluTR1 relative to t(0) are shown. (F) Arabidopsis Col-0 chloroplasts were isolated and incubated for 6 hr in darkness in the presence of hemin and intermediates of the tetrapyrrole biosynthesis pathway. Pchlide was used at 1 µM and heme and the other porphyrins at 5 µM final concentration.
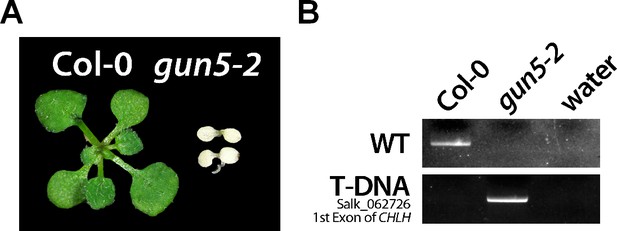
Phenotype (A) and genotyping (B) of Col-0 and the gun5-2 mutant.
Seeds of heterozygous gun5-2 progenies were surface sterilized and grown on MS plates supplemented with 1% sucrose. After 2–3 weeks, green and white seedlings were harvested separately and analyzed by PCR using primers spanning the T-DNA insertion site (WT allele, gun5-2 LP and RP). The presence of the T-DNA within the first exon of CHLH in the Salk_062726 line was confirmed using a combination of the SALK T-DNA border primer (LB1.3) and gun5-2 RP.
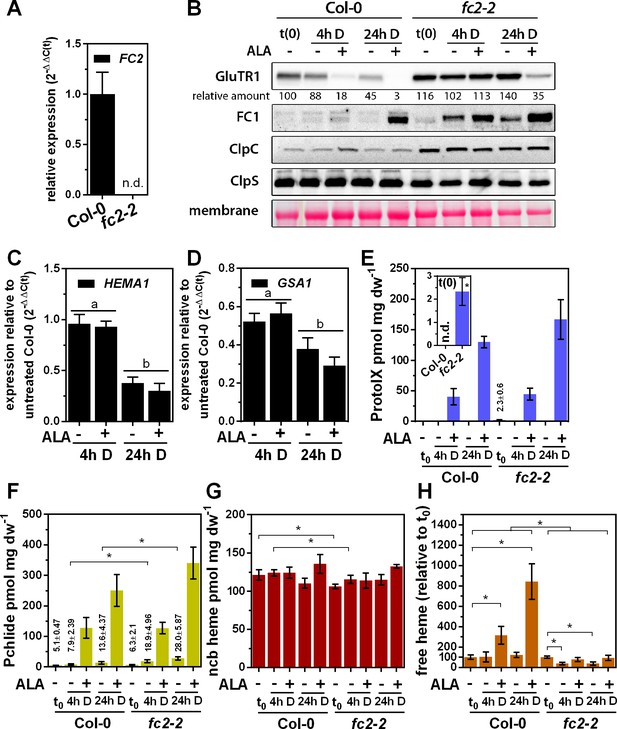
Altered ALA-induced proteolysis of GluTR1 in FC2 knockout mutants.
(A) Levels of FC2 mRNA in light-grown Col-0 and fc2-2 plants, analyzed by qRT-PCR. Data are given as mean ± sd (n = 3). n.d., not detectable (B) Levels of GluTR1, FC1, ClpC and ClpS in leaves of Col-0 and fc2-2 plants incubated without (-) or with (+) 1 mM ALA in darkness (D) for the indicated times. Signal intensities for GluTR1 relative to Col-0 seedlings harvested before the beginning of the experiment (Col-0 t(0)) are shown (D, dark). (C) and (D) Levels of HEMA1 (C) and GSA1 (D) mRNAs in fc2-2 mutants plotted relative to untreated Col-0 seedlings harvested before start of the experiment. Data are given as mean ± sd (n = 4). Letters indicate statistical groups determined by Student’s t-test (p<0.05). (E) Protoporphyrin IX (ProtoIX), (F) Pchlide, (G) ncb heme and (H) free heme contents of Col-0 and fc2-2 leaves treated as in (B). The inset in E shows the steady-state level of ProtoIX in untreated seedlings before ALA treatment (t0). Data are given as mean ± sd (n = 4). Asterisks denote statistically significant changes between samples with p<0.05 (Student’s t-test). Statistical analysis was performed before the calculation of ratios. n.d., not detectable. 21-day-old, light-grown plants were analyzed.
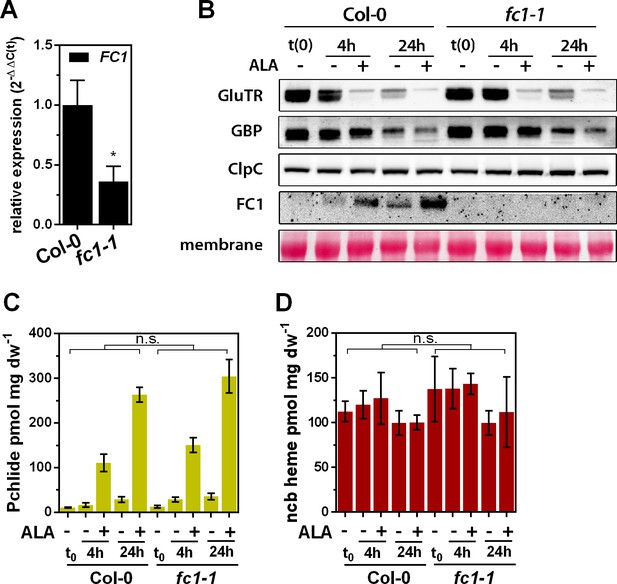
ALA-induced proteolysis of GluTR1 in FC1 knockdown mutants.
(A) FC1 mRNA levels in leaves of light-grown Col-0 and fc1-1 plants, analyzed by qRT-PCR. Data are given as mean ± sd (n = 3). The asterisk denotes the significant reduction of FC1 transcript in the fc1-1 mutant (Student’s t-test, p<0.05). (B) Levels of GluTR1, GBP, ClpC and FC1 in leaves of Col-0 and fc1-1 plants incubated without (-) or with (+) 1 mM ALA in darkness for the indicated times. (C, D) Levels of Pchlide (C) and ncb heme (D) in leaves of Col-0 and fc1-1 plants treated as in (A). Data are given as mean ± sd (n = 4). Values were tested (Student’s t-test, p<0.05) for significant differences between Col-0 and fc1-1 at each time point. The mutant accumulated the same amounts of Pchlide and heme as the WT. n.s., not significant.
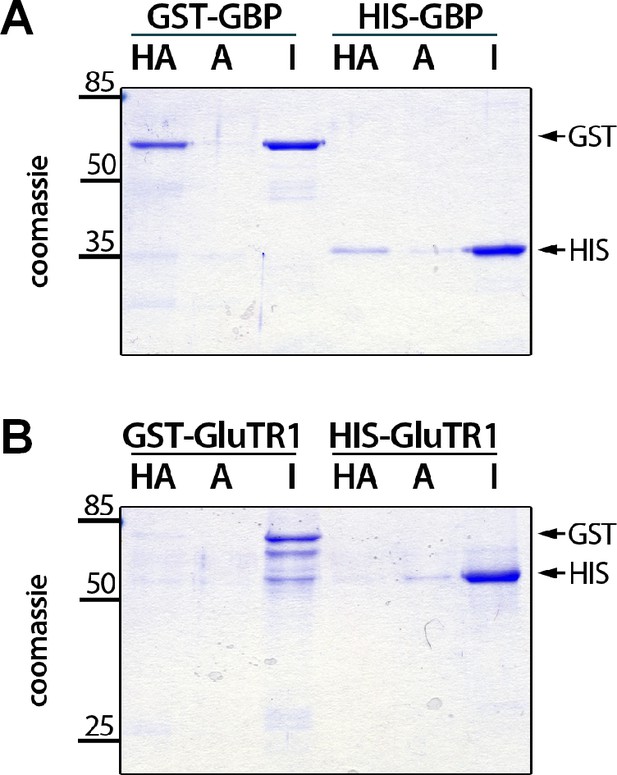
Binding of hemin to GBP.
Recombinant GST- and 6xHIS-tagged GBP (A) and GluTR1 (B), respectively, were incubated with hemin-agarose (HA) or unmodified agarose (A), washed, eluted and subjected to electrophoresis on a 12% SDS-polyacrylamide gel. One microgram of the input was loaded as a reference (I). Note that only GBP binds to the HA. Arrows on the right mark the position of the GST and 6xHIS-tagged proteins.
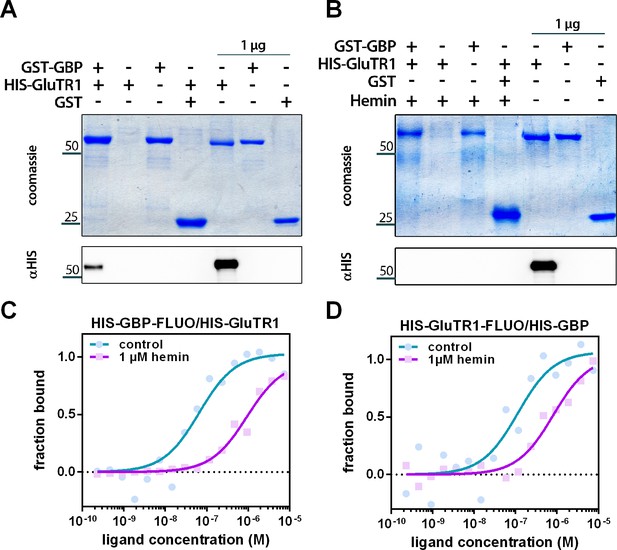
Hemin interferes with the binding of GBP to GluTR1.
(A) Pulldown of 6xHIS-GluTR1 by GST-GBP. Both proteins were incubated in the presence of Glutathione-Sepharose (GS), washed, eluted using reduced glutathione and separated on a 12% SDS-polyacrylamide gel. Eluates of reactions containing GST-GBP alone or GST and GluTR1 as well as 1 µg of the input (no GS incubation) were loaded as controls. Note that 6xHIS-GluTR1 and GST-GBP show almost identical migration behavior. Successful pulldown of 6xHIS-GluTR1 by GST-GBP was confirmed using a HIS-tag-specific antibody (αHIS) after western blotting. (B) Same experiment as in (A) but carried out in the presence of 500 µM hemin. Note that hemin prevents pulldown of 6xHIS-GluTR1 by GST-BP. (C) Microscale thermophoresis (MST) of the fluorophore-labeled (FLUO) 6xHIS-GBP titrated against increasing amounts of 6xHIS-GluTR1. The affinity of 6xHIS-GBP-FLUO for 6xHIS-GluTR1 (Kd = 65 nM) decreases upon addition of 1 µM hemin (Kd = 931 nM). n = 2 independent experiments. (D) MST of fluorophore-labeled 6xHIS-GluTR1-FLUO titrated against increasing amounts of 6xHIS-GBP. The affinity of 6xHIS-GluTR1-FLUO for 6xHIS-GBP (Kd = 113 nM) decreases upon addition of 1 µM hemin (Kd = 806 nM). n = 2 independent experiments.
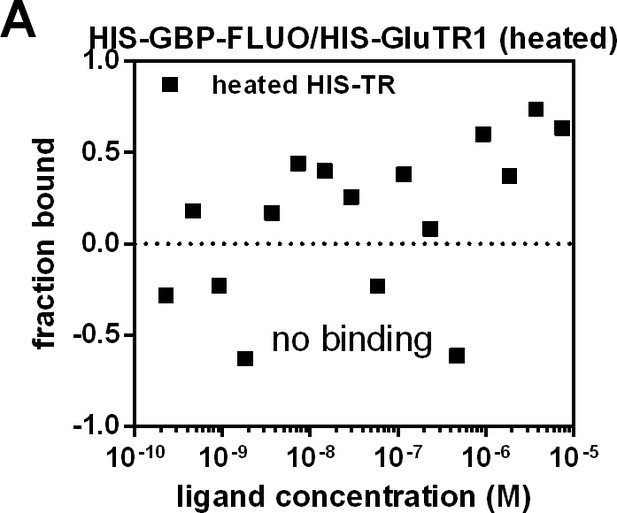
Control for MST measurement.
(A) Microscale thermophoresis (MST) of fluorophore-labeled 6xHIS-GBP titrated against increasing amounts of heat inactivated 6xHIS-GluTR1. Upon denaturation of 6xHIS-GluTR1, binding to 6xHIS-GBP-FLUO is prevented.
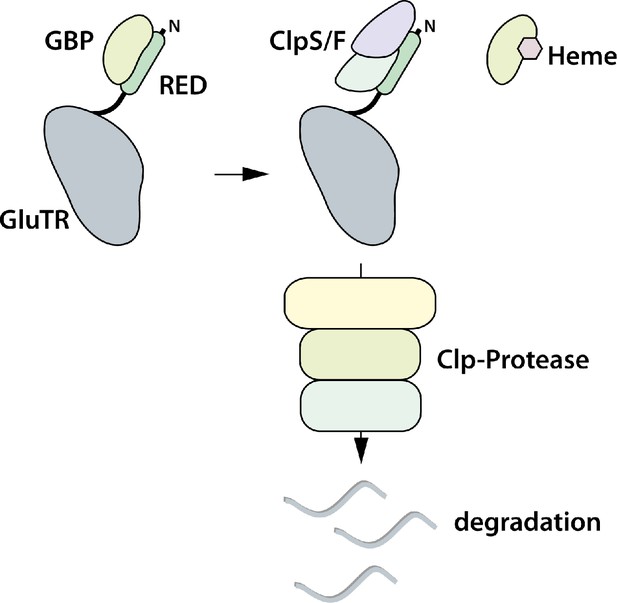
Proposed model for heme-dependent GluTR1 degradation by Clp protease.
In the absence of heme, GBP binds to the ‘regulatory domain’ (RED, formerly ‘heme binding domain’) and prevents degradation of GluTR1. When heme levels increase, release of GBP from GluTR1 enables the binding of Clp components to the latter and its concomitant proteolytic degradation.

Alignment of the amino-acid sequences of Arabidopsis GluTR1 (encoded by HEMA1), GluTR2 (HEMA2) and the regulatory domain of GluTR1 (RED, formerly heme-binding domain - HBD).
https://doi.org/10.7554/eLife.46300.013
Distribution of GBP and regulatory domain (RED, formerly heme-binding domain - HBD)-containing GluTR isoforms in photosynthetic organisms.
Protein BLAST analyses (https://blast.ncbi.nlm.nih.gov/Blast.cgi) were performed using either the full-length AtGBP (AT3G21200, left) or the RED of AtGluTR1 (right). The top 100 hits were used to build the trees. Note that both GBP and the RED-containing GluTR isoform were only found in land plant species. The query entries used are highlighted in blue.
Additional files
-
Supplementary file 1
HPLC methods.
- https://doi.org/10.7554/eLife.46300.015
-
Supplementary file 2
qPCR primers used in this study.
- https://doi.org/10.7554/eLife.46300.016
-
Transparent reporting form
- https://doi.org/10.7554/eLife.46300.017





