HIV-1 integrase tetramers are the antiviral target of pyridine-based allosteric integrase inhibitors
Figures
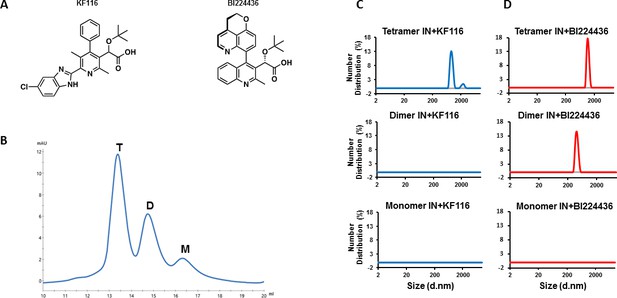
IN tetramers are preferentially targeted by pyridine and quinoline based ALLINIs.
(A) Chemical structures of pyridine-based KF116 and quinoline-based BI224436. (B) SEC based separation of IN tetramer(T), dimer(D) and monomer(M) fractions. C and D, DLS analysis of 200 nM IN fractions in the presence of 1 µM KF116 (blue lines, (C) or BI224436 (red lines, (D). DMSO controls for each DLS experiment (C and D) are shown in gray. Representative results of three independent experiments at 30 mins time point are shown.

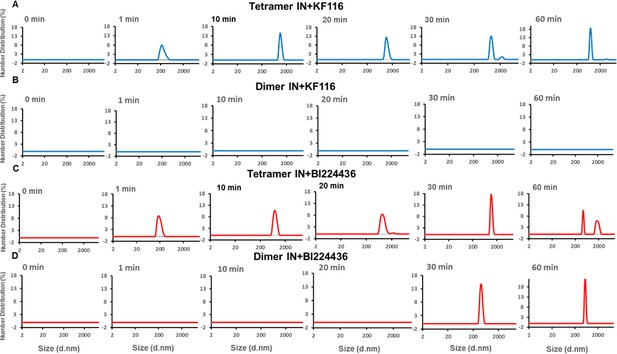
Pyridine and quinoline based ALLINIs preferentially target IN tetramers.
Kinetic analysis of KF116 (blue lines in A and B) or BI224436 (red lines in C and D) induced higher-order oligomerization of tetramer (A and C) and dimer (B and D) fractions. 1 µM inhibitor was added to 200 nM IN and higher-order oligomerization was monitored by DLS. DMSO controls for each experiment are shown in gray. Representative results of three independent experiments are shown.
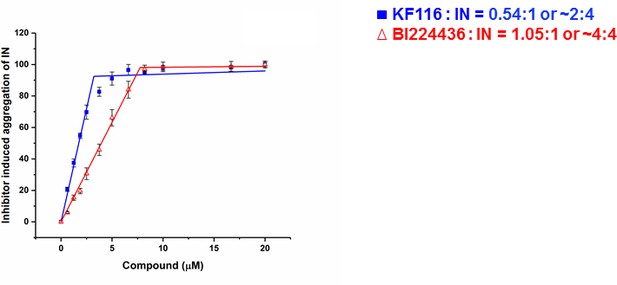
Stoichiometry of KF116 and BI224436 induced aggregation of IN.
Quantitative analysis of ALLINI induced IN aggregation. The error bars indicate the standard deviation of three independent experiments. The stoichiometry for KF116:IN and BI224436:IN were determined using piecewise linear regression.
KF116 and BI224436 induced aggregation of IN.
Increasing concentrations of the ALLINIs were added to the full length WT IN and the supernatant (S) and pellet (P) fractions were analyzed by SDS-PAGE gels with coomassie staining. Representative images of three independent experiments are shown.
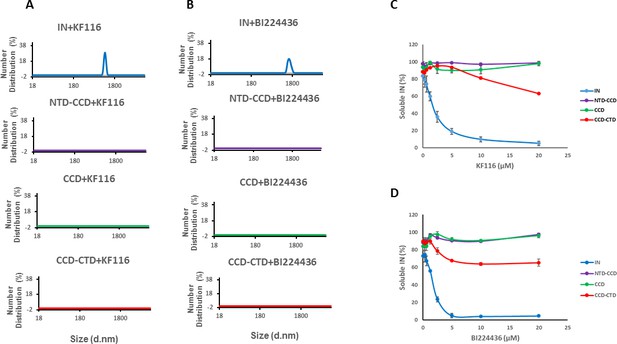
Roles of individual IN domains for ALLINI induced aggregation.
(A and B) DLS analysis of 10 µM full length IN, NTD-CCD, CCD and CCD-CTD in the presence of 1 µM KF116 or BI224436. Representative results of two independent experiments at 30 mins time point are shown. DMSO control results for each respective experiment are shown in gray. (C and D) Quantitative analysis of KF116 and BI224436 induced aggregation of 5 µM full length IN, NTD-CCD, CCD and CCD-CTD by centrifugation-based aggregation assay. The error bars indicate the standard error of two independent experiments (see Figure 3—figure supplement 1 for representative primary data).
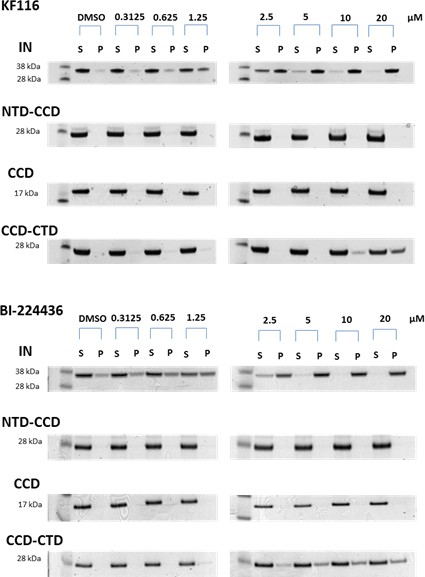
Contributions of individual IN domains for ALLINI induced aggregation.
Increasing concentrations of KF116 and BI224436 were added to 5 µM of full length IN, NTD-CCD, CCD and CCD-CTD. Following centrifugation, supernatant (S) and pellet (P) fractions were analyzed by SDS-PAGE with coomassie staining. The representative images of two independent experiments are shown.
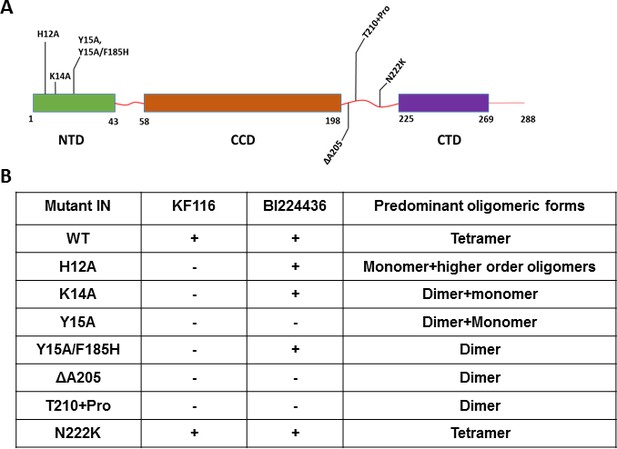
Probing the importance of the NTD and CCD-CTD linker for ALLINI induced higher-order multimerization of IN.
(A) Schematic diagram of IN with indicated mutations in NTD and CCD-CTD linker regions. (B) Summary table of all IN mutants indicating their predominant multimeric form as analyzed by SEC and effects of ALLINI induced higher-order multimerization monitored by DLS. ‘+' and '- ' indicates susceptibility and resistance of the mutant proteins to ALLINI induced higher-order multimerization, respectively.

Multimeric forms of IN mutants.
SEC profiles of WT and mutant INs were analyzed by superdex 200 10/300 GL column. X-axis indicates elution volume (mL) and y-axis indicates the intensity of absorbance (mAU). Tetramers (T), Dimers (D) and Monomers (M) are indicated.
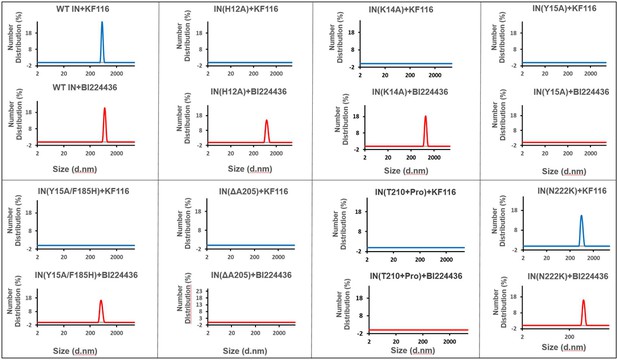
Higher-order IN multimerization induced by KF116 and BI224436.
DLS analysis of 200 nM IN mutants in the presence of 1 µM KF116 or BI224436 at 30 min time points. DMSO control runs are shown in gray. Representative results of three independent experiments are shown.
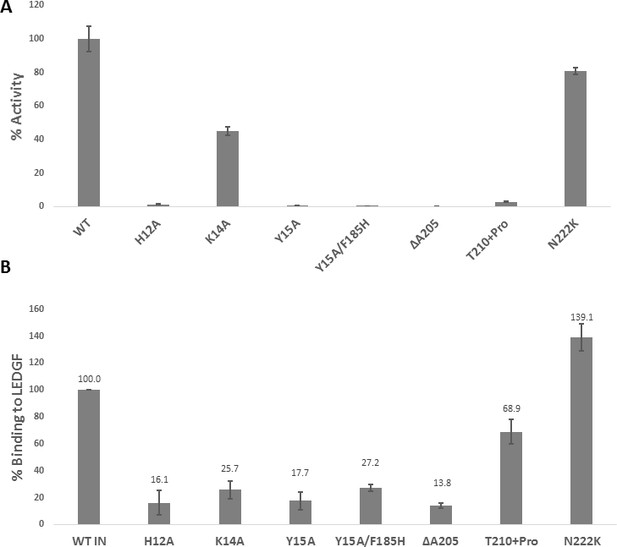
Biochemical characterization of IN mutants.
(A) Catalytic activities of mutant INs in the presence of LEDGF/p75 monitored by HTRF based assay. The bars represent the percent activity of the mutants INs relative to the WT IN. The error bars indicate the standard deviation of triplicate experiment. (B) Quantification of LEDGF/p75 binding to the mutant INs monitored by affinity pull-down of His tagged INs and tag-less LEDGF/p75 using Ni-sepharose beads. The error bars indicate the standard deviation of two independent experiments.
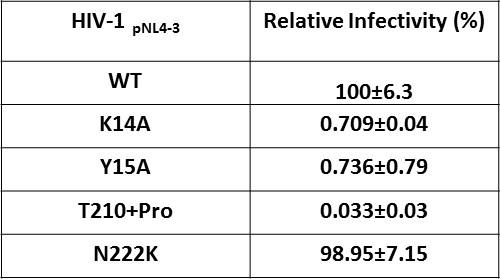
Effect of IN substitutions on the infectivity of HIV-1NL4-3.
Relative infectivity of WT and mutant viruses were tested by luciferase reporter assay in TZM-bl cells infected with 100 ng viruses. The standard deviations from three independent experiments are indicated.
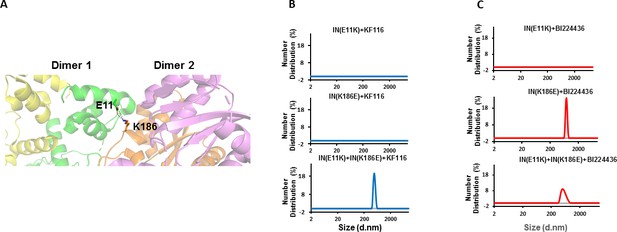
Trans-complementation of IN tetramer interface mutants to elucidate ALLINI preferences.
(A) The salt bridge between E11 and K186 residues is highlighted in the context of IN tetramer interface. (B) DLS analysis of 200 nM E11K, K186E and E11K + K186E INs in the presence of 1 µM KF116 or BI224436 after 30 mins. DMSO controls with respective INs are shown in gray. Representative results of two independent experiments are shown.

Trans-complementation of inactive mutants partially restores IN functions.
(A) Catalytic activities of the mutant INs in the presence of LEDGF/p75 were monitored by HTRF based assay. The bars represent the percent activity of the mutants INs relative to the WT IN. The error bars indicate the standard deviation of triplicate experiment. (B) Quantification of LEDGF/p75 binding to the mutant INs assayed by the affinity pull-down of His tagged INs and tag-less LEDGF/p75 using Ni-sepharose beads. The error bars indicate the standard deviation of two independent experiments.
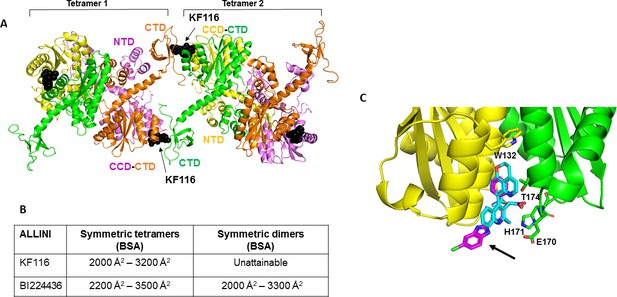
Structural analysis of KF116 and BI224436 interactions with IN.
(A) The top ranked model for symmetric tetramer-KF116-tetramer interactions. Each protomer is distinctly colored (green, yellow, violet, orange). Each domain is assigned to its respective protomer, as previously proposed for Maedi Visna IN (PDB: 3HPH)(Hare et al., 2009). (B) Buried surface areas (BSA) of KF116 and BI224436 induced higher-order IN multimers; (C) overlay of the crystal structures of KF116 (PDB: 4O55) and BI224436 bound to IN CCDF185H. KF116 and BI224436 are shown in magenta and cyan respectively. The arrow points to the benzimidazole group in KF116.
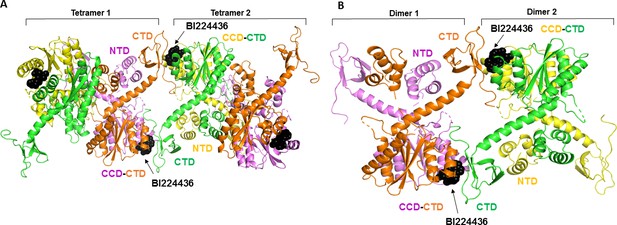
Molecular modeling of BI224436 induced higher-order IN multimerization.
The top ranked IN tetramers (A) and dimers (B) constructed with BI224436 (dark spheres).
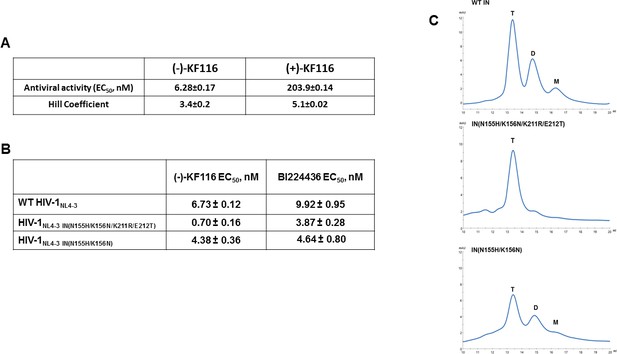
Antiviral activities of ALLINIs.
(A) Antiviral activities of (-) and (+)- KF116 against WT virus. (B) Antiviral activities of KF116 and BI224436 against DTG resistant quadruple and double mutant viruses. The error is the S.D. of three independent experiments. (C) SEC analysis of mutant INs.
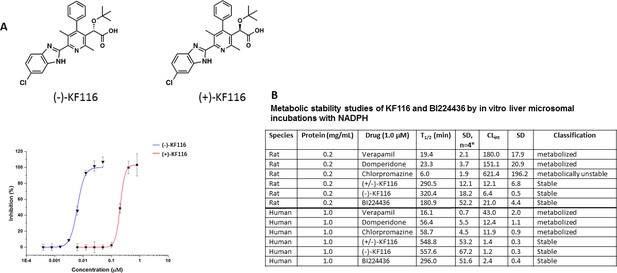
Comparative analysis of (+) and (-) enantiomers of KF116.
(A) Chemical structures and antiviral activity profiles of (+) and (-) enantiomers of KF116. (B) In vitro metabolic stabilities of KF116 and BI224436.
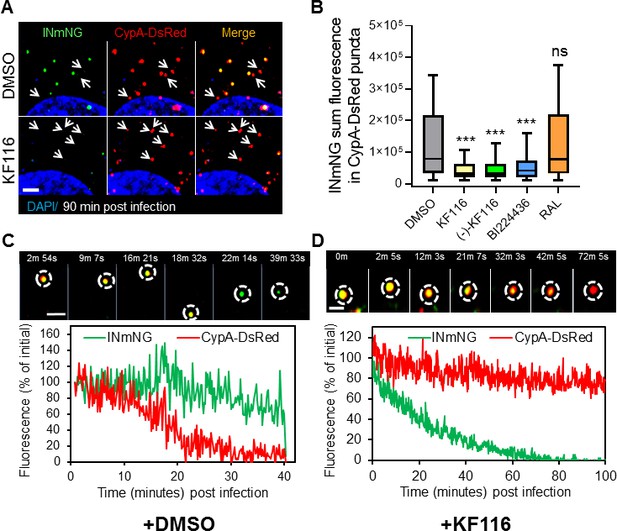
VSV-G pseudotyped fluorescent HIV-1 labeled with Vpr-IN-mNeonGreen (INmNG,a marker of viral complex) and CypA-DsRed (CA marker).
Viruses produced in the presence of 5 µM ALLINIs or raltegravir (RAL) or left untreated (DMSO) control were used to infect TZM-bl PPIA-/- cells (MOI 0.08) for 90 min. Cells were imaged on the Zeiss LSM880 using Fast-Optimal AiryScan settings, which enables sensitive detection and tracking of viral complexes with high temporal resolution and minimal photobleaching. (A) Images of CypA-DsRed labeled cores retaining INmNG signal for DMSO treated samples and loss of INmNG signal from the viral CA cores, leaving CypA-DsRed puncta for KF116 treated samples at 90 min post infection. Arrows point to single labeled CypA-DsRed complexes. (B) The sum fluorescence of INmNG within CypA-DsRed/CA puncta is plotted. (C–D) Time-lapse imaging of INmNG/CypA-DsRed labeled virus. Single particles losing the CypA-DsRed or INmNG signal were manually annotated and tracked. (C) Images and fluorescent intensity traces showing uncoating (loss of CypA-DsRed prior to INmNG loss) for single viral particle produced in the presence of DMSO. (D) Images and fluorescent intensity traces showing a gradual INmNG signal loss from a CypA-DsRed/CA puncta for viruses produced in the presence of KF116 (5 μM). Scale bar in (A, C and D) =2 µm. Statistical significance in (B) was determined by comparing respective samples with the DMSO control using a pair-wise student t-test, ***=p < 0.0001. p values > 0.5 were considered not significant (ns).
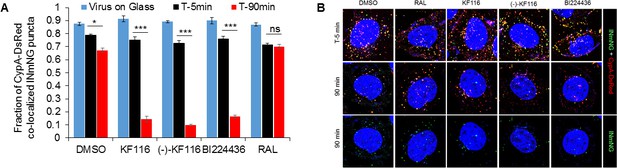
VSV-G pseudotyped HIV-1 viral particles fluorescently labeled with INmNG and CypA-DsRed in the presence of indicated treatments.
(A) The fraction of CypA-DsRed colocalized with INmNG signal for viruses bound on poly-lysine treated glass, or inside cells at 5 or 90 min post infection are shown. Object based colocalization of INmNG and CypA-DsRed puncta was determined. Data are representative of 3 independent experiments. Statistical significance was determined by comparing respective samples with the DMSO control using pair-wise student t-test, ***=p < 0.0001. p values > 0.5 were considered not significant (ns). (B) Representative images of single cells infected with viruses treated with the different ALLINIs, DMSO or RAL at time 0 min and 90 min post-infection. Images showing nuclei labeled with DAPI (blue), INmNG (green) and CypA-DsRed (red). Bottom most panel of images show the distribution of INmNG puncta of the same overlay images in middle panels at 90 min post infection. Scale bar = 5 µm.
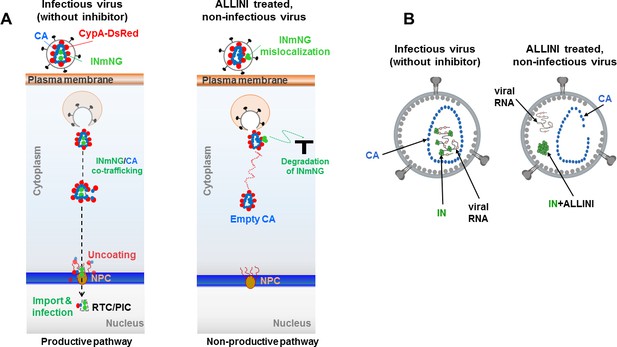
Schematic summary of the experimental results.
(A) Schematics showing the observed productive and non-productive pathways of co-trafficking of INmNG and CypA-DsRed labeled CA cores during infection of target cells with infectious (in the absence of the inhibitor) and ALLINI-treated, non-infectious virions. (B) Schematic depictions of infectious and ALLINI-treated, non-infectious virions. In infectious virions, IN binding to viral RNA ensures that RNPs are packaged within the cone-shaped CA core. In contrast, ALLINI treatment promotes higher-order, aberrant IN multimerization which in turn impairs IN binding to viral RNA. As a result, both ALLINI induced IN aggregates and RNPs lacking IN are mislocalized outside of the protective CA core.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Retroviridae) | HIV-1NL4-3 | Other | Replication competent HIV-1 virus particles produced from pNL4-3 plasmid in HEK293T producing cells; laboratory adapted HIV-1 strain | |
| Strain, strain background (Retroviridae) | HIV-1NL-lucE-R+VSVg | Other | Replication incompetent HIV-1 virus particles produced from pNL-luc. E-R+and pMD.G plasmids in HEK293T producing cells; laboratory adapted HIV-1 strain pseudotyped with VSV glycoprotein | |
| Genetic reagent (Retroviridae) | HIV-1NL4-3 IN (K14A) | Other | Replication competent HIV-1 virus particles containing the mutation K14A in the Integrase ORF | |
| Genetic reagent (Retroviridae) | HIV-1NL4-3 IN (Y15A) | Other | Replication competent HIV-1 virus particles containing the mutation Y15A in the Integrase ORF | |
| Genetic reagent (Retroviridae) | HIV-1NL4-3 IN (T210 + Pro) | Other | Replication competent HIV-1 virus particles containing the mutation T210 + Pro in the Integrase ORF | |
| Genetic reagent (Retroviridae) | HIV-1NL4-3 IN (N222K) | Other | Replication competent HIV-1 virus particles containing the mutation N222K in the Integrase ORF | |
| Genetic reagent (Retroviridae) | HIV-1NL-lucE-R+ N155H/K156NVSVg | Other | Replication incompetent, VSVg pseudotyped HIV-1 virus particles containing the following mutations in the IN ORF: N155H/K156N | |
| Genetic reagent (Retroviridae) | HIV-1NL-lucE-R+ N155H/K156N/K211R/E212TVSVg | Other | Replication incompetent, VSVg pseudotyped HIV-1 virus particles containing the following mutations in the IN ORF: N155H/K156N/K211R/ E212T | |
| Cell line (H.sapiens) | HeLa | ATCC | ATCC CCL-2 | |
| Cell line (H.sapiens) | Hek293T | ATCC | ATCC CRL-3216 | |
| Cell line (H.sapiens) | TZM-bl | NIH AIDS Reagent Program | NIH AIDS Reagent Program: 8129 | The reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: TZM-bl cells (Cat# 8129) from Dr. John C Kappes, and Dr. Xiaoyun Wu |
| Cell line (H.sapiens) | TZM-bl PPIA(-/-) | Francis and Melikyan, 2018 | ||
| Cell line (H.sapiens) | MT-4 | NIH AIDS Reagent Program | NIH AIDS Reagent Program: 120 | The reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: MT-4 from Dr. Douglas Richman |
| Biological sample (Rat) | Sprague-Dawley rat liver microsomes | Xenotech LLC | Xenotech LLC: 1510115 | Pool of 500, male; 20 mg/mL |
| Biological sample (H.sapiens) | Human Liver Microsomes | Xenotech LLC | Xenotech LLC: 1610016 | Pool of 50, mixed gender; 20 mg/mL |
| Recombinant DNA reagent | pNL4-3 (plasmid) | Adachi et al., 1986 | ||
| Recombinant DNA reagent | pNL4-3 IN (K14A) (plasmid) | This paper | Site-directed mutagenesis in pNL4-3 to generate IN K14A mutant | |
| Recombinant DNA reagent | pNL4-3 IN (Y15A) (plasmid) | This paper | Site-directed mutagenesis in pNL4-3 to generate IN Y15A mutant | |
| Recombinant DNA reagent | pNL4-3 IN (T210 + Pro) (plasmid) | This paper | Site-directed mutagenesis in pNL4-3 to generate IN T210 + Pro mutant | |
| Recombinant DNA reagent | pNL4-3 IN (T222K) (plasmid) | This paper | Site-directed mutagenesis in pNL4-3 to generate IN T222K mutant | |
| Recombinant DNA reagent | pNL-luc.E-R+ (plasmid) | Connor et al., 1995 | ||
| Recombinant DNA reagent | pNL-luc.E-R+N 155 H/K156N (plasmid) | This paper | Site-directed mutagenesis in pNL-luc.E-R+to generate IN N155H/K156N mutant | |
| Recombinant DNA reagent | pNL-luc.E-R+N155 H/K156N/K211R/E212T (plasmid) | This paper | Site-directed mutagenesis in pNL-luc.E-R+to generate IN N155H/K156N/K211R/ E212T mutant | |
| Recombinant DNA reagent | pMD.G (plasmid) | Naldini et al., 1996 | ||
| Recombinant DNA reagent | Vpr-INmNeon Green (INmNG) | Francis and Melikyan, 2018 | ||
| Recombinant DNA reagent | CypA-DsRed | Francis et al., 2016; Francis and Melikyan, 2018 | ||
| Recombinant DNA reagent | pHIVeGFP-deltaEnv | Francis and Melikyan, 2018 | ||
| Recombinant DNA reagent | Recombinant IN (pET-15b) | Larue et al., 2012 | All the mutations were carried out by site directed mutagenesis in IN coding region of pET-15b | |
| Recombinant DNA reagent | Recombinant IN domains: NTD-CCD, CCD and CCD-CTD (pET-15b) | This paper | Wild type NL4-3 IN domains were constructed by site directed mutagenesis from pET-15b truncated IN domains containing solubilizing mutants (Larue et al., 2012) | |
| Recombinant DNA reagent | Recombinant LEDGF (pLEDGF) | Tsiang et al., 2009 | ||
| Commercial assay or kit | p24 ELISA | Zeptometrix | Zeptometrix: 0801111 | |
| Commercial assay or kit | MycoscopeMycoplasm PCR detection kit | Genlantis | Genlantis: MY01100 | |
| Commercial assay or kit | CellTiter-Glo | Promega Biosciences Inc | Promega Biosciences Inc: G7571 | |
| Commercial assay or kit | Luciferase Assay System | Promega Biosciences Inc | Promega Biosciences Inc: E1500 | |
| Commercial assay or kit | QuikChange XL site directed mutagenesis kit | Agilent | Agilent: 200516 | |
| Commercial assay or kit | QuikChange XL II site directed mutagenesis kit | Agilent | Agilent: 200522 | |
| Commercial assay or kit | LANCE Europium- streptavidin for HTRF assay | PerkinElmer | PerkinElmer: AD0062 | |
| Commercial assay or kit | Reporter Lysis buffer | Promega Biosciences Inc | Promega Biosciences Inc: E3971 | |
| Commercial assay or kit | X-treme Gene HP | Roche | Roche: 6366244001 | |
| Chemical compound, drug | KF116 | Sharma et al., 2014 | ||
| Chemical compound, drug | BI224436 | MedChemExpress | MedChem Express: HY-18595 | |
| Chemical compound, drug | nicotinamide adenine dinucleotide phosphate (NADPH) | Sigma-Aldrich Chemical Company | Sigma-Aldrich: 10107824001 | |
| Chemical compound, drug | Verapamil Hydrochloride | Sigma-Aldrich Chemical Company | Sigma-Aldrich: V4629 | |
| Chemical compound, drug | Domperidone | Sigma-Aldrich Chemical Company | Sigma-Aldrich: D122 | |
| Chemical compound, drug | Chlorpromazine hydrochloride | Sigma-Aldrich Chemical Company | Sigma-Aldrich: C8138 | |
| Software, algorithm | HKL 3000 | HKL 3000 (http://hkl-xray.com) | RRID:SCR_015023 | |
| Software, algorithm | Phenix | Phenix (https://www.phenix-online.org/) | RRID:SCR_014224 | |
| Software, algorithm | Coot | Coot (http://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot) | RRID: SCR_014222 | |
| Software, algorithm | HADDOCK | Dominguez et al., 2003, van Zundert et al., 2016 | ||
| Software, algorithm | ImageJ | ImageJ (https://imagej.net) | RRID: SCR_003070 | |
| Software, algorithm | OriginLab | OriginLab software (https://www.originlab.com) | ||
| Software, algorithm | ICY image analysis software | ICY image analysis software (https://icy.bioimageanalysis.org) | RRID:SCR_010587 |
Additional files
-
Supplementary file 1
Synthesis of (+) and (-) enantiomers of KF116.
- https://doi.org/10.7554/eLife.46344.023
-
Transparent reporting form
- https://doi.org/10.7554/eLife.46344.024





