Casein kinase 1 family proteins promote Slimb-dependent Expanded degradation
Figures
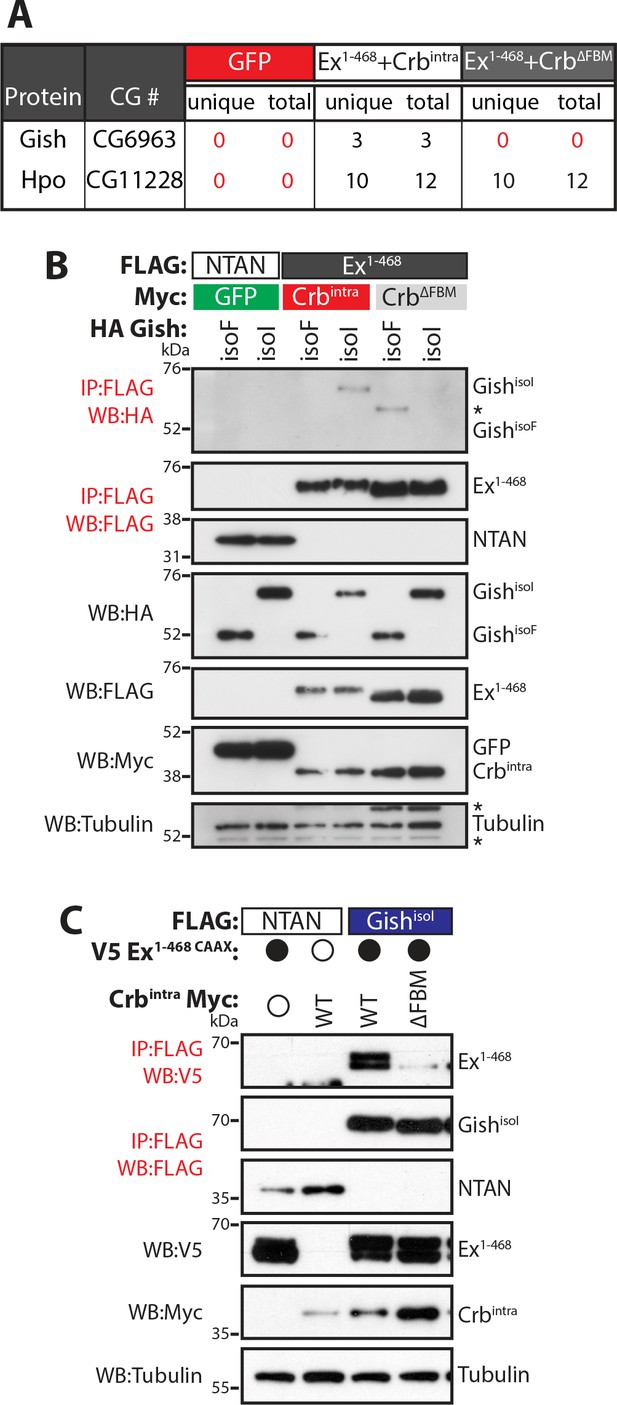
Gish, the Drosophila orthologue of CkIγ interacts with Ex in a Crb-dependent manner.
(A) An AP-MS approach identified Gish as an Ex-interacting protein. Summary table with AP-MS results for Gish and Hpo. CG# denotes Flybase CG number, while unique and total denote the number of peptides detected in the MS analysis. (B) and (C) Crbintra promotes Ex:Gish binding in a FBM-dependent manner. Reciprocal co-IPs were performed with FLAG-tagged NTAN or Ex1-468 and HA-tagged Gish (B), or with FLAG-tagged NTAN or GishisoI and V5-tagged Ex1-468 CAAX (C), in the presence of Myc-tagged GFP, Crbintra or CrbΔFBM. The expression and presence of co-purified proteins were analysed by immunoblotting with the indicated antibodies. Asterisks denote non-specific bands (IgG heavy chain in IP panel and FLAG signal in Tubulin panel). Open and full circles denote absence or presence of the indicated plasmid, respectively. Tubulin was used as loading control. Note that experiments shown in (C) were performed in the presence of proteasome inhibitors.
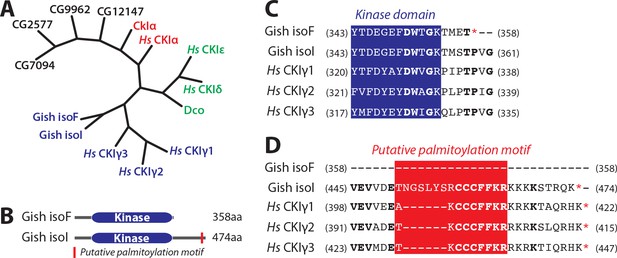
Evolutionary conservation of CKIs and features of Gish.
(A) Phylogenetic relation between human and Drosophila CKI isoforms and other Drosophila kinases with >50% overall similarity to CkIα. (B) Domain structure of isoforms F and I of Gish. Red rectangle depicts putative palmitoylation site present in all Gish isoforms except isoF. (C) and (D) Protein sequence alignments depicting conservation between Drosophila Gish isoforms and human CKIγ isoforms at the level of the kinase domain C-terminus (C) and putative palmitoylation (D). CKI sequences were aligned as indicated in Materials and methods. Numbers in parentheses indicated starting and ending aa residues of the depicted sequences. Blue indicates kinase domain sequence, whereas red denotes putative palmitoylation site.
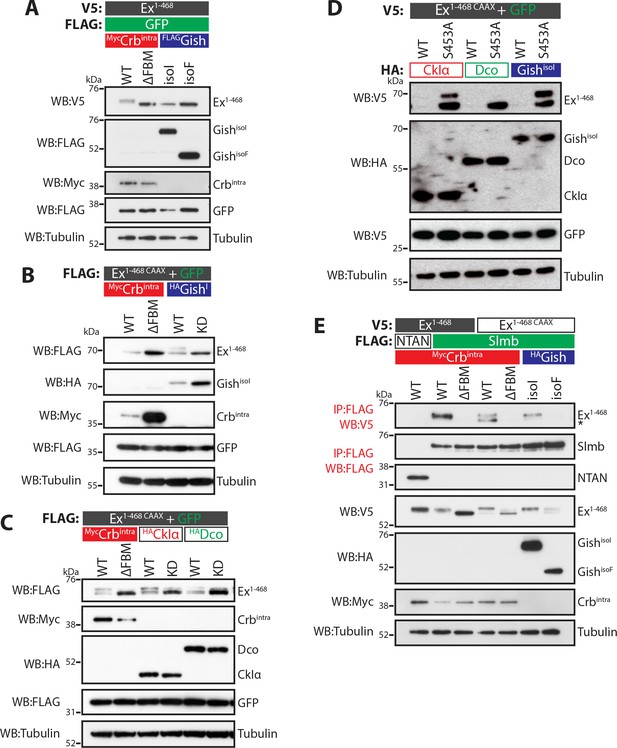
CKI kinases promote Ex phosphorylation and depletion.
(A) and (B) Gish promotes Ex phosphorylation and depletion in a kinase- and sub-cellular localisation-dependent manner. (A) S2 cells were used to co-transfect V5-tagged Ex1-468 with GFP and Crbintra, CrbΔFBM, GishisoI or GishisoF, for 48 hr before lysis. Lysates were processed for immunoblot analysis using the indicated antibodies. Note that GishisoI caused a more prominent depletion of Ex than GishisoF, the CkIγ isoform lacking a palmitoylation sequence. (B) FLAG-tagged Ex1-468 CAAX was co-transfected with the indicated plasmids for 48 hr and lysates were processed for Western blotting analysis with the indicated antibodies. Kinase-deficient (KD) GishisoI was unable to promote Ex1-468 CAAX phosphorylation and depletion. (C) CKI kinases promote Ex phosphorylation and depletion in a kinase-dependent manner. S2 cells were transfected with the indicated plasmids for 48 hr before lysis. Immunoblot analysis with the indicated antibodies revealed that CkIα and Dco promote Ex phosphorylation and depletion in a kinase-dependent manner, as the kinase-dead (KD) versions did not cause a mobility shift in Ex1-468 CAAX. (D) CKI kinases promote Ex degradation via S453. S2 cells were co-transfected with V5-tagged wt or S453A Ex1-468 CAAX and HA-tagged CkIα, Dco or GishisoI for 48 hr before lysis. Lysates were immunoblotted with the indicated antibodies. Unlike its wt counterpart, Ex1-468 S453A CAAX was refractory to the action of CKI kinases and was not degraded in the presence of the kinases. (E) Gish expression promotes Ex:Slmb binding in the absence of Crbintra. Co-IPs were performed with FLAG-tagged NTAN or Slmb and either V5-tagged Ex1-468 or Ex1-468 CAAX, in the presence of Crbintra, CrbΔFBM, GishisoI or GishisoF. Expression and presence of co-immunoprecipitated proteins was assessed by immunoblotting with the indicated antibodies. Note that, similar to Crbintra, GishisoI expression alone promoted the Ex:Slmb interaction. GFP and tubulin were used as transfection efficiency and loading controls, respectively. Asterisk denotes non-specific band (IgG heavy chain in IP panel).
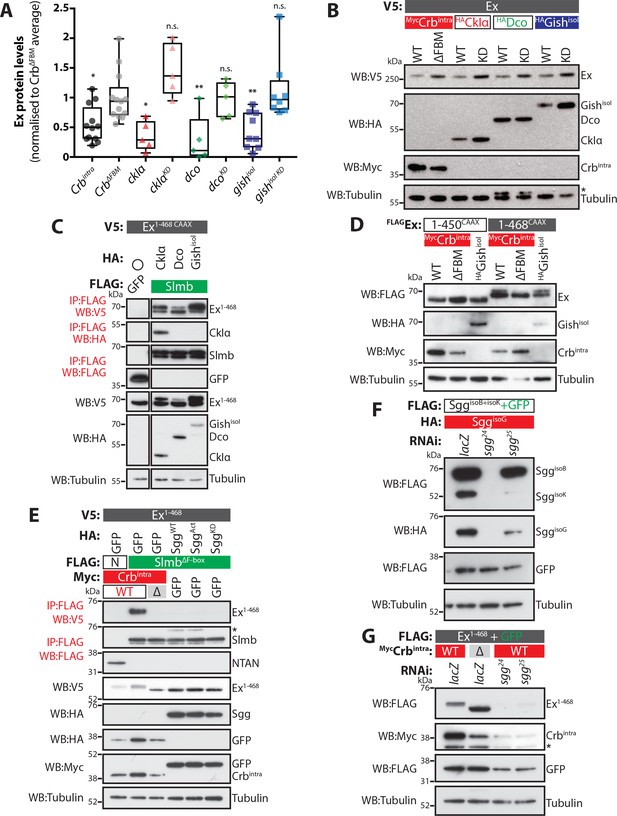
Molecular requirements for the effect of CKIs in the regulation of Ex protein stability.
(A) Expression of CKIs promotes depletion of Ex protein levels. Graph depicts results of quantification of Ex1-468 protein levels assessed by Western blotting analysis. Ex protein levels were quantified by densitometry analysis using ImageJ. Ex protein levels in all samples were normalised to the average of samples where Ex1-468 was co-expressed with the negative control CrbΔFBM, which does not promote Ex degradation. Box plots depict maximum to minimum values. *p<0.05, **p<0.01 (One-way ANOVA with Dunnett’s post hoc test). n.s. denotes non-significant. (B) Full-length Ex is phosphorylated and depleted in a CKI kinase-dependent manner. S2 cells were transfected with the indicated plasmids for 48 hr before lysis. Western blot analysis with the indicated antibodies revealed that CkIα, Dco and GishisoI promote Ex phosphorylation and depletion in a kinase-dependent manner, as the kinase-dead (KD) versions did not cause depletion of ExFL. (C) CKI kinases promote Ex:Slmb binding. FLAG-tagged GFP or Slmb were used to co-IP V5-tagged Ex1-468 CAAX in the presence of HA-tagged CkIα, Dco or GishisoI. Expression and presence of co-purified proteins was detected by immunoblot analysis using the indicated antibodies. (D) An Ex truncation lacking the Slmb/β-TrCP consensus sequence is refractory to Gish-mediated changes. The electrophoretic mobility of FLAG- and CAAX-tagged Ex1-450 or Ex1-468 was assessed upon co-transfection with Crbintra, CrbΔFBM or GishisoI. Note that while Gish promotes a mobility shift in Ex1-468, the 1–450 construct was not affected. (E) Ectopic Sgg fails to promote the Ex:Slmb interaction. FLAG-tagged NTAN (N) or SlmbΔF-box were used to co-IP V5-tagged Ex1-468 in the presence of Crbintra, CrbΔFBM, and either wt Sgg, activated Sgg (SggAct) and inactive Sgg (SggKD). Expression and presence of co-purified proteins was detected by immunoblot analysis using the indicated antibodies. Note that, unlike Crbintra, Sgg over-expression failed to promote the Ex:Slmb interaction. (F) Efficiency of Sgg RNAi sequences. S2 cells were transfected with the indicated plasmids 24 hr after treatment with lacZ or sgg RNAi. Protein expression was assessed by immunoblotting using the indicated antibodies. Note that sgg24 efficiently targets all isoforms, while sgg25 is more active toward SggisoK. (G) RNAi-mediated depletion of sgg does not affect Crb-induced Ex degradation. The levels of Ex1-468 were monitored in S2 cells co-transfected with Crbintra or CrbΔFBM, in the presence of control RNAi (lacZ) or RNAi targeting sgg. Asterisks denote non-specific bands (IgG heavy chain in IP panel, HA signal in Tubulin panel and FLAG signal in Myc panel). Tubulin was used as loading control.
-
Figure 2—figure supplement 1—source data 1
Source data for quantification of Expanded protein levels from Western blot analyses performed in Drosophila S2 cells.
- https://doi.org/10.7554/eLife.46592.006
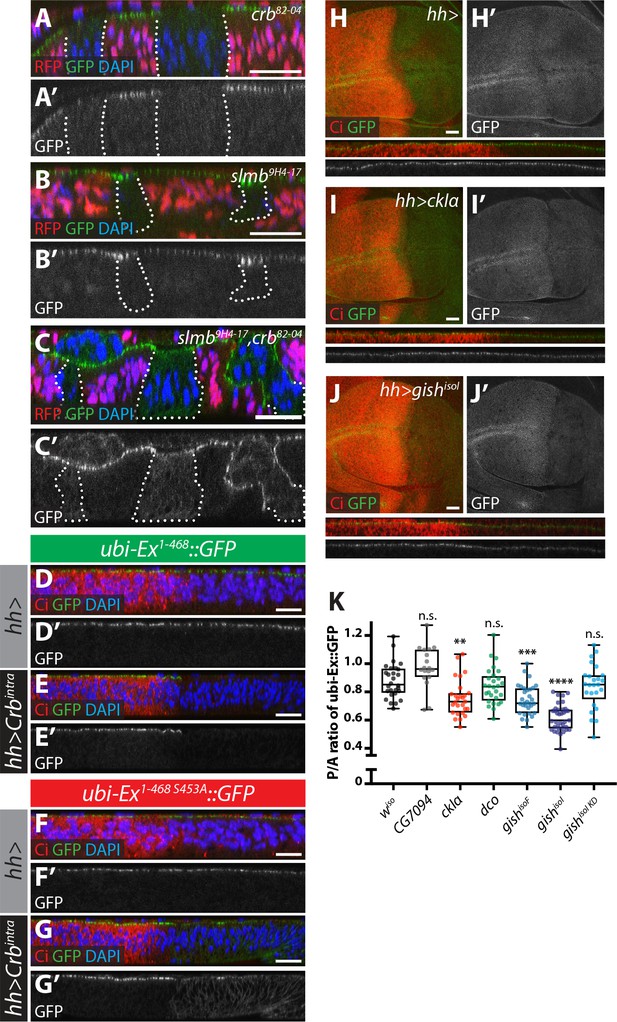
Crb-, Slmb- and CKI-mediated regulation of an in vivo Ex protein stability reporter.
(A–C) Crb and Slmb regulate localisation and in vivo levels of the ubi-Ex1-468::GFP reporter. Confocal micrographs of transverse sections of wing imaginal discs from wandering third instar larvae containing clones mutant for crb82-04 (A), slmb9H4-17 (B), or doubly mutant for slmb9H4-17 and crb82-04 (C). Clones are marked by absence of RFP and highlighted by white dashed lines. The ubi-Ex1-468::GFP reporter (green in A-C, grey in A’-C’) is lost from the apical cortex of crb clones, accumulates apically in slmb clones, and accumulates in the cytoplasm of slmb, crb clones. DAPI staining (blue) marks nuclei. (D–G) Effect of Crbintra on the in vivo Ex protein stability reporter. Confocal micrographs of transverse sections of wing discs from wandering third instar larvae expressing a wt (D and E) or a S453A mutant version of the ubi-Ex1-468::GFP reporter (F and G) (green in D-G and grey in D’-G’), in the absence (D and F) or presence (E and G) of hh-Gal4-driven Crbintra over-expression. Ci immunostaining (red) indicates the anterior compartment, where hh is not expressed. DAPI nuclear staining is shown in blue. In the absence of Crbintra, both versions of the reporter localise at the apical surface. Expression of Crbintra causes depletion of the wt reporter, while it promotes mislocalisation of the S453A variant, in agreement with previously published data (Ribeiro et al., 2014). (H–K) Over-expression of CKI kinases promotes depletion of the Ex stability reporter. XY and transverse sections of third instar wing imaginal discs expressing ubi-Ex1-468::GFP (green in H-J, grey in H’-J’) and either no transgene (H), UAS-ckIα (I) or UAS-gishisoI (J) under the control of hh-Gal4. Ci immunostaining (anterior compartment, lacking hh expression) is shown in red. Expression of CkIα or Gish reduces the levels of the Ex in vivo stability reporter in vivo. (K) Quantification of the ratio between the levels of the ubi-Ex1-468::GFP reporter in the posterior versus the anterior compartment in wing discs of the indicated genotypes (transgene expression driven by hh-Gal4 as in H-J). Data are shown in box (median and 25th-75th percentile) and whiskers (minimum to maximum) plots with all data points represented. n > 18 for all genotypes. Significance was assessed using a one-way ANOVA comparing all genotypes to wiso, with Dunnett’s multiple comparisons test. **, p<0.01; ***, p<0.001, ****, p<0.0001. n.s. non-significant. In XY sections, ventral is up, whereas apical is up in transverse sections. Scale bars, 10 μm in A-C and 20 μm in D-J.
-
Figure 3—source data 1
Source data for quantification of relative protein levels of Ex::GFP in vivo reporter.
- https://doi.org/10.7554/eLife.46592.010
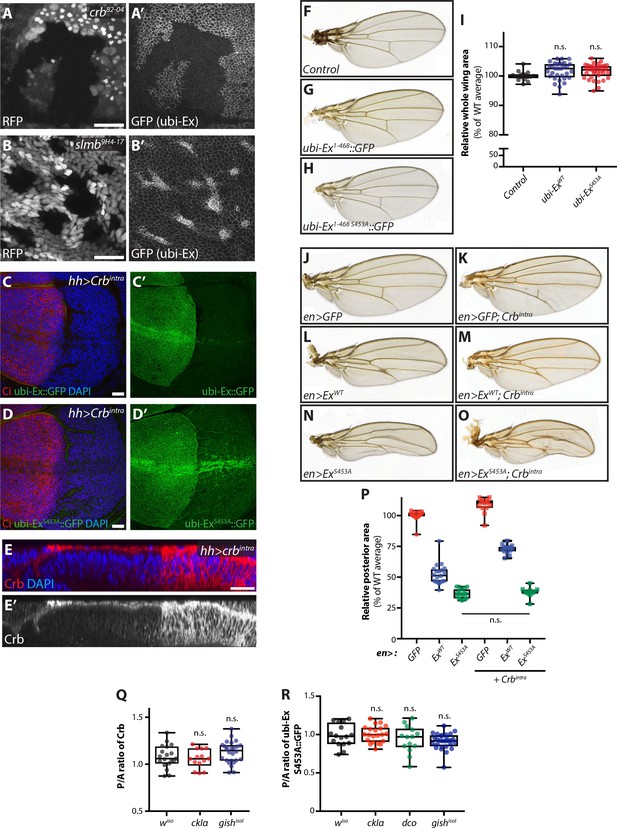
Characterisation of the Ex stability reporter and in vivo regulation of Ex stability and tissue growth.
(A) and (B) Crb and Slmb regulate localisation and levels of the ubi-Ex1-468::GFP reporter. XY confocal micrograph of wing imaginal discs from wandering third instar larvae carrying clones mutant for crb82-04 (A) or slmb9H4-17 (B). Clones are marked by absence of RFP (grey in A and B). The Ex1-468::GFP in vivo reporter (grey in A’ and B’) is absent from the apical cortex of crb mutant cells and accumulates in slmb clones. Note that XY micrographs correspond to transverse images shown in Figure 3A and B, respectively. (C) and (D) Effect of Crbintra on the in vivo Ex protein stability reporters. XY confocal micrographs of wing discs expressing ubi-Ex1-468::GFP (C) or ubi-Ex1-468 S453A::GFP (D) in the presence of hh-Gal4-driven Crbintra. Tissues were stained with Ci and DAPI (red and blue, respectively), while green is direct GFP fluorescence. The wt version of the reporter is completely degraded in the presence of Crbintra, whereas the mutant version can still be partially seen at the apical surface but is mostly relocalised to internal membranes. (E) In vivo localisation of ectopic Crbintra. Transverse sections of third instar wing imaginal disc expressing UAS-crbintra under the control of hh-Gal4, stained for Crb (red in E and grey in E’) and DAPI (blue in E). Note that Crb spreads from the apical surface to internal membranes when Crbintra is over-expressed. (F–I) The Ex protein stability reporter does not overtly affect tissue growth in vivo. Compared with control adult wings (F), constitutive expression of ubi-Ex1-468::GFP (G) or ubi-Ex1-468 S453A::GFP (H) does not affect overall wing size. (I) Quantification of adult wing size. Data are represented as % of the average area of wt adult wings. Box (median and 25th-75th percentile) and whisker (minimum to maximum) plots depict the data, which includes all data points. n.s. denotes non-significant. (J–P) In vivo role of Ex S453 in the regulation of tissue growth. Shown are adult wings from flies raised at 18°C expressing the indicated transgenes in the posterior compartment of the wing under the control of en-Gal4. Compared to control adult wings expressing GFP (J), Crbintra-expressing wings were larger (K). In contrast, expression of full-length ExWT caused a size reduction (L), which was partially rescued by co-expression of Crbintra (M). Interestingly, expression of ExS453A resulted in a further reduction in wing size (N), which was refractory to Crbintra (O). (P) Quantification of adult wing size in flies expressing the indicated transgenes under the control of en-Gal4. Data are represented as % of the average of relative area of the posterior compartment of wt adult wings. Data is shown as box (median, 25th-75th percentile) and whiskers (minimum to maximum) plots, with all data points represented. (Q) and (R) hh-Gal4-mediated over-expression of CKI kinases does not significantly alter apical levels of Crb or Ex1-468 S453A::GFP. Levels of endogenous Crb (Q) or apical Ex1-468 S453A::GFP (R) were quantified in wing imaginal discs expressing CKI kinases under the control of hh-Gal4. Shown are the ratios between the posterior and anterior levels. Data is represented in box (median, 25th-75th percentile) and whiskers (minimum to maximum) plots, with all data points indicated. n.s. denotes non-significant. Scale bars, 10 μm in A and B, and 20 μm in C-E.
-
Figure 3—figure supplement 1—source data 1
Source data for quantification of adult wing sizes and relative protein levels of Crb and Ex S453A::GFP reporter.
- https://doi.org/10.7554/eLife.46592.009
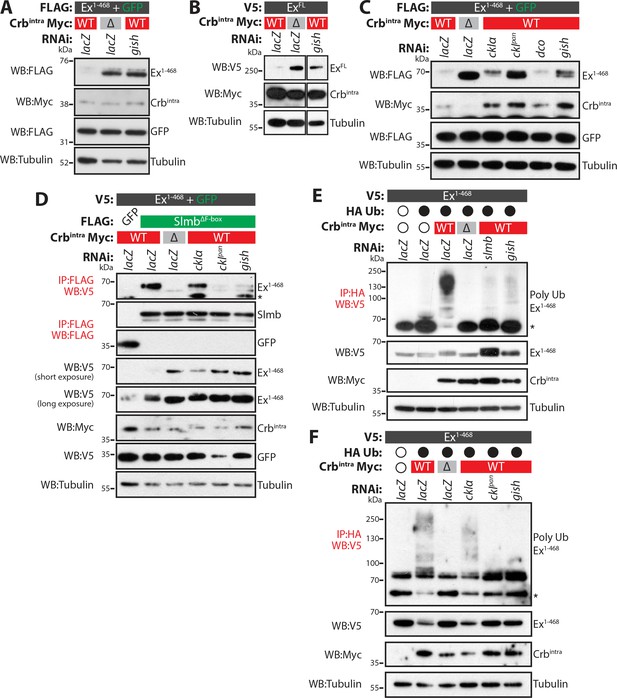
CkIα and Gish are required for Crb-induced Ex degradation.
(A) and (B) RNAi-mediated depletion of gish abrogates Crb-induced Ex degradation. FLAG-tagged Ex1-468 (A) or V5-tagged ExFL (B) were co-expressed with Crbintra or CrbΔFBM, in the presence of dsRNA targeting lacZ (control) or gish. Lysates were processed for immunoblot analysis using the indicated antibodies. In both cases, gish depletion blocked Ex degradation induced by Crbintra expression. (C) CKI kinase knockdown blocks Ex degradation induced by expression of Crbintra. S2 cells treated with dsRNA targeting lacZ or CKI kinases were co-transfected with FLAG-tagged Ex1-468 and GFP, Crbintra or CrbΔFBM. Immunoblot analysis of lysates using the indicated antibodies revealed that depleting ckIα and gish alone or all CKIs (ckIpan) dramatically impaired the ability of Crbintra to promote Ex1-468 degradation. (D) Crb requires CKI kinase function to promote Ex:Slmb binding. Co-IPs were performed between FLAG-tagged GFP or SlmbΔF-box and V5-tagged Ex1-468 in the presence of Crbintra or CrbΔFBM and depletion of lacZ or CKI kinases. Lysates were analysed by immunoblot using the indicated antibodies for detection of protein expression and co-purification. Note that depletion of all CKIs or gish prevented the Ex:Slmb interaction induced by Crbintra. (E) and (F) CKI kinases are required for Crbintra-induced Ex ubiquitylation. S2 cells were treated with the indicated dsRNAs for 24 hr before transfection with the indicated constructs. Following lysis under denaturing conditions, ubiquitylated proteins were isolated using anti-HA antibodies. The presence of Ex and Crbintra was assessed with the indicated antibodies. Knockdown of gish (E and F) or of all CKIs with ckIpan RNAi (F) significantly reduced Ex ubiquitylation, similar to depletion of slmb. GFP was used as transfection efficiency control. Tubulin was used as loading control. Open and full circles denote absence or presence of the indicated plasmid, respectively. Asterisks denote non-specific bands (IgG heavy chain in IP panels).
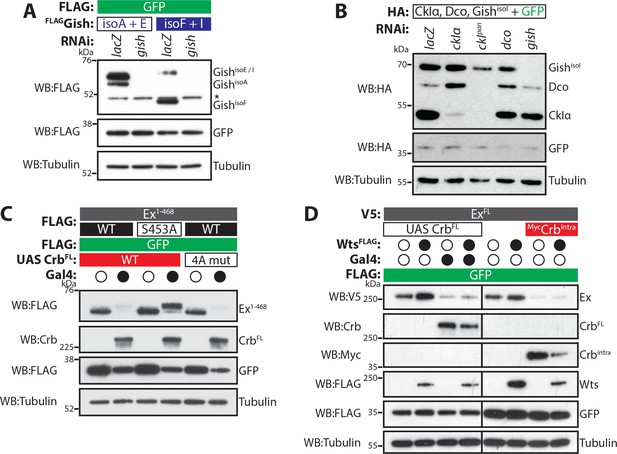
Role of CKI kinases and alternative kinases in the regulation of Ex stability.
(A) Validation of gish RNAi tools. S2 cells were treated with the indicated dsRNA 24 hr before transfection with the indicated constructs. Expression of FLAG-tagged Gish and RNAi efficiency were assessed by immunoblot analysis of lysates using the indicated antibodies. Note that gish RNAi efficiently targets all tested isoforms. (B) Validation of CKI kinase RNAi efficiency. S2 cells were treated with dsRNAs 24 hr before co-transfection with the indicated constructs. RNAi specificity and efficiency were analysed by Western blotting using the indicated antibodies. Note that both ckIα and gish dsRNA were specific for their respective targets, while dco RNAi was unable to target dco. Treating cells with the ckIpan dsRNA revealed that this RNAi targets all CKI kinases. (C) aPKC-mediated phosphorylation of Crb does not modulate the ability of Crb to promote Ex degradation. The stability of FLAG-tagged Ex1-468 (wt or S453A mutant) was assessed by Western blotting with the indicated antibodies in lysates from cells co-transfected with CrbFL or Crb4A mut (which carries mutations in putative aPKC phosphorylation sites). Crb plasmids were under the control of a UAS promoter and, therefore, expression was controlled by Gal4. Note that Crb4A mut is equally adept in promoting Ex1-468 degradation as wt CrbFL. (D) Crb-mediated degradation of Ex supersedes Wts-mediated regulation of a C-terminal Slmb/β-TrCP phosphodegron. The protein stability of V5-tagged full-length Ex (ExFL) was monitored after co-transfection with FLAG-tagged Wts, in the presence or absence of CrbFL or Crbintra. S2 cells were lysed 48 hr after transfection and lysates were processed for immunoblot analysis using the indicated antibodies. In agreement with previous results (Zhang et al., 2015), expression of Wts resulted in the stabilisation of ExFL. However, when ExFL was co-expressed with both Wts and either CrbFL or Crbintra, it was efficiently degraded, and levels were indistinguishable from those seen when ExFL was co-expressed with CrbFL or Crbintra alone. GFP was used as transfection efficiency control. Asterisk denotes non-specific band (FLAG non-specific band present in S2 cell lysates). Tubulin was used as loading control. Open and full circles denote the absence and presence of the indicated plasmid, respectively.
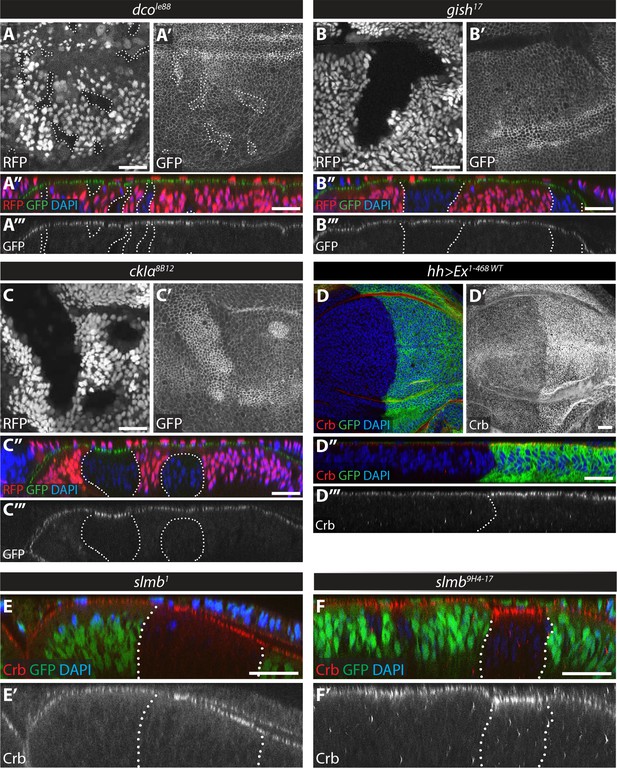
Loss of function of ckIα, but not gish or dco, modulates levels of an Ex protein stability reporter in vivo.
(A) dco mutant clones do not affect the Ex in vivo stability reporter. XY (A, A’) and transverse sections (A’’, A’’’) of ubi-Ex1-468::GFP-expressing third instar wing imaginal discs containing dcole88 mutant clones (marked by absence of RFP and highlighted by white dashed lines), showing direct fluorescence from GFP (green in A’’ and grey in A’ and A’’’) or RFP (red in A’’ and grey in A), and DAPI staining (blue). (B) gish mutant clones do not affect Ex1-468::GFP levels. XY (B, B’) and transverse sections (B’’, B’’’) of a third instar wing imaginal disc expressing ubi-Ex1-468::GFP (green in B’’ and grey in B’ and B’’’) and carrying gish17 mutant clones (marked by absence of RFP and highlighted by white dashed lines) stained with DAPI (blue). (C) ckIα loss-of-function induces higher levels of Ex1-468::GFP. XY (C, C’) and transverse sections (C’’, C’’’) of third instar wing imaginal discs expressing ubi-Ex1-468::GFP and carrying ckIα8B12 mutant clones (marked by absence of RFP and highlighted by white dashed lines). GFP reporter is shown in green (C’’) or grey (C’, C’’’). RFP fluorescence is shown in red (C’’) or grey (C) and the nuclear marker DAPI is shown in blue. (D–F) Over-expression of Ex1-468 or loss of slmb function induce higher apical levels of endogenous Crb. Shown are XY (D, D’) and transverse sections (D’’, D’’’, E, E’, F and F’) of third instar wing imaginal discs expressing UAS-Ex1-468 under the control of hh-Gal4 (D), or carrying loss-of-function clones for the β-TrCP alleles slmb1 (E) or slmb9H4-17 (F). GFP marks hh-Gal4-expressing domain in D, while absence of GFP expression marks slmb mutant clones in E and F (both highlighted by white dashed lines). Crb staining is shown in red (D–F and D’’) or grey (D’–F’ and D’’’) and the nuclear marker DAPI is shown in blue. Dorsal and apical are up in XY and transverse sections, respectively. Scale bars, 20 μm.
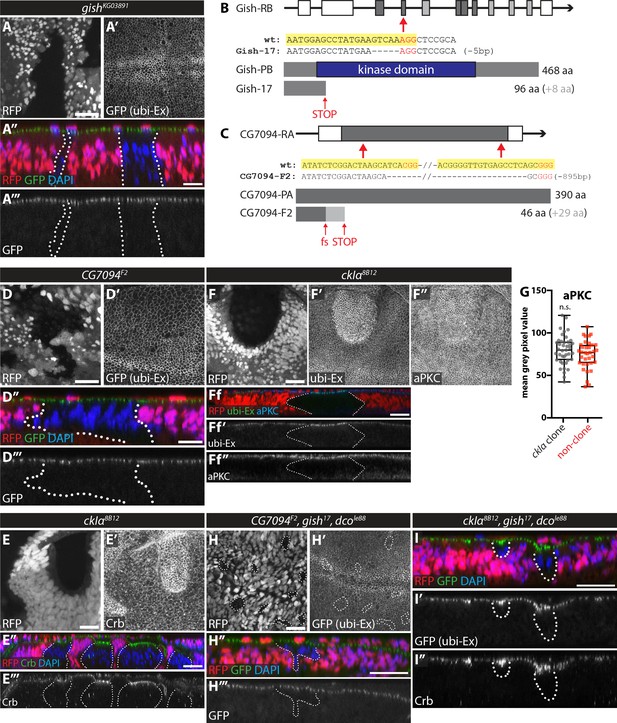
In vivo clonal analysis of the role of CKI kinases in the regulation of the Ex stability reporter.
(A) gishKG03891 mutant clones do not affect Ex1-468::GFP levels. XY (A and A’) and transverse (A’’ and A’’’) confocal micrographs of third instar wing discs carrying gishKG03891 mutant clones (marked by absence of RFP). GFP fluorescence is shown as grey (A’ and A’’’) or green (A’’), RFP is shown as grey (A) or red (A’’), and DAPI staining is shown in blue. (B) and (C) Schematic representation of CRISPR/Cas9-mediated strategies used to generate novel mutant alleles for gish (B) and CG7094 (C). Shown are the gene structures for each gene (coding sequence is depicted by grey rectangles, while white rectangles depict 5’ and 3’ UTR sequences), the targets for CRISPR gRNA sequences (indicated by red arrows), the sequencing results from mutant alleles confirming the mutation generated (large truncation for gish17 and early frameshift and truncation for CG7094F2) and a schematic depiction of the wt and predicted protein in the respective mutants. (D) CG7094 mutant clones do not affect Ex1-468::GFP levels. XY (D and D’) and transverse (D’’ and D’’’) confocal micrographs of third instar wing discs carrying CG7094F2 mutant clones (marked by absence of RFP). GFP reporter is shown in grey (D’ and D’’’) or green (D’’), RFP fluorescence is shown in grey (D) or red (D’’) and DAPI staining is shown in blue. (E) ckIα mutant clones have increased apical levels of Crb. XY (E and E’) and transverse (E’’ and E’’’) confocal micrographs of third instar wing discs containing ckIα8B12 mutant clones (marked by absence of RFP and outlined with dashed lines) and immunostained for Crb (grey in E’ and E’’’ and green in E’’). RFP is in grey (E) or red (E’’) and DAPI is shown in blue. (F) Apical levels of aPKC are unchanged in ckIα mutant clones. XY (F–F’’) and transverse (Ff-Ff’’) sections of third instar wing discs containing ckIα8B12 mutant clones (marked by absence of RFP and outlined with dashed lines, (F and Ff) showing ubi-Ex1-468::GFP direct fluorescence (F’ and Ff’) and stained for aPKC (F’’ and Ff’’). (G) Quantification of aPKC levels in discs carrying ckIα8B12 mutant clones. aPKC levels are not significantly different between ckIα8B12 mutant clones and surrounding wt tissue. Data are represented as box (median, 25th-75th percentile) and whiskers (minimum to maximum) plot, with all data points shown. (H) Combined loss-of-function of CG7094, gish and dco does not affect the levels of the in vivo Ex protein stability reporter. XY (H and H’) and transverse (H’’ and H’’’) confocal micrographs of third instar wing discs carrying CG7094F2, gish17 and dcole88 triple mutant clones (marked by absence of RFP, grey in H and red in H’’) and stained for DAPI (blue). GFP reporter fluorescence is shown in grey (H’ and H’’’) or green (H’’). (I) Combined loss-of-function of ckIα, gish and dco increases Ex1-468::GFP and Crb levels. Transverse confocal micrographs of third instar wing discs containing ckIα8B12, gish17 and dcole88 triple mutant clones (marked by absence of RFP; red in I) and stained for Crb (grey in I’’) and DAPI (blue in I). Shown are images of direct fluorescence of GFP (green in I and grey in I’). n.s. denotes non-significant. Scale bars, 20 μm in E, F, H and I, and 10 μm in A and D.
-
Figure 5—figure supplement 1—source data 1
Source data for quantification of aPKC protein levels in wing imaginal discs carrying clones mutant forckIα.
- https://doi.org/10.7554/eLife.46592.016
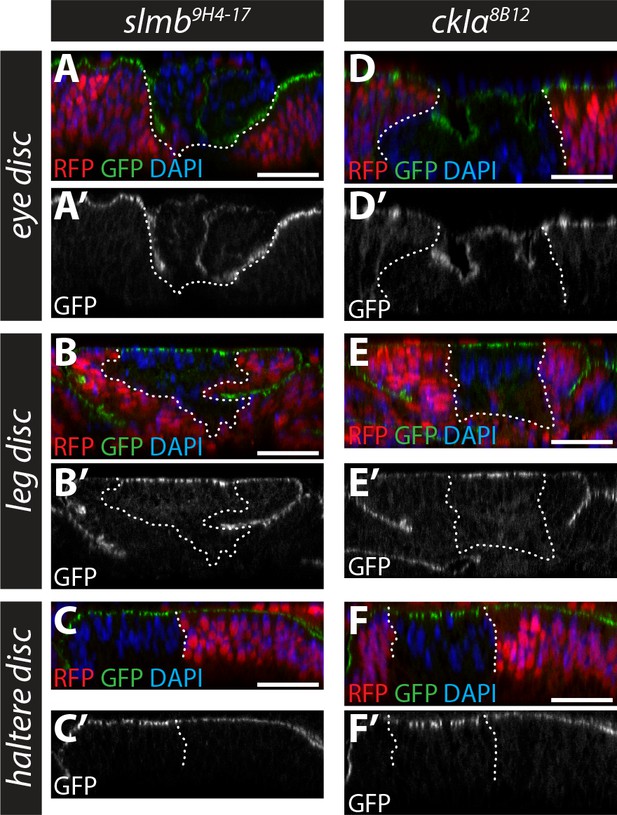
Role of Slmb and CkIα in regulating an in vivo Ex protein stability reporter in eye, leg and haltere discs.
(A–F) Slmb and CkIα regulate levels of the ubi-Ex1-468::GFP reporter in leg and haltere discs, but not eye discs. Confocal micrographs of transverse sections of eye (posterior to the morphogenetic furrow, (A and D), leg (B and E) and haltere (C and F) discs from wandering third instar larvae containing clones mutant for slmb9H4-17 (A–C) or ckIα8B12 (D–F). The ubi-Ex1-468::GFP reporter is shown in green (A–F) or grey (A’–F’). Clones are marked by absence of RFP (red) and highlighted by white dashed lines, and DAPI staining (blue) marks nuclei. Loss of slmb or ckIα in eye discs results in clone extrusion (A and D), and accumulation of the ubi-Ex1-468::GFP reporter at the apical cortex in leg (B and E) and haltere (C and F) discs. Scale bars, 20 μm.
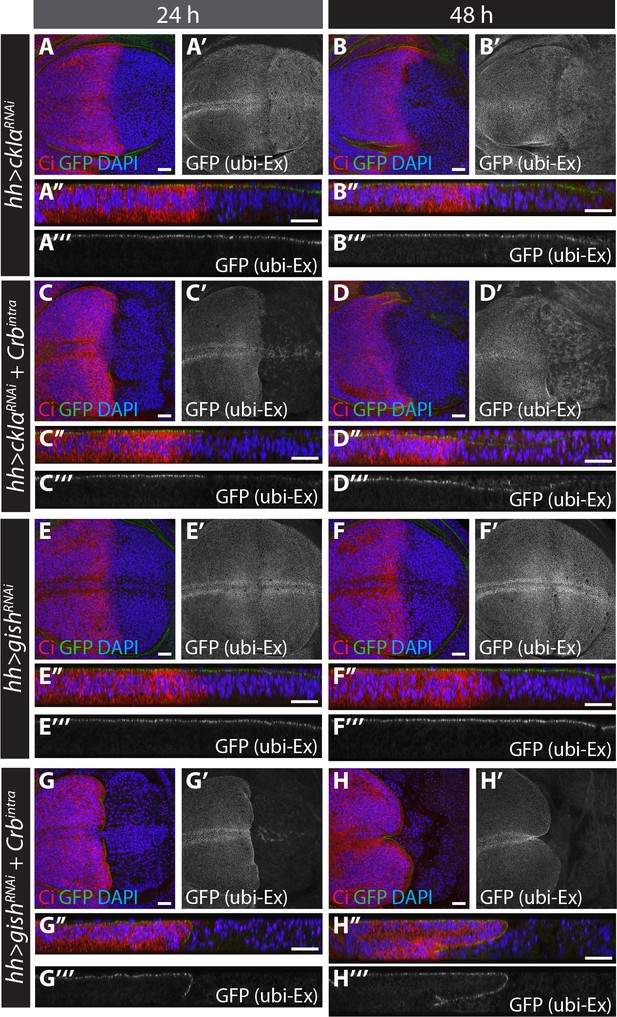
RNAi-mediated depletion of ckIα, but not gish, suppresses Crbintra-induced degradation of an in vivo Ex protein stability reporter.
(A) and (B) ckIα knockdown has a minimal effect on Ex1-468::GFP levels.(C) and (D) ckIα RNAi blocks Crbintra-mediated depletion of Ex1-468::GFP. (E–H) gish RNAi-mediated depletion does not significantly affect in vivo Ex1-468::GFP levels in the absence or presence of Crbintra. XY and transverse sections of third instar wing imaginal discs containing ubi-Ex1-468::GFP, in which hh-Gal4 was used to drive expression of UAS-ckIαRNAi alone (A and B), UAS-ckIαRNAi and UAS-Crbintra (C and D), UAS-gishRNAi alone (E and F) or UAS-gishRNAi and UAS-Crbintra (G and H). Temporal control of Gal4 activity was achieved with a tub-Gal80ts transgene, raising the larvae at 25°C and shifting them to 29°C for the indicated times. ubi-Ex1-468::GFP is shown in green (A–H and A’’–H’’) or grey (A’–H’ and A’’’–H’’’). Ci immunostaining (red) indicates anterior compartment where transgenes are not expressed. DAPI (blue) stains nuclei. Ventral and apical are up in XY and transverse sections, respectively. Scale bars, 20 μm.
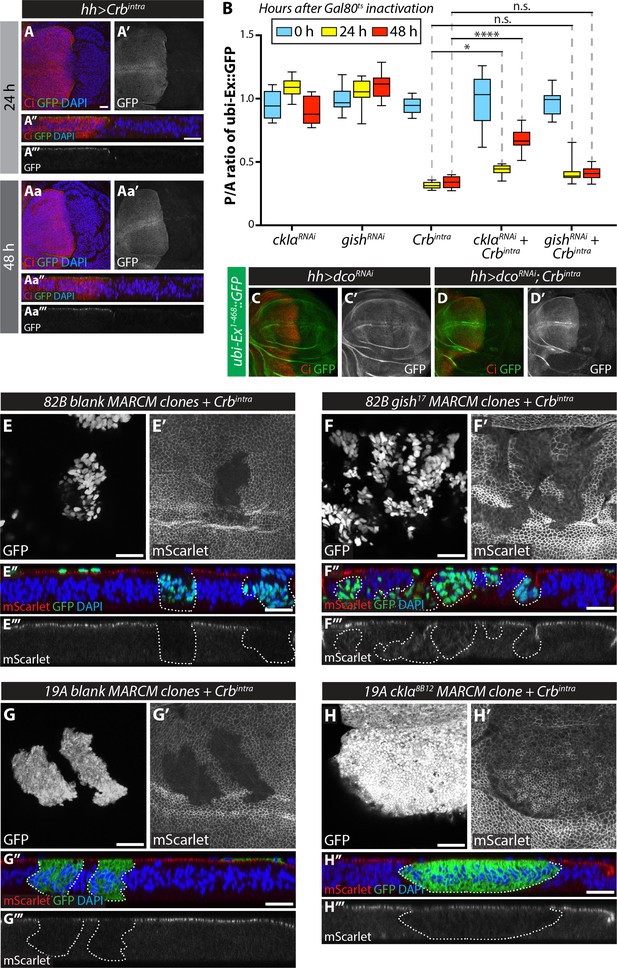
Analysis of the effect of RNAi-mediated depletion and clonal mutation of CKI kinases in the regulation of Crbintra-induced Ex degradation.
(A) Temporally-controlled expression of Crbintra promotes Ex1-468::GFP degradation in vivo. XY (A, A’, Aa and Aa’) and transverse sections (A’’, A’’’, Aa’’ and Aa’’’) of third instar wing imaginal discs expressing ubi-Ex1-468::GFP and UAS-Crbintra for 24 hr (A) or 48 hr (Aa) under the control of hh-Gal4 and tub-Gal80ts. The reporter is shown in green (A, A’’, Aa and Aa’’) or grey (A’, A’’’, Aa’ and Aa’’’). Discs were stained for Ci (red) and DAPI (blue). Note that Ex1-468::GFP is lost from the hh-Gal4-expressing compartment (marked by absence of Ci expression) within 24 hr. (B) Quantification of ratio between the levels of Ex1-468::GFP in the posterior versus anterior compartment. Data for 0 hr, 24 hr and 48 hr after Gal80ts inactivation are represented in blue, yellow and red, respectively. Data are shown as interleaved box (median, 25th-75th percentile) and whiskers (minimum to maximum) plots. *, p<0.05; ****, p<0.0001. n.s. denotes non-significant. (C) and (D) In vivo depletion of dco does not modulate Ex1-468::GFP levels in the absence or presence or Crbintra. Confocal micrographs of third instar wing discs expressing ubi-Ex1-468::GFP and UAS-dcoRNAi alone (C) or in combination with UAS-Crbintra (D), under the control of hh-Gal4. Tissues were stained with Ci (red). Note that there are no differences in Ex1-468::GFP levels when dco was depleted by RNAi (C) and that dcoRNAi was not sufficient to block Crbintra-mediated Ex degradation (D). (E–H) MARCM clone analysis of the effect of loss-of-function of CKI kinases on Crb-mediated Ex degradation. XY (E–H and E’–H’) and transverse confocal micrograph sections (E’’–H’’ and E’’’–H’’’) of wing imaginal discs expressing ubi-Ex1-468::mScarlet and carrying MARCM clones expressing Crbintra in FRT82B blank clones (E), gish17 mutant clones (F), FRT19A blank clones (G) or ckIα8B12 mutant clones. Clones are marked by the presence of GFP (grey in E-H and green in E’’-H’’). mScarlet fluorescence is shown in grey (E’–H’) or red (E’’–H’’), whereas DAPI nuclear staining is shown in blue. Note that loss of ckIα partially rescues the apical levels of Ex in the presence of Crbintra. Scale bars, 20 μm.
-
Figure 6—figure supplement 1—source data 1
Source data for quantification of relative levels of the Ex::GFPin vivoreporter in wing imaginal discs expressing RNAi targeting CKI kinases alone or in combination with expression of Crbintra.
- https://doi.org/10.7554/eLife.46592.019
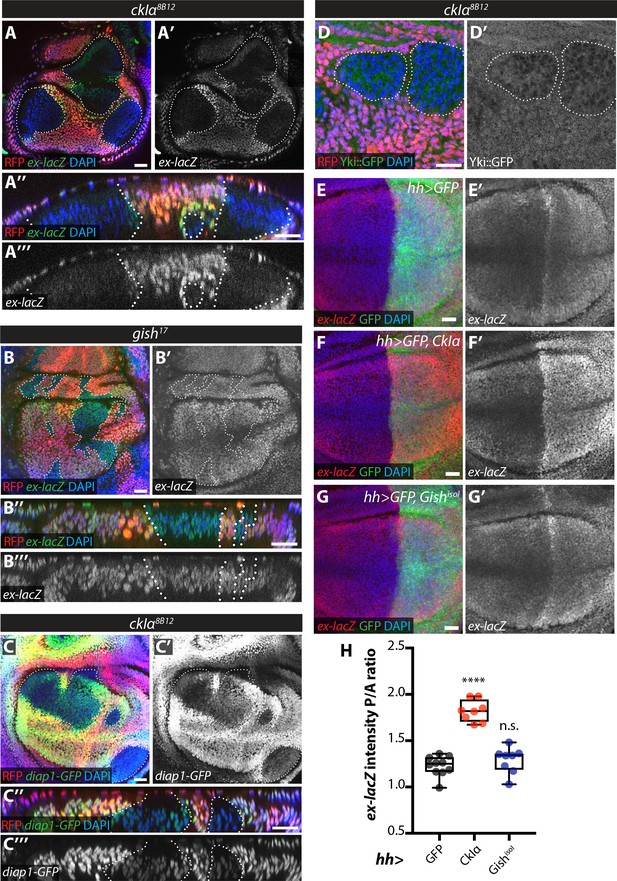
Effect of CkIα and Gish loss- and gain-of-function on Yki target gene expression.
(A–D) Yki transcriptional activity is reduced by loss of ckIα, but unaffected by loss of gish. XY and XZ confocal images of third instar wing imaginal discs bearing clones mutant for ckIα8B12 (A, C, D), or gish17 (B), co-expressing the Yki transcriptional reporter genes ex-lacZ (A, B), diap1-GFP3.5 (C), or a Yki::GFP fusion protein (a knock-in at the endogenous locus, (D). ex-lacZ is visualised by immunostaining for β-galactosidase (green in A, A’’, B and B’’, grey in A’, (A’’’, B’ and B’’’); diap1-GFP and Yki::GFP are visualised by direct GFP fluorescence (green in C, C’’ and D, grey in C’, (C’’’ and D’). Clones are marked by absence of RFP (red) and highlighted by white dashed lines; DAPI (blue) stains nuclei. Reporter gene expression is drastically reduced in ckIα8B12 (A, C), but not gish17 (B), mutant clones. Yki::GFP appears excluded from the nucleus of ckIα8B12 mutant cells (D). Scale bars 20 μm. (E–H) Overexpression of CkIα, but not GishisoI, results in upregulation of ex-lacZ. Maximum intensity projections of z-stacks of the pouch region of wing imaginal discs from third instar larvae overexpressing no transgene (E), UAS-CkIα (F), or UAS-GishisoI (G) under the control of hh-Gal4. Crosses were raised at 25 °C and larvae were dissected at wandering L3 stage. ex-lacZ expression was detected by immunostaining for β-galactosidase (red in E-G, grey in E’-G’); the posterior compartment is marked by expression of GFP (green); DAPI (blue) stains nuclei. (H) Quantification of the posterior to anterior ratio of ex-lacZ signal intensity in the pouch region; CkIα expression significantly upregulates this Yki target gene (p=0.0001, one-way ANOVA comparing all means to hh >control, with correction for multiple comparisons; n ≥ 8 for all genotypes), while GishisoI does not (p=0.4808). Scale bars 20 μm.
-
Figure 7—source data 1
Source data for quantification of relative levels of the Ex::GFPin vivoreporter in wing imaginal discs expressing RNAi targeting CKI kinases alone or in combination with expression of Crbintra.
- https://doi.org/10.7554/eLife.46592.021
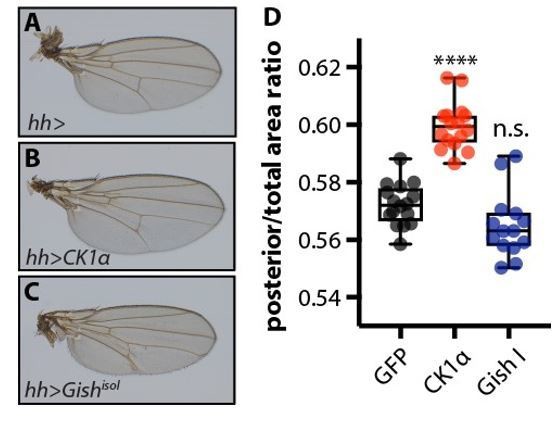
Overexpression of CkIα, but not GishisoI, causes a mild overgrowth of the posterior compartment of the wing.
(A-C) Adult wings from female flies overexpressing no transgene (A), UAS-CkIα (B), or UAS-GishisoI (C), in the posterior compartment of the developing wing under the control of hhGal4. Crosses were raised at 25 °C. (D) Quantification of the ratio of the area of the posterior compartment to total wing area in adult wings. Overexpression of CkIα causes a significant overgrowth (p=0.0001, one way ANOVA comparing all means to hh> controls, with correction for multiple comparisons). GishisoI overexpressing wings tend to be slightly smaller than controls, though this is not statistically significant (p=0.0547), and sometimes exhibit blisters. N≥14 for all genotypes.
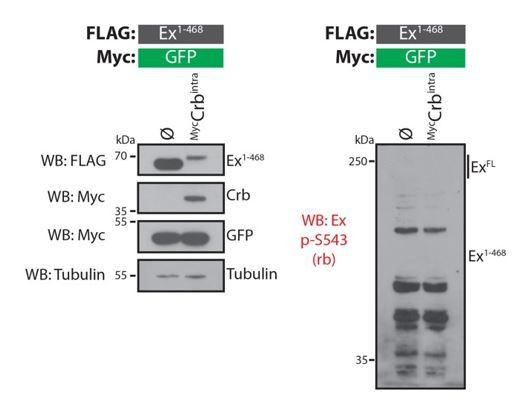
Evaluation of rabbit Ex phospho-S453 antibody in Drosophila S2 cells.
S2 cells were transfected with the indicated plasmids 48h prior to lysis and processing for Western blot analysis. Note that the phospho-S453 Ex antibody does not recognise any protein in the vicinity of the predicted sizes of full-length Ex or Ex1468. ø denotes empty vector. GFP and tubulin were used as transfection and protein loading control, respectively.
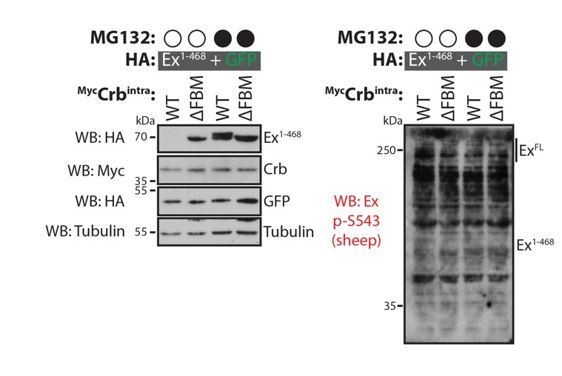
Evaluation of sheep Ex phospho-S453 antibody in Drosophila S2 cells.
S2 cells were transfected with the indicated plasmids 48h prior to lysis and processing for immunoblotting. Note that the phospho-S453 Ex antibody does not seem to specifically recognise full length Ex or Ex1-468. Open and closed circles denote absence or presence of MG132 treatment, respectively. GFP and tubulin were used as transfection and protein loading control, respectively.
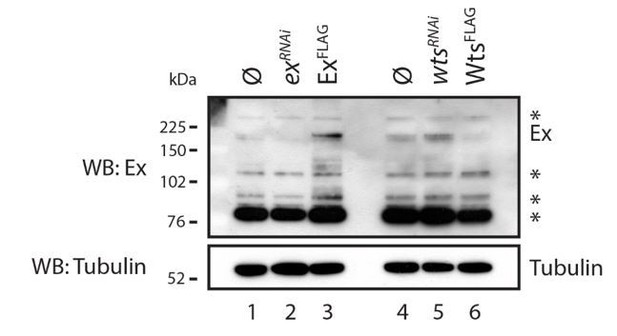
Evaluation of Ex antibody in the recognition of endogenous Ex protein.
S2 cells were transfected with the indicated plasmids 48h prior to lysis and dsRNA treatment was performed 24h before transfection. Note that there are several bands recognised by the Ex antibody. One of the weaker slow migrating bands is consistent with it being endogenous Ex since it is increased when exogenous Ex was transfected and when wts was depleted by RNAi. ø denotes empty vector; * indicate non-specific bands recognised by the Ex antibody. Tubulin was used as loading control.
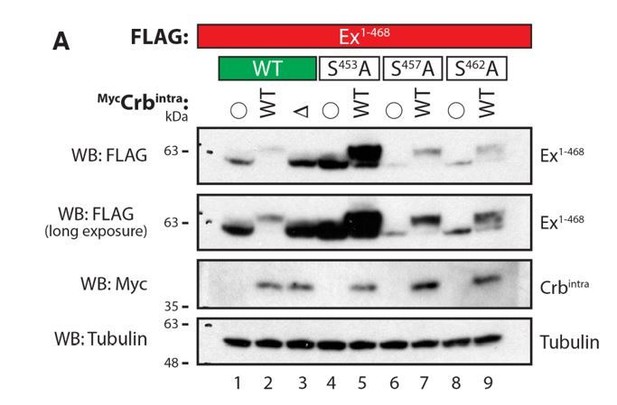
Evaluation of Ex electrophoretic mobility shift using Phos-tag gels.
S2 cells were transfected with the indicated plasmids 48h prior to lysis. Lysates were run in Phos-tag gels to assess phosphorylation pattern of Ex1-468 (WT, S453A, S457A or S462A) in the presence of Crbintra WT or a variant carrying a mutated FERM-binding motif (DFBM). Immunoblot analysis was performed using the indicated antibodies. Open circle indicates empty vector. Tubulin was used as loading control.
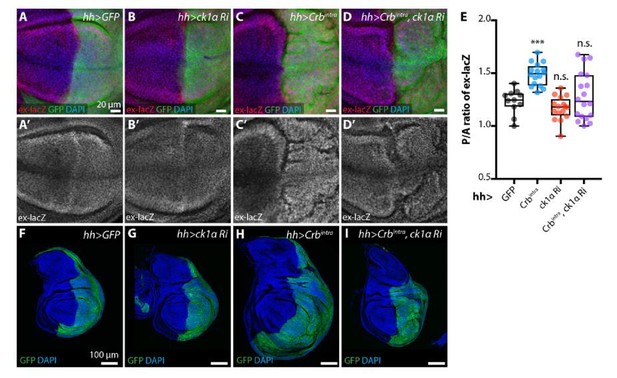
ckIαRNAi partially reverts Crbintra-induced upregulation of Yki target genes and overgrowth.
(A-E) ckIαRNAi partially reverts Crbintra-induced upregulation of the Yki activity reporter ex-lacZ. Sum slices projections of z-stacks of the pouch region of third instar wing imaginal discs expressing ex-lacZ and either no transgene (A), UAS-ckIαRNAi (B), UAS-Crbintra (C), or UASckIαRNAiplus UAS-Crbintra (D). Transgene expression was driven by hh-Gal4, and the time of onset of expression was controlled using tub-Gal80ts; crosses were raised at 25 °C then shifted to 29 °C 48 h prior to dissection as wandering L3 larvae. The posterior compartment is marked by CD8::GFP (green); DAPI (blue) stains nuclei. Scale bars 20 μm. (A’-D’) ex-lacZ channels alone. (E) Quantification of posterior to anterior ratio of nuclear ex-lacZ intensity in sum slices projections. Expression of Crbintra for 48 h induces a significant increase in ex-lacZ expression levels, which is partially reverted by co-expression of ckIαRNAi (p=0.7240, one way ANOVA comparing all means to hh>GFP control, with correction for multiple comparisons, n≥11 for all genotypes) (F-I) ckIαRNAi partially reverts Crbintra-induced overgrowth of wing imaginal discs. XY sections of whole wing imaginal discs from third instar larvae, which have expressed the indicated transgenes under the control of hh-Gal4 for 48 h prior to dissection. The posterior compartment is marked by CD8::GFP (green); DAPI (blue) stained nuclei. hh>Crbintra discs tend to be larger overall, and the posterior compartment of the pouch develops ectopic folds as the disc attempts to accommodate the additional cells. Coexpression of ckIαRNAi results in less severe overgrowth, and less pronounced ectopic folding of the pouch region. Scale bars 100 μm. Genotypes for Rebuttal Figure 8: (A, A’, F) w; ex-lacZ/+; hh-Gal4, UAS-CD8::GFP/+; (B, B’, G) w; UAS-ckIαRNAi (110768KK)/ ex-lacZ; hh-Gal4, UAS-CD8::GFP/ tub-Gal80ts; (C, C’, H) w; ex-lacZ/+; hh-Gal4, UAS-CD8::GFP/ UAS-Crbintra, tub-Gal80ts; (D, D’, I) w; UASckIαRNAi (110768KK)/ ex-lacZ; hh-Gal4, UAS-CD8::GFP/ UAS-Crbintra, tub-Gal80ts
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.46592.022





