A Drosophila model of neuronal ceroid lipofuscinosis CLN4 reveals a hypermorphic gain of function mechanism
Figures
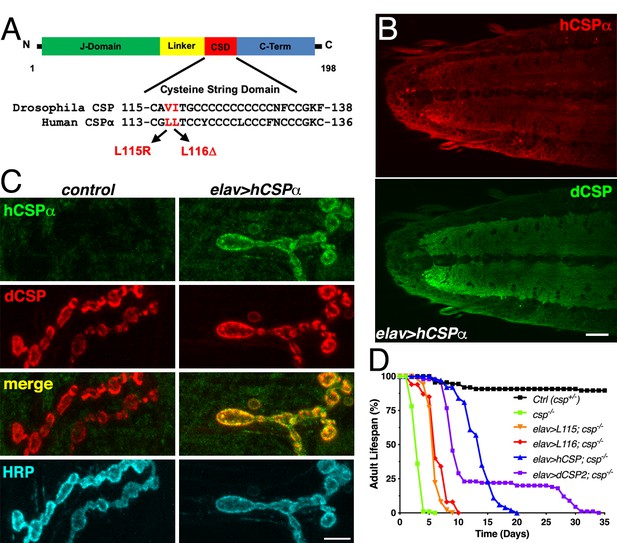
Generation of a Drosophila CLN4 model.
(A) Structure of CSP and position of CLN4 mutations in the cysteine-string (CS) domain of hCSPα and dCSP. CSP’s N-terminal J domain, linker domain and C-terminus are indicated. (B) Larval VNC of animals expressing WT hCSPα in neurons from a single transgene with an elav driver immunostained for hCSPα and endogenous dCSP. Scale bar, 20 μm. (C) Larval NMJs of control and animals expressing WT hCSPα immunostained for hCSPα, dCSP, and HRP marking the presynaptic plasma membrane. Scale bar, 5 μm. (D) Adult lifespan of control (dcspX1/+, black), cspX1/R1 deletion mutants (green) and cspX1/R1 mutants expressing WT hCSPα (blue), hCSP-L115, (orange), hCSPα-L116 (red), or dCSP2 (purple) with an elav driver.
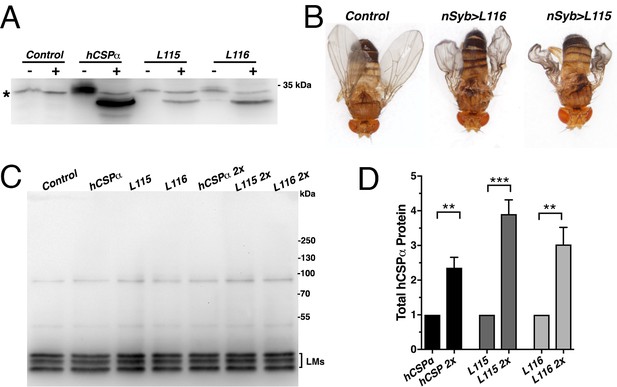
Phenotypic effects of CLN4 mutations.
(A) Lipidation of WT hCSPα, hCSP-L115 and -L116. Immunoblot of larval VNC extracts from control (w1118) and animals expressing elav-driven hCSPα, hCSP-L115 or -L116 were probed for hCSPα. Samples were treated overnight with either 0.5 M hydroxylamine (+) or equimolar Tris (-). Asterisk denotes unspecific signal. (B) Adult control (w1118) and animals expressing nSyb-driven hCSP-L115 or -L116 in neurons. (C) Immunoblot probed for dCSP of larval brain extracts from control (w1118) and animals expressing WT hCSPα, hCSPα-L115 or -L116 from either one or two (2x) transgenes with an elav driver. The lipidated monomeric dCSP isoforms (LMs) are indicated. (D) Increase in total protein levels of WT hCSPα, hCSP-L115 and -L116 induced by expressing either one or two transgenes (2x). Signals were normalized to loading control and plotted as n-fold change to respective expression levels from one transgene (mean ± SEM; N = 6, two-tailed unpaired t test; **, p<0.01; ***, p<0.001.
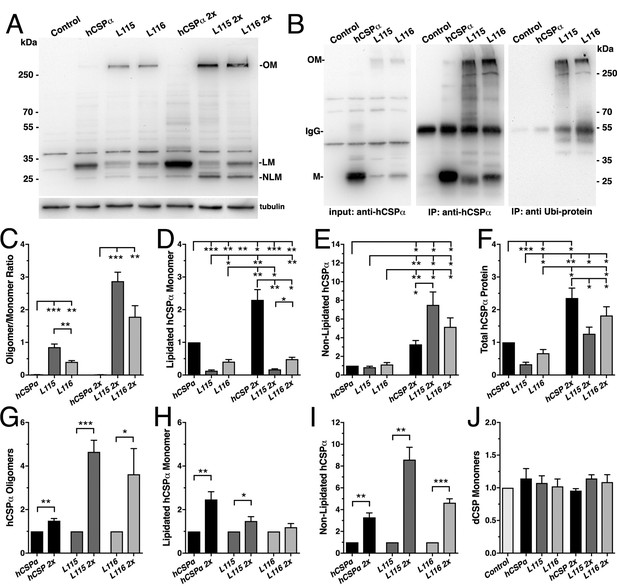
CLN4 mutations cause dose-dependent oligomerization of hCSPα in neurons.
WT and mutant hCSPα (L115/L116) were expressed in larval neurons of white1118 animals (control) with an elav driver from one or two transgenes (2x). (A) Western blot of larval brain protein extracts probed for hCSPα. Signals for SDS-resistant hCSPα oligomers (OM), lipidated monomeric hCSP (LM), and non-lipidated hCSP (NLM) are indicated. β-tubulin was used as loading control. (B) Western blots probed for hCSPα or lysine-linked-ubiquitin of hCSPα-immunoprecipitated extracts from adult heads of indicated genotypes. Signals for IgG heavy chain are indicated. (C) Average oligomer/monomer ratios (N = 5). (D–F) Levels of lipidated (D), non-lipidated (E), and total hCSPα (F) normalized to WT hCSPα (N = 6). (G–I) Dosage-dependent increase of hCSPα oligomer (G), lipidated (H), and non-lipidated monomer levels (I). Signals were normalized to loading control and plotted as n-fold change from 1-copy expression of WT hCSPα (N = 6). (J) Levels of monomeric dCSP shown as n-fold change from control (N = 3). Graphs display mean ± SEM. Statistical analysis used one-way ANOVA (C–F, J) or two-tailed unpaired t test (G–I); *, p<0.05; **, p<0.01; ***, p<0.001.
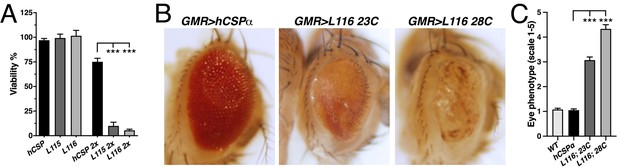
CLN4 mutations cause dose-dependent lethality and eye degeneration.
(A) Viability of animals expressing WT, L115, or L116 mutant hCSPα pan-neuronally from one or two transgenes (2x) with an elav driver (N ≥ 3, n ≥ 740). (B) Images of adult fly eyes expressing WT hCSPα or hCSPα-L116 with a GMR-Gal4 driver at 23°C and 28°C. (C) Semi-quantitative assessments of CLN4-induced eye phenotypes (N ≥ 9). Graphs display mean ± SEM. Statistical analysis used one-way ANOVA (A) and Kruskal-Wallis test (C); *, p<0.05; **, p<0.01; ***, p<0.001.
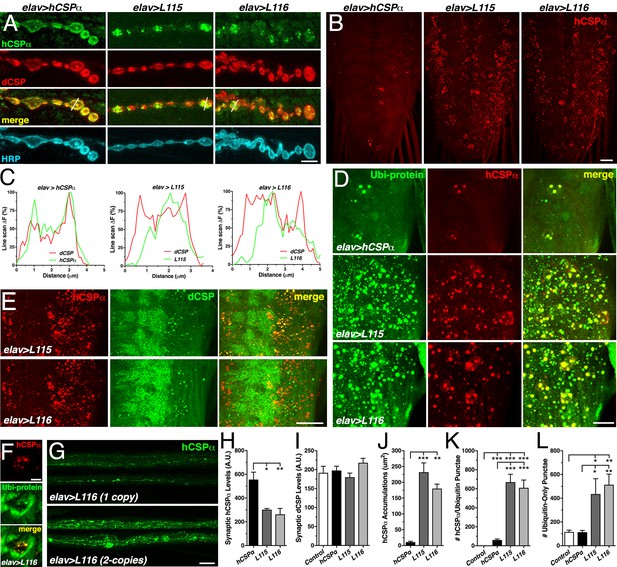
CLN4 mutations reduce synaptic hCSPα levels and cause abnormal accumulations with endogenous dCSP and ubiquitinated proteins in axons and somata.
WT, L115 or L116 mutant hCSPα were expressed in larval neurons with an elav-Gal4 driver from one (D–E, J–L) or two (A, H–I) transgenes. Genotypes are indicated. (A) Larval NMJs immunostained for hCSPα, endogenous dCSP, and the neuronal membrane marker HRP. White lines denote line scans shown in C. (B) Larval VNCs stained for hCSPα. (C) Plots of hCSPα and dCSP fluorescence from single line scans through synaptic boutons (white lines in A). (D) Larval VNC segments stained for hCSPα (red) and lysine-linked-ubiquitin visualizing ubiquitinated proteins (Ubi-protein). (E) Larval brain segments stained for hCSPα and dCSP. (F) Synaptic bouton of larval NMJ stained for hCSPα and Ubi-proteins. (G) Proximal larval segmental nerves stained for hCSPα. (H–I) Average levels of hCSPα (H) and dCSP (I) at synaptic boutons of larval NMJs (N > 4). (J) Cumulative area of abnormal hCSPα accumulations in larval brains (N ≥ 3). (K–L) Average number of accumulations immunopositive for both hCSPα and ubiquitin (K), or only positive for ubiquitin (L) but not hCSPα (N ≥ 4). Scale bars: 5 μm (A), 20 μm (B, G), 15 μm (D), 10 μm (E), 5 μm (F). Graphs display mean ± SEM. Statistical analysis used one-way ANOVA (H–L); *, p<0.05; **, p<0.01; ***, p<0.001.
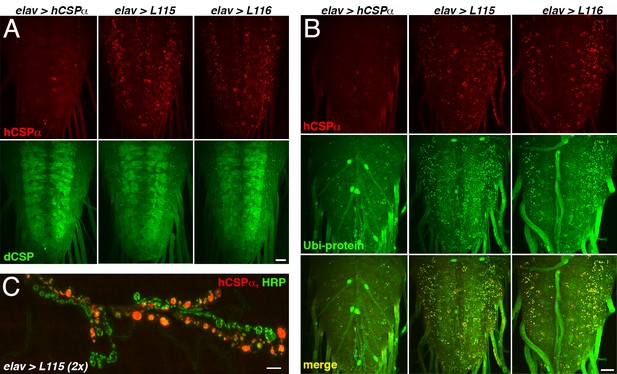
hCSP-L115 and -L116 abnormally accumulate with endogenous dCSP and ubiquitinated proteins.
WT and mutant hCSPα (L115 and L116) were expressed in larval neurons with an elav-Gal4 driver from 1 (A–B) or two (C) transgenes (2x). (A–B) Larval VNCs of indicated genotypes immunostained for hCSPα and dCSP (A) or lysine-linked-ubiquitin (B). (C) Larval NMJ stained for hCSPα and HRP showed occasionally extreme accumulations of hCSP-L115 in synaptic boutons. Scale bar, 20 μm (A–B), 5 μm (C).
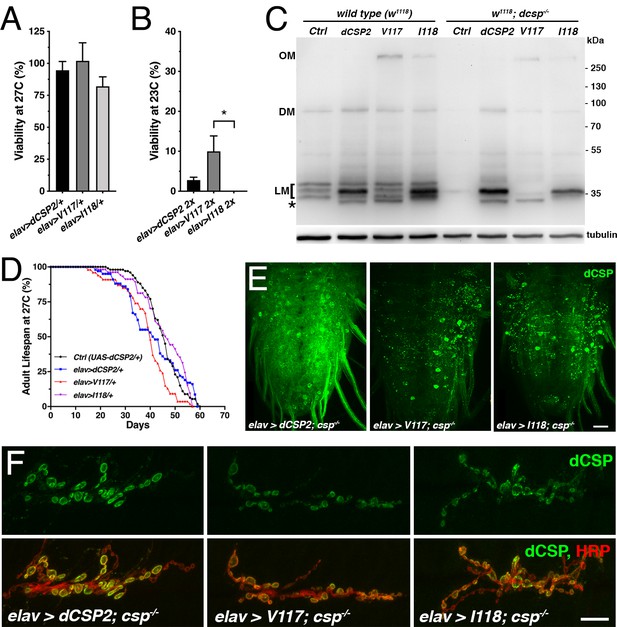
Effects of CLN4-analogous mutations in dCSP.
WT dCSP2, dCSP-V117R (analogous to human L115R) and dCSP-I118Δ (analogous to L116Δ) were expressed in larval neurons with an elav-Gal4 driver from one or two transgenes (2x) in otherwise wild type control (w1118) or homozygous dcsp null mutants (dcspX1/R). (A–B) Viability of animals expressing dCSP2, dCSP-V117R, or -I118Δ pan-neuronally from one transgene at 27°C (A) and two transgenes (2X) at 23°C (B, N = 4). (C) Immunoblot of larval VNC extracts of indicated genotypes probed for dCSP. Signals corresponding to lipidated dCSP monomers (LM), dimers (DM), and high-molecular weight oligomers (OM) are indicated. Asterisk denotes partially lipidated dCSP. β-tubulin was used as loading control. (D) Adult lifespan at 27°C of control (w1118; UAS-dCSP2) and mutants expressing WT dCSP2, dCSP-V117, or -I118 from one copy with an elav driver. (E) Larval VNCs of indicated genotypes immunostained for dCSP. (F) Larval NMJs of indicated genotypes stained for dCSP and HRP. Scale bar, 20 μm (A), 10 μm (C). Graphs display mean ± SEM. Statistical analysis used one-way ANOVA (A) or a Kruskal-Wallis test (B); *, p<0.05; **, p<0.01; ***, p<0.001.
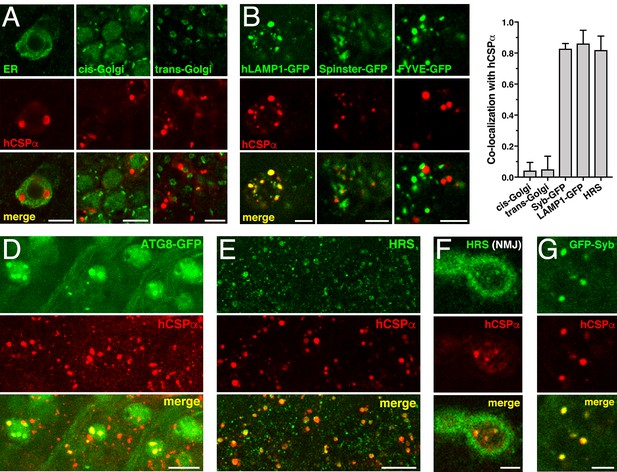
CLN4 mutant hCSPα accumulates on LAMP1- and HRS-positive endosomes.
hCSP-L116 was expressed in larval neurons with an elav-Gal4 driver from one transgene. As indicated, respective reporter transgenes were co-expressed. (A) Neurons of larval VNCs co-immunostained for hCSPα and the ER marker GFP-KDEL, the cis-Golgi marker GMAP, or the trans-Golgi marker Golgin 245. (B) Neurons co-immunostained for hCSPα (red) and co-expressed hLAMP1-GFP, Spinster-GFP or the PI3P marker FYVE-GFP. (C) Fraction of organelle markers colocalizing with hCSPα accumulations (mean ± SEM; n ≥ 65, N ≥ 4). (D–E) Segments of larval brains costained for hCSPα and ATG8/LC3-GFP (D) or HRS (E). (F) Synaptic boutons at larval NMJs co-stained for hCSPα and HRS. (G) Neuron co-immunostained for hCSPα and coexpressed GFP-nSyb. Scale bars: 10 μm (D–E), 5 μm (A–B, F–G).
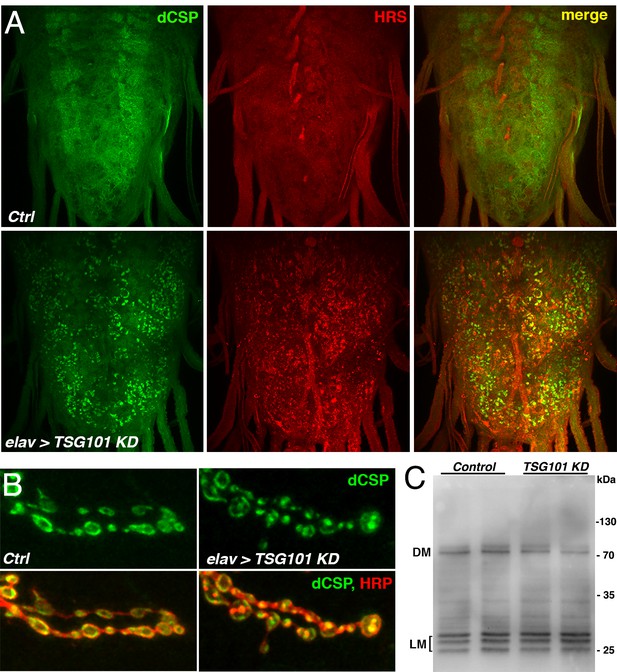
RNAi-mediated KD of TSG101 causes accumulation of dCSP on HRS-positive endosomes.
(A) Larval VNCs immunostained for dCSP and HRS of driverless control (w1118; UAS-TSG101 KD/+) and elav-driven KD of TSG101 using a hairpin transgene (w1118, elav-Gal4; UAS-TSG101 KD/+). Note the mislocalization of endogenous dCSP from its typical diffuse neuropil localization to HRS-positive accumulations in neuropil and neuronal somata. (B) Larval NMJs of indicated genotypes stained for dCSP and HRP. Note the abnormal localization of dCSP from the periphery to more centrally located HRP-positive endosomes. (C) Immunoblot probed for dCSP of larval brain extracts from control and elav-driven TSG101 KD. Note that TSG101 KD does not cause high-molecular weight oligomerization of dCSP despite the induced mislocalization to HRS-positive endosomes.
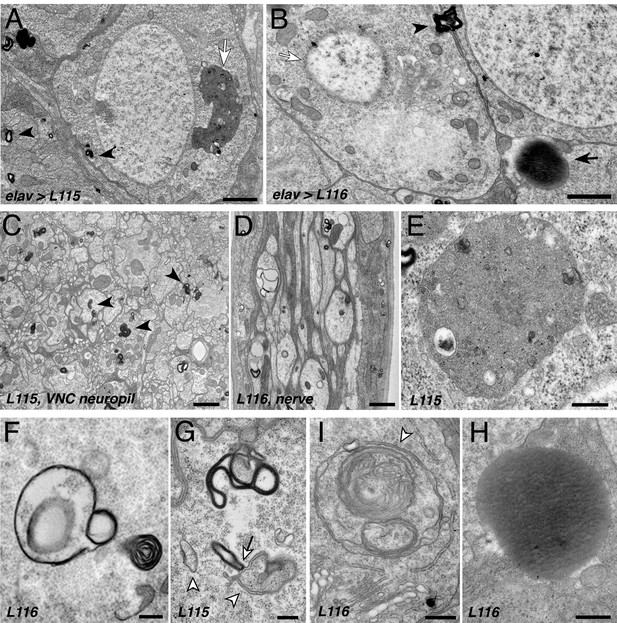
CLN4 mutations cause abnormal endomembrane structures and EM-dense accumulations.
TEM micrographs of ultrathin (70 nm) sections from larval VNCs expressing hCSP-L115 or -L116 with an elav driver from two transgenic copies. Genotypes are indicated. (A–B) Neuronal somata containing electron-dense membrane whirls (black arrowheads), large electron-dense extracellular deposits (black arrow, (B), and occasionally ‘residual lysosomes’ (white arrow, (A), bloated Golgi Apparati (B), and degenerating nuclear membranes (white arrow, (B). (C) Neuropil of larval VNC containing membrane whirls in neuronal processes (black arrowheads). (D) Sagittal section of larval segmental nerve containing membrane whirls and abnormal autophagosome-like structures in axons of sensory and motor neurons. (E–H) High magnification images showing residual lysosome with a diverse variety of intraluminal vesicles (E), various forms of EM-dense membrane whirls (F–G) and autophagosome-like structures (white arrowheads, (G–I) that may interact with EM-dense structures (white arrow, (G), and an electron-dense extracellular deposit (H). Scale bars: 1 μm (A–C), 200 nm (D–E).
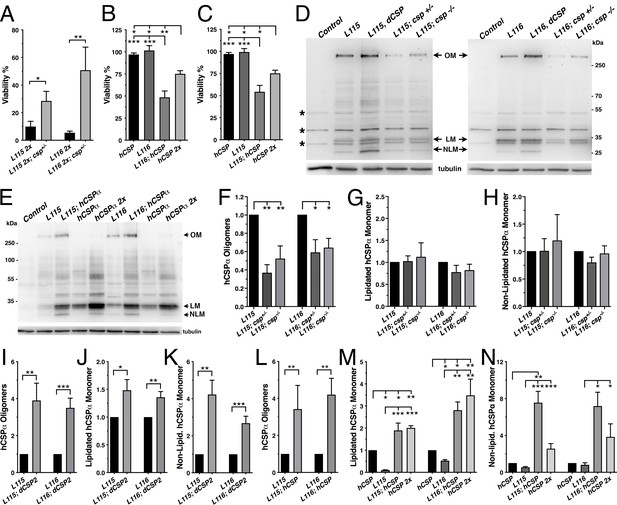
Altering wild type CSP levels modifies CLN4 phenotypes.
WT hCSPα, hCSP-L115 or -L116 were expressed in neurons from one or two transgenes (2x) with an elav driver in control (w1118), heterozygous cspX1/+, and homozygous cspX1/R1 deletion mutants, or co-expressed with WT hCSPα or dCSP. Genotypes are indicated. (A–C) Effects of reducing endogenous dCSP (A) or co-expressing WT hCSPα (B–C) on the viability of hCSP-L115 and -L116 mutant flies (N > 3; n > 144). (D–E) Immunoblots of protein extracts from larval VNC of indicated genotype probed for hCSPα and β-tubulin (loading control). hCSPα oligomers (OM), lipidated (LM), non-lipidated hCSPα monomers (NLM), and unspecific signals (*) are denoted. (F–N) Effects of reduced endogenous dCSP (F–H), increased dCSP (I–K), and increased WT hCSPα (L–N) levels on hCSPα oligomers (F, I, L), lipidated monomers (G, J, M), and non-lipidated monomers (H, K, N). Signals were normalized to loading control and plotted as n-fold change of L115 or L116 levels when expressed in a WT background (N = 5). Graphs display mean ± SEM. Statistical analysis used unpaired t test (A, I–L) or one-way ANOVA (B–C, F–H, M–N); *, p<0.05; **, p<0.01; ***, p<0.001.
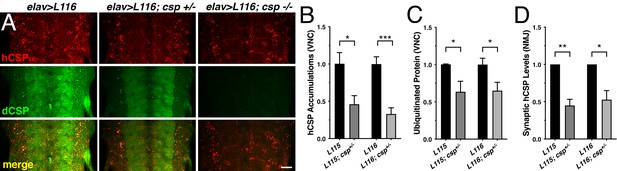
Effects of reduced dCSP levels on CLN4 mutant hCSPα localization and protein ubiquitination.
WT hCSPα, hCSP-L115 or -L116 were expressed in neurons with an elav driver in control (w1118), heterozygous cspX1/+, or homozygous cspX1/R1 deletion mutants. (A) Larval VNC stained for hCSPα and dCSP. Genotypes are indicated. Scale bar, 20 μm. (B–D) Effects of reduced dcsp gene dosage on the accumulation of hCSP-L115 and -L116 in larval VNC (B, N ≥ 3), ubiquitinated protein levels in larval VNCs (C, N ≥ 4), and synaptic levels of hCSP-L115 and -L116 at larval NMJs (D, N ≥ 4). Signals were normalized and plotted as n-fold change to levels of elav-L115 and -L116 expression in WT control background. Graphs display mean ± SEM. Statistical analysis used a paired (D) and unpaired t test (B–C); *, p<0.05; **, p<0.01; ***, p<0.001.
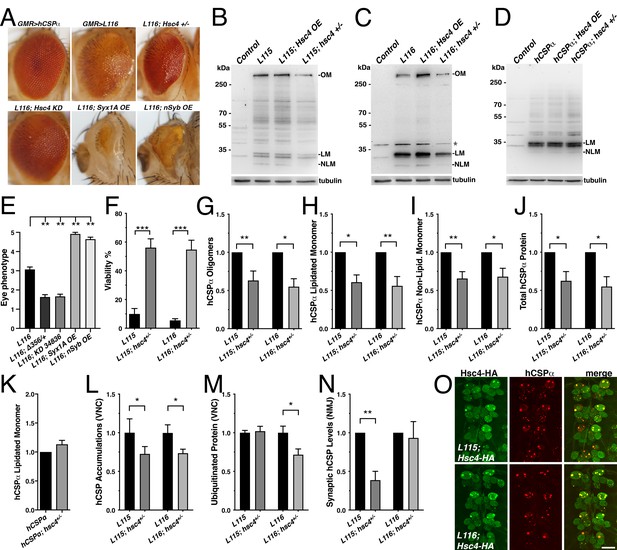
Reducing the gene dosage of Hsc4 attenuates CLN4 phenotypes.
(A) Adult eyes of flies expressing WT hCSPα (control) or hCSP-L116 with a GMR-Gal4 driver in the absence or presence of a heterozygous Hsc4+/- (hsc4∆356) deletion, a Hsc4 KD (34836), OE of Syx1A or Syb. (B–D) Immunoblots of extracts from larval VNCs of indicated genotypes were probed for hCSPα. Lipidated monomeric hCSP (LM), non-lipidated hCSP (NLM) and hCSPα oligomers (OM) are indicated. Respective transgenes were expressed with an elav-driver; control was w1118. β-tubulin was used as loading control.( E) Semi-quantitative assessments of genetic modifier effects on L116-induced eye phenotypes (N ≥ 12).( F) Viability of animals expressing hCSPα-L115 or -L116 from two transgenes driven by elav-Gal4 in a control (w1118) or a heterozygous hsc4∆356 deletion background (N ≥ 3; n > 74). (G-K) Effects of reduced hsc4 gene dosage on levels of hCSPα oligomers (G), lipidated monomers (H), non-lipidated monomers (I) and total protein levels (J). Signals were normalized to loading control and plotted as n-fold change to levels induced by elav-driven expression of hCSP-L115 or -L116 in a WT control background (N > 6). (K) Effect of reduced hsc4 gene dosage on lipidated WT hCSPα monomer levels (N = 4). (L) Effects of reduced hsc4 gene dosage on endosomal accumulations of hCSP-L115/L116 in larval VNC (N ≥ 4). (M–N) Effects of reduced hsc4 gene dosage on ubiquitinated protein levels in larval VNCs (M, N = 5) and synaptic hCSP-L115/L116 expression levels at larval NMJs (N, N = 6). Signals were normalized and plotted as n-fold change from levels of elav-L115/L116 expression in WT control background. (O) Motor neuron somata of larval VNCs co-expressing HA-tagged Hsc4 with mutant hCSPα-L115 or -L116 stained for HA and hCSPα. Scale bars: 20 μm (K), 15 μm (M). Graphs display mean ± SEM. Statistical analysis used an unpaired t test (F, M), paired t test (L, N), or one-way ANOVA (E, G–J); *, p<0.05; **, p<0.01; ***, p<0.001.
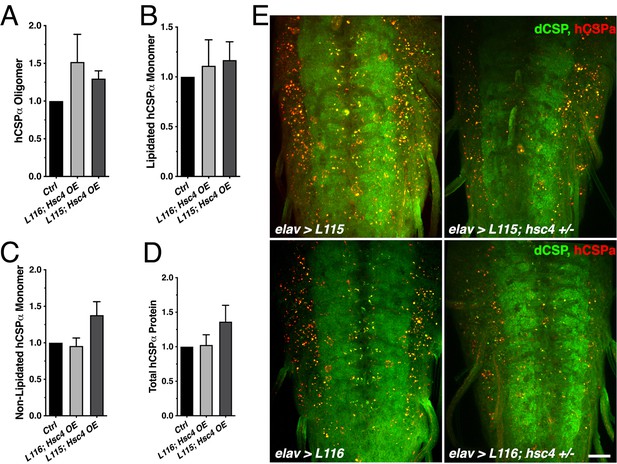
Effects of altered hsc4 gene dosage on CLN4 mutant hCSPα protein expression and localization.
(A-D) Effects of Hsc4 overexpression (OE) on hCSP-L115 and -L116 oligomers (A, N ≥ 8), lipidated monomers (B, N ≥ 6), non-lipidated monomers (C, N = 8), and overall protein levels (D, N = 8). Signals were normalized to loading control and plotted as n-fold change from L115/L116 levels (control) when expressed in a WT background (N = 5). (E) Larval VNCs for indicated genotypes immunostained for dCSP (green) and hCSPα (red). hCSP-L115 and -L116 were expressed with an elav driver from one transgene in WT control (white1118) or heterozygous hsc4∆356 deletion mutants. Scale bar, 20 μm. Graphs display mean ± SEM. Statistical analysis used RM one-way ANOVA (A–D); *, p<0.05; **, p<0.01; ***, p<0.001.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Drosophila melanogaster) | csp | Flybase | FLYB:FBgn0004179 | Cysteine string protein |
| Gene (human) | DNAJC5 | NCBI | GeneID:80331 | Encodes CSPα |
| Transcript (Drosophila melanogaster) | dCSP-2/CSP-PC | NCBI Genbank | NM_168950.4 | Reference Sequence used for synthesis of dCSP constructs, ORF: 186–872 |
| Recombinant DNA reagent | pBID-UASC | Wang et al., 2012 | RRID:Addgene_35200 | PhiC31 attB 10xUAS vector for site specific recombinase insertions |
| Recombinant DNA reagent | pUC57-dCSP2.WT | Genscript; This paper | Full length wildtype dCSP2 ORF with Kozak sequence and NotI/KpnI restrictions sites cloned into pUC57 | |
| Recombinant DNA reagent | pUC57-dCSP2.V117R | Genscript; This paper | As above with V117R mutation | |
| Recombinant DNA reagent | pUC57-dCSP2.I118Δ | Genscript; This paper | As above with I118Δ mutation | |
| Recombinant DNA reagent | PGEX-KG_rCSPα | Zhang and Chandra, 2014 | Wildtype rat CSPα cDNA sequence with V112F point mutation to match human aa sequence | |
| Recombinant DNA reagent | PGEX-KG_hCSPα.L115R | Zhang and Chandra, 2014 | As above with L115R mutation | |
| Recombinant DNA reagent | PGEX-KG_hCSPα.L116Δ | Zhang and Chandra, 2014 | As above with L116Δ mutation | |
| Recombinant DNA reagent | pBID-UASC_CSPα.WT | This study | full length wildtype ORF encoding rat/human CSPα cloned into pBID-UASC transformation vector | |
| Recombinant DNA reagent | pBID-UASC_CSPα.L115R | This study | As above with L115R mutation | |
| Recombinant DNA reagent | pBID-UASC_CSPα.L116Δ | This study | As above with L116Δ mutation | |
| Recombinant DNA reagent | pBID-UASC_dCSP2.WT | This study | Full length wildtype ORF encoding dCSP2 cloned into pBID-UASC transformation vector | |
| Recombinant DNA reagent | pBID-UASC_dCSP2.V117R | This study | As above with V117R mutation | |
| Recombinant DNA reagent | pBID-UASC_dCSP2.I118Δ | This study | As above with I118Δ mutation | |
| Sequenced-based reagent | rCSP NotI for | Sigma | 5’-GAGCGGCCGCCAAAATGGCTGACCAGAGGCAGCGCTC-3’ | |
| Sequenced- based reagent | rCSP KpnI rev | Sigma | 5’-CATGGTACCTTAGTTGAACCCGTCGGTGTGATAGCTGG-3’ | |
| Genetic reagent (D. melanogaster) | w[1118] | Caltech collection: Seymour Benzer, Caltech | FlyB:FBal0018186 | Genotype: w[1118]; isogenized genetic background |
| Genetic reagent (D. melanogaster) | elav::Gal4[C155] | Bloomington Drosophila Stock Center | FlyB:FBst0000458; RRID:BDSC_458 | Genotype: P{w[+mW.hs]=GawB}elav[C155] |
| Genetic reagent (D. melanogaster) | nSyb::Gal4 | Hugo Bellen, Baylor College of Medicine | FlyB:FBst0051635; RRID:BDSC_51635 | Genotype: y[1] w[*]; P{w[+m*]=nSyb-GAL4.S}3 |
| Genetic reagent (D. melanogaster) | GMR::Gal4 | Bloomington Drosophila Stock Center | FlyB:FBst0001104; RRID:BDSC_1104 | Genotype: w[*]; P{w[+mC]=GAL4 ninaE.GMR}12 |
| Genetic reagent (D. melanogaster) | UAS::nsyb-EGFP | Bloomington Drosophila Stock Center | FlyB:FBst0006921; RRID:BDSC_6921 | Genotype: w[*]; P{w[+mC]=UAS nSyb.eGFP}2 |
| Genetic reagent (D. melanogaster) | UAS::GFP-myc-2xFYVE | Bloomington Drosophila Stock Center | FlyB:FBst0042712; RRID:BDSC_42712 | Genotype: w[*]; P{w[+mC]=UAS-GFP-myc-2xFYVE}2 |
| Genetic reagent (D. melanogaster) | UAS::TSG101 RNAi | Bloomington Drosophila Stock Center | FlyB:FBst0035710; RRID:BDSC_35710 | Genotype: y[1] sc[*] v[1] sev[21]; P{y[+t7.7] v[+t1.8]=TRiP.GLV21075}attP2 |
| Genetic reagent (D. melanogaster) | UAS:GFP-KDEL | Bloomington Drosophila Stock Center | FlyB:FBst0009898; RRID:BDSC_9898 | Genotype: w[*]; P{w[+mC]=UAS GFP.KDEL}11.1 |
| Genetic reagent (D. melanogaster) | UAS::hLAMP1-GFP | Bloomington Drosophila Stock Center | FlyB:FBti0150347; RRID:BDSC_42714 | Genotype: w[*]; P{w[+mC]=UAS-GFP-LAMP}2; P{w[+m*]=nSyb-GAL4.S}3/T(2;3)TSTL, CyO: TM6B, Tb[1] |
| Genetic reagent (D. melanogaster) | UAS::GFP-Rab5 | Bloomington Drosophila Stock Center | FlyB:FBst0043336; RRID:BDSC_43336 | Genotype: w[*]; P{w[+mC]=UAS-GFP-Rab5}3 |
| Genetic reagent (D. melanogaster) | UAS::spin-myc.EGFP | Bloomington Drosophila Stock Center | FlyB:FBst0039668; RRID:BDSC_39668 | Genotype: w[*]; P{w[+mC]=UAS spin.myc-EGFP}B |
| Genetic reagent (D. melanogaster) | UAS::GFP-LC3(ATG8) | Bloomington Drosophila Stock Center | FlyB:FBst0008730; RRID:BDSC_8730 | Genotype: w[*]; P{w[+mC]=UASp-eGFP-huLC3}1; P{w[+mC]=GAL4::VP16-nos.UTR}CG6325[MVD1] |
| Genetic reagent (D. melanogaster) | UAS::syx-1A | Bloomington Drosophila Stock Center | FlyB:FBst0051619; RRID:BDSC_51619 | Genotype: y[1] w[*]; P{w[+mC]=UAS-Syx1A.B}6 |
| Genetic reagent (D. melanogaster) | UAS::hsc70-4 KD1 | Bloomington Drosophila Stock Center | FlyB:FBst0054810; RRID:BDSC_54810 | Genotype: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.HMJ21529}attP40 |
| Genetic reagent (D. melanogaster) | UAS::hsc70-4 KD2 | Bloomington Drosophila Stock Center | FlyB:FBst0028709; RRID:BDSC_28709 | Genotype: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF03136}attP2 |
| Genetic reagent (D. melanogaster) | UAS::hsc70-4 KD3 | Bloomington Drosophila Stock Center | FlyB:FBst0034836; RRID:BDSC_34836 | Genotype: y[1] sc[*] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.HMS00152}attP2/TM3, Sb[1] |
| Genetic reagent (D. melanogaster) | attP-22A | Bloomington Drosophila Stock Center | FlyB:FBst0024481; RRID:BDSC_24481 | Genotype: y[1] M{vas-int.Dm}ZH-2A w[*]; M{3xP3-RFP.attP'}ZH-22A PhiC31 Insertion background, all generated lines outcrossed into w[1118] |
| Genetic reagent (D. melanogaster) | UAS::venus-Rab7.WT | R Hisinger (Freie Universität, Berlin, Germany); Cherry et al., 2013 | FlyB:FBal0294208 | |
| Genetic reagent (D. melanogaster) | hsc70-4[Δ356] | Bronk et al., 2001 | FlyB:FBal0124174 | Genotype: w[1118];;hsc70-4[Δ356] |
| Genetic reagent (D. melanogaster) | UAS::Hsc70-4 | Karen Palter, Temple University; Elefant and Palter, 1999, | FlyB:FBst0005846; RRID:BDSC_5846 | Genotype: w[126]; P{w[+mC]=UAS-Hsc70-4.WT}B |
| Genetic reagent (D. melanogaster) | UAS:HA-Hsc70-4.WT | P Verstreken; Uytterhoeven et al., 2015 | FlyB:FBal0318413 | Genotype: w[1118]; P{w[+mC]=UAS-Hsc70-4.HA} |
| Genetic reagent (D. melanogaster) | UAS::SNAP25 | Bloomington Drosophila Stock Center | FlyB:FBst0051997; RRID:BDSC_51997 | Genotype: y[1] w[*]; P{w[+mC]=UAS-Snap25.L}9 |
| Genetic reagent (D. melanogaster) | UAS::SNAP25 KD | Bloomington Drosophila Stock Center | FlyB:FBst0027306; RRID:BDSC_27306 | Genotype: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF02615}attP2 |
| Genetic reagent (D. melanogaster) | csp[R1] | Zinsmaier et al., 1994 | FlyB:FBst0032035; RRID:BDSC_32035 | Genotype: w[1118]; csp[X1]/TM6Tb[1]Sb[1]; partial deletion of csp (genetic null) |
| Genetic reagent (D. melanogaster) | csp[X1] | Zinsmaier et al., 1994 | FlyB:FBst0051998; RRID:BDSC_51998; | Genotype: w[1118]; csp[R1]/TM6Tb[1]Sb[1]; complete deletion of csp locus, maintained in this lab |
| Genetic reagent (D. melanogaster) | UAS::dCSP2 | this study | Genotype: w[1118];M{UAS-dCSP2}ZH-22A | |
| Genetic reagent (D. melanogaster) | UAS::dCSP2.V117R | this study | Genotype: w[1118];M{UAS-dCSP2.V117R}ZH-22A | |
| Genetic reagent (D. melanogaster) | UAS::dCSP2.I118Δ | this study | Genotype: w[1118];M{UAS-dCSP2.I118Δ}ZH-22A | |
| Genetic reagent (D. melanogaster) | UAS::hCSPα | this study | Genotype: w[1118];M{UAS-hCSPα}ZH-22A | |
| Genetic reagent (D. melanogaster) | UAS::hCSPα.L115R | this study | Genotype: w[1118];M{UAS-hCSPα.L115R}ZH-22A | |
| Genetic reagent (D. melanogaster) | UAS::hCSPα.L116Δ | this study | v: w[1118];M{UAS-hCSPα.L116R}ZH-22A | |
| Antibody | anti-CSPα (rabbit polyclonal) | Enzo Life Sciences | Cat#: VAP-SV003E; RRID:AB_2095057 | Immunostaining (IS, 1:2000); Western Blot (WB, 1:20000); IP (1:250) |
| Antibody | anti-dCSP (mouse monoclonal) | Zinsmaier et al., 1990 | Cat# DSHB:DCSP-1 (ab49); RRID:AB_2307345 | IS (1:20); WB (1:250) |
| Antibody | anti-GFP (mouse monoclonal) | Developmental Studies Hybridoma Bank | Cat# DSHB: GFP-12A6; RRID:AB_2617417 | IS (1:1000) |
| Antibody | anti-HRS (guinea-pig polyclonal) | HJ Bellen, Baylor College of Medicine, Houston, TX; Lloyd et al., 2002 | IS (1:2000); WB (1:2000) | |
| Antibody | anti-HA (rat monoclonal) | Roche | Cat#: 3F10; RRID:AB_2314622 | IS (1:200) |
| Antibody | anti-GM130 (rabbit polyconal) | Abcam | Cat# ab31561, RRID:AB_2115328 | IS (1:200) |
| Antibody | anti-Golgin245 (goat polyclonal) | Developmental Studies Hybridoma Bank | Cat#: Golgin245; RRID:AB_2618260 | IS (1:2000) |
| Antibody | anti-GMAP (goat polyclonal) | Developmental Studies Hybridoma Bank | Cat#: GMAP; RRID:AB_2618259 | IS (1:2000) |
| Antibody | anti-Rab7 (mouse monoclonal) | Developmental Studies Hybridoma Bank | Cat#: Rab7; RRID:AB_2722471 | IS (1:100) |
| Antibody | anti-Syx1A (mouse monoclonal) | Developmental Studies Hybridoma Bank | Cat#: 8C3; RRID:AB_528484 | IS (1:200) |
| Antibody | anti-β-tubulin (mouse monoclonal) | Developmental Studies Hybridoma Bank | Cat#: E7; RRID:AB_528499 | WB (1:1000) |
| Antibody | anti-conjugated-ubiquitin (mouse monoclonal) | Enzo Life Sciences | Cat#: BML-PW8810; RRID:AB_10541840; Clone FK2 | IS (1:2000); WB (1:2000) |
| Antibody | anti-HRP Alexa Fluor 647-conjugated (goat polyclonal) | Jackson ImmunoResearch Labs | Cat#: 123-605-021; RRID:AB_2338967 | IS (1:500) |
| Antibody | anti-mouse IgG1 AlexaFluor 488-conjugated (goat polyclonal) | Thermo Fisher Scientific | Cat#: A-21121; RRID:AB_2535764 | IS (1:500) |
| Antibody | anti-rabbit IgG (H+L) Cy3-conjugated (donkey polyclonal) | Jackson ImmunoResearch Labs | Cat#: 711-165-152; RRID:AB_2307443 | IS (1:500) |
| Antibody | anti-guinea pig IgG (H+L) Alexa Fluor 488-conjugated (goat polyclonal) | Thermo Fisher Scientific | Cat#: A-1107; RRID:AB_2534117 | IS (1:1000) |
| Antibody | anti-rat IgG (H+L) Alexa Fluor 488-conjugated (goat polyclonal) | Jackson ImmunoResearch Labs | Cat#: 112-545-167; RRID:AB_2338362 | IS (1:500) |
| Antibody | anti-rabbit IgG (H+L) HRP-conjugated (goat polyclonal) | Thermo Fisher Scientific | Cat#: A16096; RRID:AB_2534770 | WB (1:10000) |
| Antibody | anti-mouse IgG HRP-conjugated (goat polyclonal) | Thermo Fisher Scientific | Cat#: 32430; RRID:AB_1185566 | WB (1:5000) |
| Commercial assay or kit | Western Clarity ECL kit | BioRad | Cat#: 1705061 | |
| Commercial assay or kit | A/G PLUS-Agarose Beads | Santa Cruz Biotechnology | Cat#: sc-2003 | |
| Software, algorithm | FIJI/ImageJ | NIH | RRID:SCR_002285 | |
| Software, algorithm | Prism | Graphpad | RRID:SCR_002798 | |
| Software, algorithm | Adobe Photoshop CC | Adobe | RRID:SCR_014199 | |
| Software, algorithm | Quantity One 1-D Analysis Software | BioRad | RRID:SCR_014280 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.46607.016





