Critical roles of ARHGAP36 as a signal transduction mediator of Shh pathway in lateral motor columnar specification
Figures
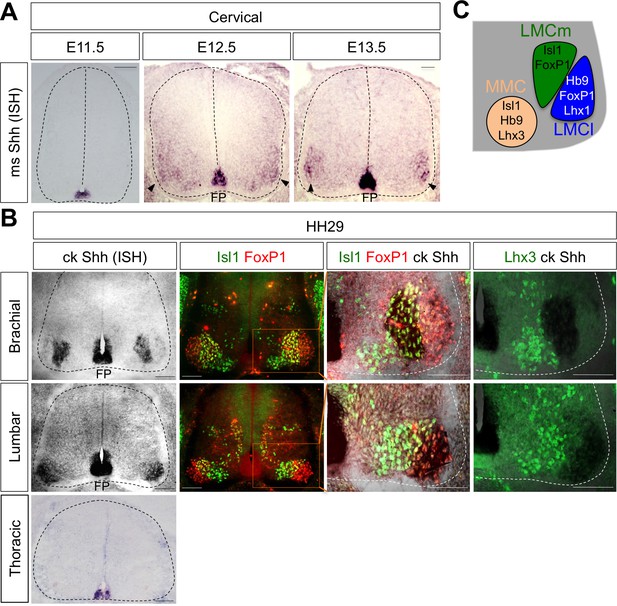
Shh is expressed in LMC neurons in developing mouse and chick spinal cord.
(A) ISH analysis of mouse spinal cord (dotted outline) showed the expression of Shh in FP and LMC region (arrowhead) at later stages when motor columns are specified. (B) ISH analysis of chick Shh combining with IHC of Isl1/FoxP1 and Lhx3 in chick spinal cord. At HH St.29, Shh is mainly detected in LMCm (Isl1+/FoxP1+) neurons at brachial levels and LMCl (Isl1-/FoxP1+) neurons at lumbar levels but not in motor neurons at thoracic levels of chick spinal cord (dotted outline). Scale bars: 100 μm. (C) Schematic drawing shows the LMCm, LMCl, and MMC motor columns in the ventral spinal cord with representative markers.
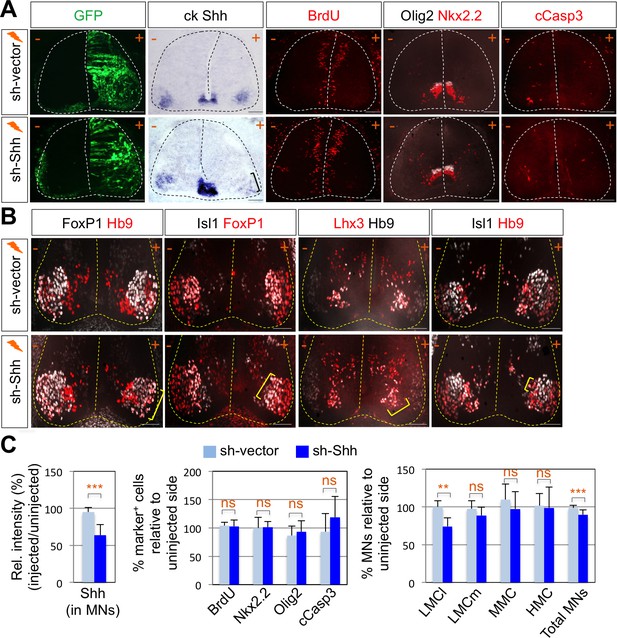
Shh signaling is required for LMC specification in chick spinal cord.
(A) IHC analyses in chick spinal cords electroporated with sh-Shh and sh-vector construct. ISH analysis showed the reduced expression of chick Shh in sh-Shh electroporated chick embryo but not in control sh-vector injected chick embryo. Knock-down of chick Shh did not affect the proliferation (BrdU) or survival (cCasp3) of neural progenitor cells and ventral neural patterning (Olig2 and Nkx2.2). (B) Knock-down of chick Shh reduced the number of LMCl (Hb9+/FoxP1+) neurons but had no effect on other motor columns such as LMCm, MMC, and HMC and consequently reduced the number of total MNs compared to the uninjected control side. +, electroporated side; -, non-electroporated control side. (C) Quantification of the relative intensity of Shh ISH signal in motor neurons, % marker+ (BrdU, Nkx2.2, Olig2, and cCasp3) cells relative to uninjected side and % motor columns relative to uninjected side of the spinal cord. Each set of chick electroporation experiments in this figure was repeated independently at least three times with 6 to 10 embryos. Embryos were harvested 4 days post electroporation (dpe). Data are mean ± s.d. **p<0.001, ***p<0.0001; ns, non-significant (Student’s t-test). n = 6 ~ 15 independent images per each sample. Scale bars: 100 μm.
-
Figure 2—source data 1
Source data for Figure 2C.
- https://doi.org/10.7554/eLife.46683.004
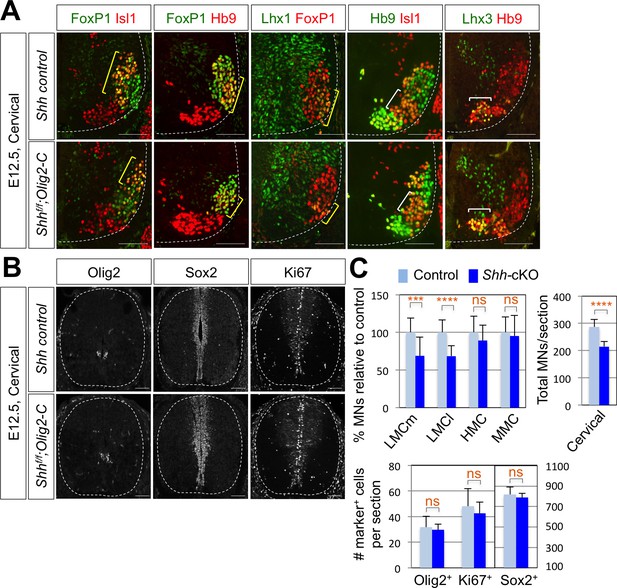
Shh is required for LMC formation in developing mouse spinal cord.
(A) IHC analyses of E12.5 Shh-cKO (Shhf/f;Olig2-Cre) mutant embryos (n = 4) (lower panel) and control littermates (n = 4) (upper panel). The cervical level of ventral spinal cord is shown. LMCm (Isl1+/FoxP1+) neurons and LMCl (Hb9+/FoxP1+ or Lhx1+/FoxP1+) neurons (yellow bracket) in Shh conditional knock-out (Shh-cKO) were significantly reduced. On the other hand, the number of MMC (Hb9+/Lhx3+) and HMC (Hb9+/Isl1+) neurons did not change (white bracket). (B) IHC analyses of Olig2, Sox2, and Ki67 in E12.5 Shh-cKO mutant embryo and control littermates (cervical level). No significant difference in the expression of Sox2, Olig2 and Ki67 within the spinal cord. Scale bars: 100 μm. (C) Quantification of the number of LMCm (Isl1+/FoxP1+), LMCl (Hb9+/FoxP1+ or Lhx1+/FoxP1+), MMC (Hb9+/Lhx3+) and HMC (Hb9+/Isl1+) neurons, Olig2+, Sox2+, Ki67+ cells and total MNs at cervical level in E12.5 mouse embryonic spinal cord. Data are mean ± s.d. ***p<0.0001, ****p<0.00001; ns, non-significant (Student’s t-test). n = 5 ~ 28 independent images per each sample.
-
Figure 3—source data 1
Source data for Figure 3C.
- https://doi.org/10.7554/eLife.46683.008
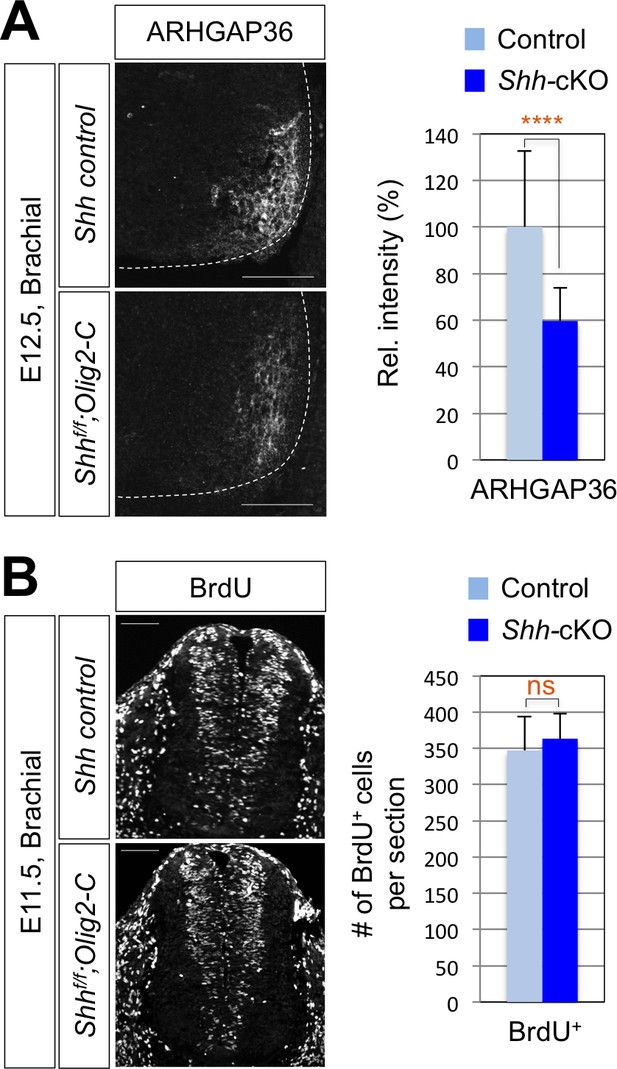
Reduced expression of ARHGAP3 does not affect progenitor cell proliferation in Shh-cKO mouse spinal cord.
(A) IHC analysis for ARHGAP36 in E12.5 Shh-cKO (n = 4) and littermate control (n = 4) embryos showing the reduced expression of ARHGAP36 in ventro-lateral region of the spinal cord at cervical level. (B) IHC analysis for BrdU incorporation in E11.5 Shh-cKO (n = 4) and littermate control (n = 5) embryos showing no defect in proliferation of progenitor cells within the spinal cord. Scale bars: 100 μm. Data are mean ± s.d. ****p<0.00001; ns, non-significant (Student's t-test). n = 14 ~ 32 independent images per each sample.
-
Figure 3—figure supplement 1—source data 1
Source data for Figure 3—figure supplement 1.
- https://doi.org/10.7554/eLife.46683.007
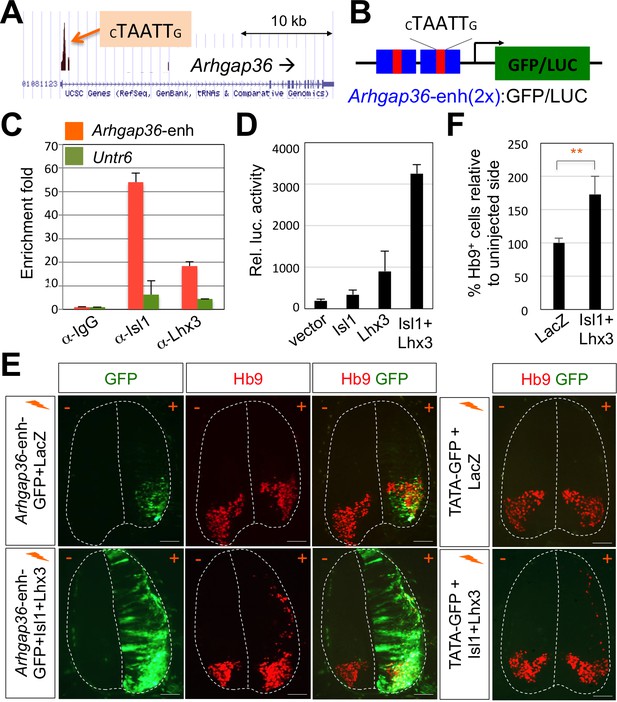
ChIP-seq peaks for the Isl1-Lhx3 complex in Arhgap36 and their in vivo recruitment of the Isl1-Lhx3 complex.
(A) Isl1-Lhx3 complex binding sites in Arhgap36. The peak has HxRE motif. (B) A schematic representation of reporter constructs linked to two copies of Arhgap36-enhancer genomic DNA fragment. (C) Both Isl1 and Lhx3 were recruited to Isl1-Lhx3-bound ChIP-seq peak in Arhgap36 gene. ChIP was performed with anti-IgG antibody (control), anti-Isl1 and anti-Lhx3 antibodies using E12.5 mouse embryonic spinal cord extracts. Quantitative PCR amplification of the binding region of Arhgap36 and negative control region, Untr6. ChIP experiments were repeated independently twice. Data are presented as the mean of duplicate values and error bars represent standard deviation. (D) Luciferase assay for a reporter directed by two copies of Arhgap36-enhancer. Transfections were repeated independently at least three times. Data are presented as the mean of triplicate values and error bars represent standard deviation. (E) In ovo electroporation of LacZ (to measure electroporation efficiency) and a GFP reporter directed by two copies of Arhgap36-enhancer without or with co-expression of Isl1 and Lhx3. TATA-GFP vector with no HxRE was used as a negative control and this reporter was not activated even when Isl1 +Lhx3 expression induces ectopic MNs in dorsal spinal cord. Each set of DNA was injected and electroporated in chick neural tube and embryos (n = 5 ~ 10) were harvested 3 days post electroporation (three dpe). Hb9 staining labels endogenous and ectopically induced motor neurons in the spinal cord. +, electroporated side, –, non-electroporated side. White dotted lines indicate the outline of the spinal cord. Experiments were repeated independently at least three times. Scale bars: 100 μm. (F) Quantification of the number of Hb9+ cells relative to uninjected side of the spinal cord. Data are mean ± s.d. **p<0.001 (Student’s t-test). n = 5 ~ 8 independent images per each sample.
-
Figure 4—source data 1
Source data for Figure 4C.
- https://doi.org/10.7554/eLife.46683.010
-
Figure 4—source data 2
Source data for Figure 4D.
- https://doi.org/10.7554/eLife.46683.011
-
Figure 4—source data 3
Source data for Figure 4F.
- https://doi.org/10.7554/eLife.46683.012
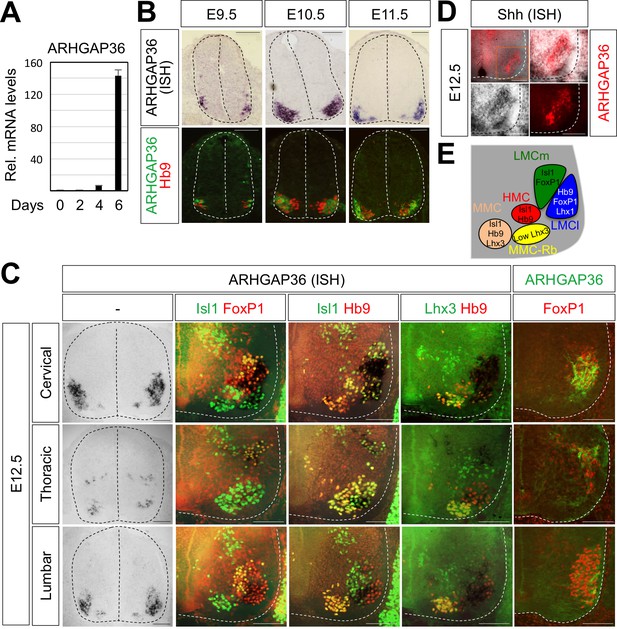
Expression of ARHGAP36 in the developing spinal MNs.
(A) Induction of ARHGAP36 expression in MNs differentiated from mouse ESCs was determined by qRT-PCR. Relative expression levels are shown as the mean of duplicate values obtained from representative experiments. Error bars represent standard deviation. (B,C) ARHGAP36 was specifically expressed in MNs of mouse embryos at E9.5, E10.5, E11.5 and E12.5 stages, as shown by ISH with a probe detecting ARHGAP36 and IHC for ARHGAP36, Isl1/FoxP1, Isl1/Hb9, Lhx3/Hb9 and FoxP1. From E12.5 and onward, ARHGAP36 expression was highly enriched in LMCl (Isl1-/FoxP1+) region, some in MMC-rhomboideus (Rb) (Hb9+/Lhx3low) and a very little in the most medial part of MMC but not in LMCm (Isl1+/FoxP1+) at cervical level. ARHGAP36 is also expressed in PGC (FoxP1+/Isl1+) and HMC (Isl1+/Hb9+) neurons at thoracic level but with relatively lower expression compared to the cervical level. At lumbar level, ARHGAP36 is enriched in LMCl (Isl1-/FoxP1+) of the spinal cord. Scale bars: 100 μm. (D) Co-localization of ARHGAP36 with Shh shown by ISH of Shh and IHC of ARHGAP36 in mouse E12.5 spinal cord at cervical level. Shh is co-localized with ARHGAP36 mostly in LMCl region in mouse spinal cord. Scale bars: 100 μm. (E) Schematic drawing shows the LMCm, LMCl, HMC, MMC and MMC-rhomboideus (Rb) motor columns in the ventral spinal cord with representative markers.
-
Figure 5—source data 1
Source data for Figure 5A.
- https://doi.org/10.7554/eLife.46683.014
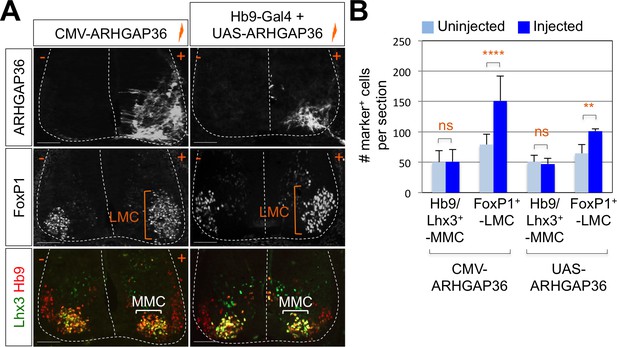
Expression of ARHGAP36 promotes LMC specification in developing chick spinal cord.
(A) ARHGAP36 constructs were injected and electroporated in chick neural tube and embryos (n = 8 ~ 15) were harvested 4 days post electroporation (four dpe). Ectopic expression of ARHGAP36 driven by CMV promoter in most injected cells induced robust expression of FoxP1+ LMC neurons (orange bracket) in ventral spinal cord but had no effect on MMC (Hb9+/Lhx3+) neurons (white bracket). Targeting the expression of ARHGAP36 specifically in motor neurons using Hb9-Gal4/UAS-ARHGAP36 system also lead to the robust induction of FoxP1+ LMC neurons (orange bracket) but had no effect on MMC (Hb9+/Lhx3+) neurons (white bracket). +, electroporated side; -, non-electroporated control side. Experiments were repeated independently at least three times. Scale bars: 100 μm. (B) Quantification of the number of FoxP1+ neurons and MMC (Hb9+/Lhx3+) neurons on the electroporated (+) and non-electroporated (-) sides of the spinal cord. Data are mean ± s.d. **p<0.001, ****p<0.00001; ns, non-significant (Student’s t-test). n = 6 ~ 20 independent images per each sample.
-
Figure 6—source data 1
Source data for Figure 6B.
- https://doi.org/10.7554/eLife.46683.019
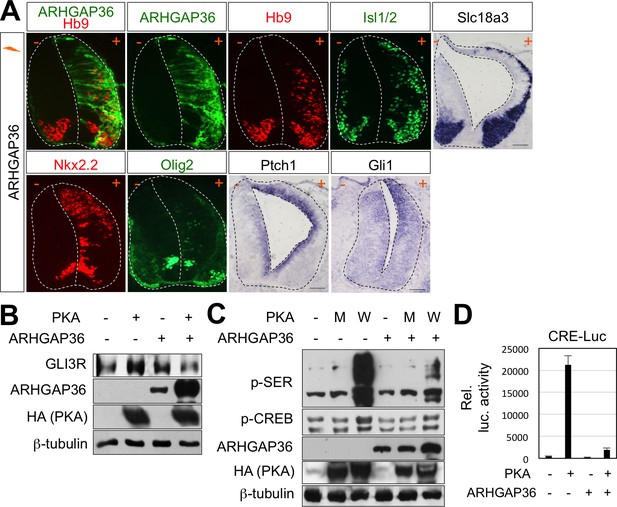
Activation of Shh pathway by ARHGAP36 expression in spinal cord.
(A) In ovo electroporation of ARHGAP36 construct. ARHGAP36 construct was injected and electroporated in chick neural tube and embryos (n = 8 ~ 12) were harvested 3 days post electroporation (three dpe). Ectopic expression of ARHGAP36 induced expression of ventral spinal neuronal genes including motor neuron genes (Hb9, Isl1/2, Slc18a3), ventral progenitor genes (Nkx2.2 and Olig2) and downstream target genes of Shh pathway (Ptch1 and Gli1) as shown by ISH and IHC. +, electroporated side; -, non-electroporated control side. Experiments were repeated independently at least three times. Scale bars: 100 μm. (B) Western blot analysis showed that ARHGAP36 inhibits PKA activity. PKA expression resulted in producing Gli3R, repressor form of Gli3 (lane 2) in HEK293T cells. Co-expression of ARHGAP36 blocks the formation of Gli3R by PKA (lane 4). β-tubulin was used as a loading control. (C) ARHGAP36 inhibited the level of phosphorylation of putative PKA substrates shown by western blot with anti-p-SER antibody and the level of phospho-CREB (p-CREB), a known PKA target in NIH3T3 cells. W indicates PKA wild type and M indicates PKA kinase dead mutant (K73H). β-tubulin was used as a loading control. (D) Luciferase reporter assay in HEK293T cells. The activation of CRE-luc is directed by PKA-phosphorylated CREB and this activity was blunted by co-expression of ARHGAP36, indicating that ARHGAP36 inhibits the kinase activity of PKA. Data are mean ± s.d.
-
Figure 6—figure supplement 1—source data 1
Source data for Figure 6—figure supplement 1D.
- https://doi.org/10.7554/eLife.46683.017
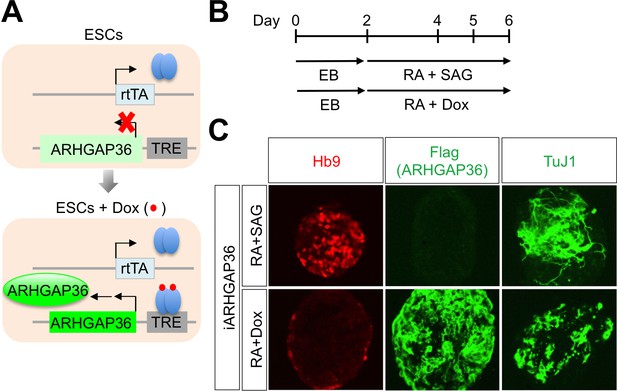
ARHGAP36 is not sufficient to induce MN differentiation in mESCs.
(A) Schematic representation of inducible ARHGAP36-ESCs before and after treatment with Dox (Doxycycline). TRE, tetracycline response element; rtTA, reverse tetracycline transactivator. (B) Illustration of experimental design to differentiate ESCs to MNs. RA and SAG or RA and Dox were treated at day two and EBs were harvested at day six for further analyses. EB, embryoid body; RA, retinoic acid; SAG, Smoothened agonist. (C) IHC analyses in ESC-derived MNs cultured with RA +SAG or RA +Dox at differentiation day 6. Hb9 labels MNs and it was induced only by RA +SAG. Treatment of RA +Dox induced ARHGAP36 and pan-neuronal marker TuJ1, but not MN marker Hb9.
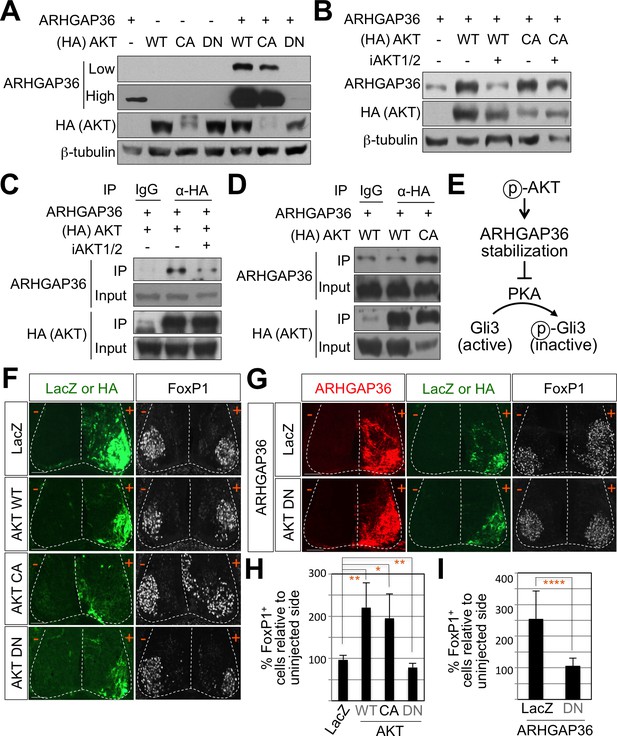
AKT potentiates Shh signaling through stabilization of ARHGAP36 proteins and AKT-ARHGAP36 axis is required for LMC specification.
(A) ARHGAP36 was stabilized dramatically by AKT WT and CA, but not by DN in HEK293T cells. ARHGAP36 was transiently transfected with AKT constructs in HEK293T cells and the protein levels were monitored by western blotting. β-tubulin was used as a loading control. (B) 10 μM of AKT inhibitor (iAKT1/2) was treated for 20 hr and the protein level of ARHGAP36 was monitored. AKT inhibitor reversed the effect of AKT WT in stabilizing ARHGAP36 protein but had no effect on constitutively active form of AKT. (C) Co-immunoprecipitation assay with HEK293T cells transiently transfected with the expression vectors for HA-tagged AKT and ARHGAP36 showed that AKT WT co-purified ARHGAP36, and this interaction was decreased by iAKT1/2, the AKT inhibitor. (D) The CA form of AKT interacted with ARHGAP36 more robustly than AKT WT. ARHGAP36 with either HA-tagged AKT WT or AKT CA was transfected into HEK293T cells and immunoprecipitated with anti-HA antibody that pull-downs AKT. Anti-IgG antibody was used as a negative control. (E) Illustration of the modulatory pathway showing that activated AKT stabilizes ARHGAP36 proteins, which in turn blocks the kinase activity of PKA, which results in Gli-dependent transcriptional activation via dephosphorylation of Gli. (F) IHC analyses in the chick neural tube electroporated with AKT WT, CA and DN. Embryos (n = 8–10) were harvested 4dpe. AKT WT or CA increased the number of FoxP1+ cells by almost two fold in the electroporated side (+) compared to the non-electroporated control side (-). Experiments were repeated independently at least three times. Scale bars: 100 μm. (G) The analysis of ectopic FoxP1+ neuron formation by ARHGAP36 in the presence of either AKT DN or LacZ in the chick neural tube. Embryos (n = 8–10) were harvested 4dpe. +, electroporated side; -, non-electroporated control side. AKT DN completely blocked the effect of ARHGAP36 in inducing ectopic FoxP1 expression in the electroporated cells. Experiments were repeated independently at least three times. Scale bars: 100 μm. (H,I) Quantification of the number of FoxP1+ neurons on the electroporated (+) and non-electroporated (-) sides of the spinal cord. Data are mean ± s.d. *p<0.01, **p<0.001, ****p<0.00001 (Student’s t-test). n = 6 ~ 27 independent images per each sample.
-
Figure 7—source data 1
Source data for Figure 7H and 7I.
- https://doi.org/10.7554/eLife.46683.025
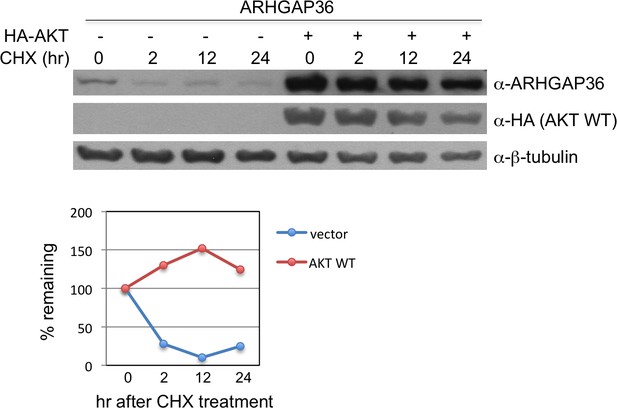
AKT stabilizes protein level of ARHGAP36.
Measurement of half-life of ARHGAP36 in HEK293T cells transfected with empty vector or vector encoding AKT WT, following treatment with 200 ng/ml of the protein synthesis inhibitor cycloheximide (CHX). The half-life of ARHGAP36 was roughly estimated to ~1 hr for vector alone, while it was not degraded within 24 hr for AKT WT. These experiments were repeated multiple times, and we obtained similar results.
-
Figure 7—figure supplement 1—source data 1
Source data for Figure 7—figure supplement 1.
- https://doi.org/10.7554/eLife.46683.022
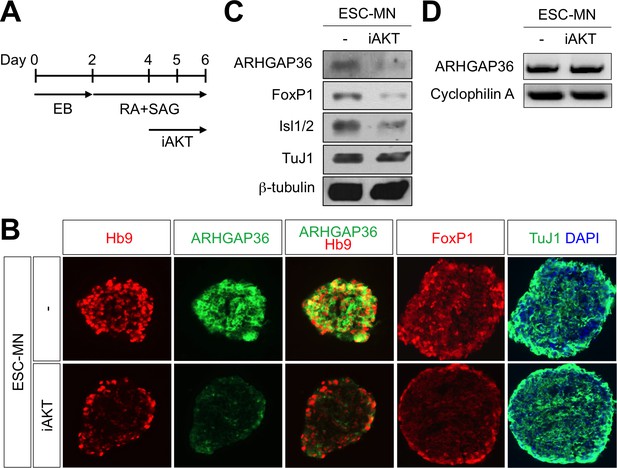
AKT inhibition blocks MN differentiation from mouse ESCs.
(A) Illustration of experimental design to differentiate ESCs to MNs. RA and SAG were treated at day two and AKT inhibitor was treated from day four and EBs were harvested at day six for further analyses. EB, embryoid body; RA, retinoic acid; SAG, Smoothened agonist. (B) IHC analyses in ESC-derived MNs cultured with RA +SAG with or without iAKT at differentiation day 6. Hb9 labels MNs. ARHGAP36 was induced in MN differentiation condition. Treatment of iAKT reduced expression of ARHGAP36, FoxP1 and Hb9 but not pan-neuronal marker TuJ. (C) Western blotting analysis showed reduced expression of ARHGAP36, FoxP1 and Isl1/2 but not pan-neuronal marker TuJ by AKT inhibitor treatment. (D) RT-PCR analysis revealed that ARHGAP36 mRNA level was not affected by AKT inhibition. Cyclophilin A was used as a loading control.
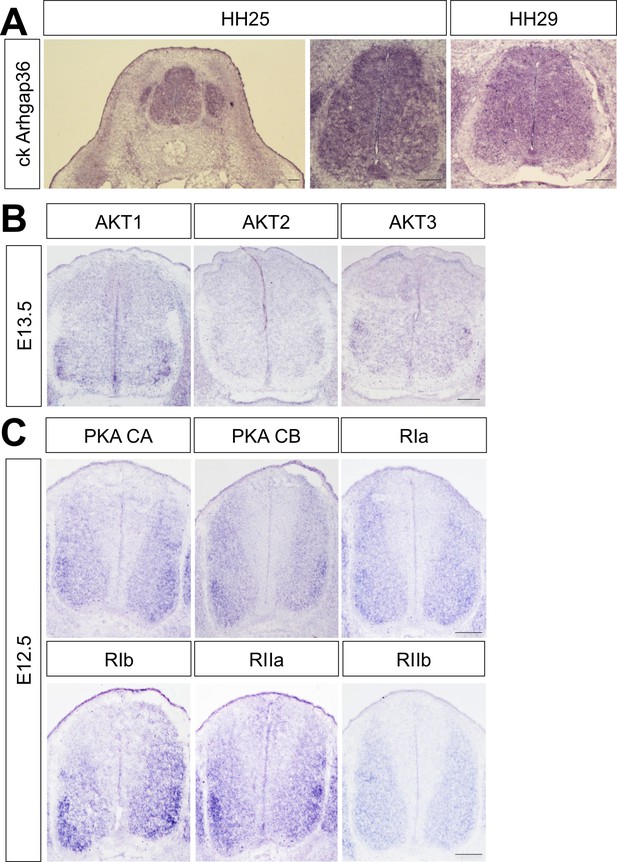
Expression of Arhgap36 in chick spinal cord and AKTs and PKAs in developing mouse spinal cord.
(A) ISH for Arghap36 in HH25 and HH29 chick embryos showing its broad expression in the spinal cord at brachial level. (B) ISH for AKT1, AKT2 and AKT3 in E13.5 mouse embryos showing their expression in ventro-lateral region of the spinal cord at cervical level. (C) ISH for the PKA catalytic subunits CA and CB and the regulatory subunits RIa, RIb, RIIa and RIIb in E12.5 mouse embryos showing their expression in ventro-lateral region of the spinal cord at cervical level. Scale bars: 100 μm.
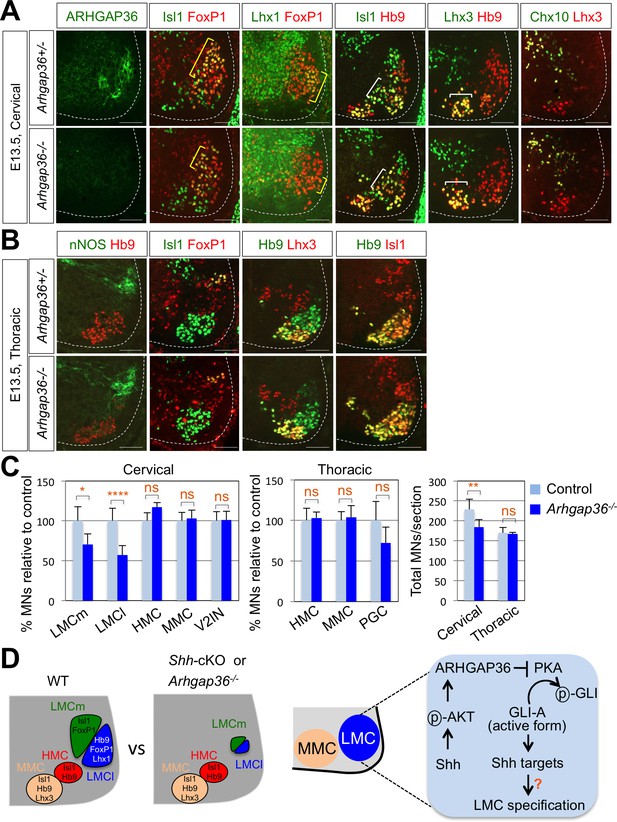
ARHGAP36 is required for LMC formation in mice.
(A) IHC analyses of E13.5 Arhgap36-/- mutant embryo (n = 4) (lower panel) and their littermate controls (n = 5) (upper panel). Ventrolateral quadrants of the cervical level of spinal cord are shown in all panels. IHC with anti-ARHGAP36 antibody confirms the absence of ARHGAP36 expression in Arhgap36-null embryos. LMCm (Isl1+/FoxP1+) and LMCl (Lhx1+/FoxP1+) neurons were significantly reduced in Arhgap36-/-. On the other hand, the numbers of MMC (Hb9+/Lhx3+), HMC (Isl1+/Hb9+) neurons and V2-INs (Lhx3+/Chx10+) did not change. Scale bars: 100 μm. (B) At thoracic level, there was no difference in PGC (nNOS+ or Isl1+/FoxP1+), HMC (Hb9+/Isl1+), and MMC (Hb9+/Lhx3+) neurons compared to control littermates. Scale bars: 100 μm. (C) Quantification of the number of LMCm (Isl1+/FoxP1+), LMCl (Lhx1+/FoxP1+), MMC (Hb9+/Lhx3+), HMC (Isl1+/Hb9+) and V2-INs (Lhx3+/Chx10+) at cervical level in E13.5 mouse embryonic spinal cord. Data are mean ± s.d. *p<0.01, **p<0.001, ****p<0.00001; ns, non-significant; n = 6 ~ 12 independent images per each sample. (D) Proposed model. In Shh-cKO or Arhgap36-/- mutant embryos, LMCm and LMCl neurons are reduced with no expansion of other motor columns, and thus this results in the reduction of total MNs compared to WT control. AKT, activated in response to Shh, stabilizes ARHGAP36 protein, which in turn inhibits the kinase activity of PKA. This results in Gli-dependent transcriptional activation and LMC formation in MNs at cervical level of the spinal cord.
-
Figure 8—source data 1
Source data for Figure 8C.
- https://doi.org/10.7554/eLife.46683.031
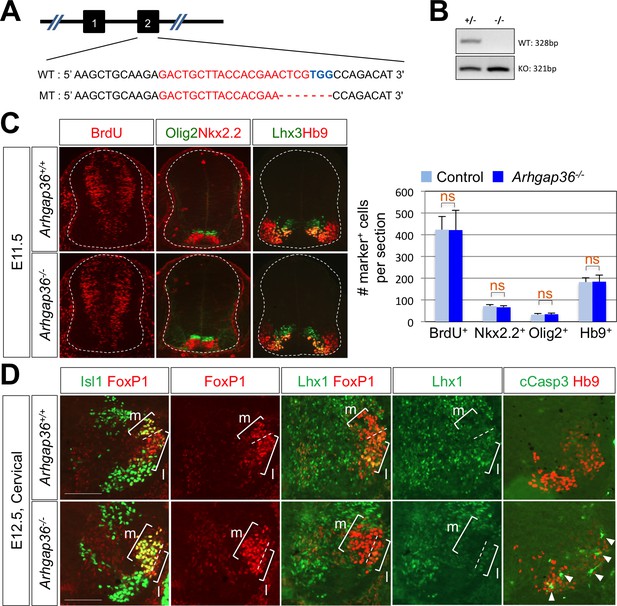
ARHGAP36 is required for proper LMC formation in mice.
(A) Schematic showing the CRISPR/Cas9-mediated ARHGAP36 knockout mouse generation. Sequences for target exon2 of WT allele (upper) and ARHGAP36 KO allele (lower). The target sequence of sgRNA is marked in red, and the PAM sequence is in blue. (B) Genotyping PCR results show one DNA band for KO alleles from KO mouse genomic DNA and two DNA bands of WT and KO alleles from heterozygous. (C) IHC analyses of E11.5 Arhgap36-/- mutant embryos (lower panels, n = 3) and control littermates (upper panels, n = 5). There is no defect in cell proliferation (BrdU), ventral patterning (Nkx2.2and Olig2) and motor neuron generation (Hb9) at early developmental stage. Data are mean ± s.d. ns, non-significant; n = 7 ~ 14 independent images per each sample. (D) IHC analyses of E12.5 Arhgap36-/- mutant embryos (lower panels, n = 4) and control littermates (upper panels, n = 4). At cervical level, LMCm neurons are increased and LMCl neurons are decreased in Arhgap36-/- mutant embryo. At the same time, there is an increase in apoptotic marker (cCasp3) in Arhgap36-/- mutant spinal cord (arrowheads). Scale bars: 100 μm.
-
Figure 8—figure supplement 1—source data 1
Source data for Figure 8—figure supplement 1C.
- https://doi.org/10.7554/eLife.46683.028
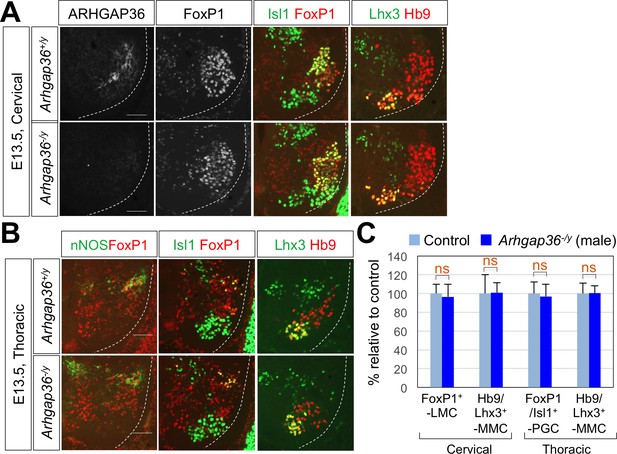
ARHGAP36 is not required for LMC formation in male mouse embryos.
(A) IHC analyses of E13.5 Arhgap36-/y mutant male embryos (n = 4) (lower panel) and their littermate controls (n = 5) (upper panel). At cervical level, the numbers of FoxP1+ LMC MNs and Hb9+/Lhx3+ MMC neurons did not change in Arhgap36-/y. (B) At thoracic level, there was no difference in nNOS+ PGC neurons, Hb9+/Isl1+ HMC neurons, and Hb9+/Lhx3+ MMC neurons compared to control littermates. Scale bars: 100 μm. (C) Quantification of the number of FoxP1+ LMC and Hb9+/Lhx3+ MMC at cervical level and FoxP1+/Isl1+PGC and Hb9+/Lhx3+ MMC at thoracic level in E13.5 male mouse embryonic spinal cord. Data are mean ±s.d. ns, non-significant; n = 6 ~ 12 independent images per each sample.
-
Figure 8—figure supplement 2—source data 1
Source data for Figure 8—figure supplement 2C.
- https://doi.org/10.7554/eLife.46683.030
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (M. musculus) | Shhf/f | Jackson Laboratories | RRID:MGI:2165468 | |
| Genetic reagent (M. musculus) | Olig2-Cre | PMID: 18046410 | Dr. Bennett G. Novitch (University of California, Los Angeles) | |
| Genetic reagent (M. musculus) | Arhgap36-/- | This paper | Exon2 targetting sgRNA made by ToolGEN, injected by KRIBB | |
| Transfected construct (M. musculus) | Arhgap36-(enhancer)2:LUC | This paper | Arhgap36 enhancer containing HxRE sequence cloned into TK-LUC vector | |
| Transfected construct (M. musculus) | Arhgap36-(enhancer)2:GFP | This paper | Arhgap36 enhancer containing HxRE sequence cloned into TATA-GFP vector | |
| Transfected construct (R. norvegicus) | Isl1 | PMID: 22343290 | ||
| Transfected construct (M. musculus) | Lhx3 | PMID: 22343290 | ||
| Transfected construct (E. coli) | β-galactosidase | PMID: 22343290 | ||
| Transfected construct (M. musculus) | Arhgap36 | Open Biosystems | Accession: BC145645 | |
| Transfected construct (M. musculus) | PKA WT | PMID: 23644383 | ||
| Transfected construct (M. musculus) | PKA K73H | PMID: 23644383 | ||
| Transfected construct (M. musculus) | AKT2 CA | Addgene | RRID:Addgene_9016 | myristoylated form |
| Transfected construct (M. musculus) | AKT2 DN | Addgene | RRID:Addgene_60128 | |
| Sequence-based reagent | Shh shRNA_sense strand | This paper | PCR primers | GAT CCA AGC TCT TCT ACG TCA TCG TTC AAG AGA CGA TGA CGT AGA AGA GCT TTT TTT A |
| Sequence-based reagent | Shh shRNA_antisense strand | This paper | PCR primers | AGC TTA AAA AAA GCT CTT CTA CGT CAT CGT CTC TTG AAC GAT GAC GTA GAA GAG CTT G |
| Sequence-based reagent | Arhgap36 enhancer_F | This paper | PCR primers | ACTGCCTATTCGCATCGGCCTTTGA, for cloning |
| Sequence-based reagent | Arhgap36 enhancer_R | This paper | PCR primers | TTCTGCGGAGCCATTAGTGCGATTG, for cloning |
| Sequence-based reagent | mouse Arhgap36_F | This paper | PCR primers | TGG GAT CCA AGA GGA AGA TG, for RT-PCR |
| Sequence-based reagent | mouse Arhgap36_R | This paper | PCR primers | CAG CCA CAT CAT GGA CAT TC, for RT-PCR |
| Sequence-based reagent | mouse Cyclophilin A_F | This paper | PCR primers | GTC TCC TTC GAG CTG TTT GC, for RT-PCR |
| Sequence-based reagent | mouse Cyclophilin A_R | This paper | PCR primers | GAT GCC AGG ACC TGT ATG CT, for RT-PCR |
| Sequence-based reagent | mouse Arhgap36 enhancer_F | This paper | PCR primers | ACC TTG TAG CAG GAC TGG GGT, for ChIP |
| Sequence-based reagent | mouse Arhgap36 enhancer_R | This paper | PCR primers | AGC CAT TAG TGC GAT TGC TCT, for ChIP |
| Sequence-based reagent | Untr6_F | PMID: 18854042 | PCR primers | TCA GGC ATG AAC CAC CAT AC, for ChIP |
| Sequence-based reagent | Untr6_R | PMID: 18854042 | PCR primers | AAC ATC CAC ACG TCC AGT GA, for ChIP |
| Antibody | anti-Hb9/MNR2 (Mouse) | DSHB | DSHB Cat# 81.5C10, RRID:AB_2145209 | IHC, 1:500 |
| Antibody | anti-Isl1 (Rabbit monoclonal) | Abcam | Abcam Cat# ab109517, RRID:AB_10866454 | IHC, 1:2000 |
| Antibody | anti-FoxP1 (Rabbit polyclonal) | Abcam | Abcam Cat# ab16645, RRID:AB_732428 | IHC, 1:1000 |
| Antibody | anti-Nkx2.2 (Mouse monoclonal) | DSHB | DSHB Cat# 74.5A5, RRID:AB_531794 | IHC, 1:100 |
| Antibody | anti-Pax6 (Mouse monoclonal) | DSHB | DSHB Cat# pax6, RRID:AB_528427 | IHC, 1:500 |
| Antibody | anti-Olig2 (Rabbit polyclonal) | Abcam | Millipore Cat# AB15328, RRID:AB_2299035 | IHC, 1:1000 |
| Antibody | anti-β-gal (Chicken polyclonal) | Abcam | Abcam Cat# ab9361, RRID:AB_307210 | IHC, 1:5000 |
| Antibody | anti-Lhx3 (Rabbit polyclonal) | Abcam | Abcam Cat# ab14555, RRID:AB_301332 | IHC, 1:500 |
| Antibody | anti-nNOS (Rabbit polyclonal) | Immunostar | ImmunoStar Cat# 24287, RRID:AB_572256 | IHC, 1:1000 |
| Antibody | anti-Chx10 (Guinea pig polyclonal) | PMID: 18539116 | IHC, 1:1000 | |
| Antibody | anti-GFP (Rabbit polyclonal) | Life Technologies | Thermo Fisher Scientific Cat# A-11122, RRID:AB_221569 | IHC, 1:1000 |
| Antibody | anti-Hb9 (Guinea pig polyclonal) | PMID: 30177510 | IHC, 1:1000 rat Hb9 C-terminus (234–403 aa | |
| Antibody | anti-ARHGAP36 (Rabbit polyclonal) | This paper | IHC, 1:2000 mouse ARHGAP36(201–590 aa) | |
| Antibody | anti-HA (Mouse monoclonal) | Covance | Covance Research Products Inc Cat# MMS-101R-500, RRID:AB_10063630 | IP, IB, 1:5000 |
| Antibody | anti-Gli3 (Goat polyclonal) | R and D Systems | R and D Systems Cat# AF3690, RRID:AB_2232499 | IB, 1:250 |
| Antibody | anti-ARHGAP36 (Rabbit polyclonal) | Sigma-Aldrich | Sigma-Aldrich Cat# HPA002064, RRID:AB_1078891 | IB, 1:2000 |
| Antibody | anti-β-tubulin (Rabbit polyclonal) | Santa Cruz | Santa Cruz Biotechnology Cat# sc-9104, RRID:AB_2241191 | IB, 1:2000 |
| Antibody | anti-pSER | Cell Signaling | Cat. #9651 | IB, 1:5000 |
| Antibody | anti-TuJ1 (Mouse monoclonal) | Covance | Covance Research Products Inc Cat# MMS-435P, RRID:AB_2313773 | IB, 1:5000 IHC, 1:5000 |
| Antibody | anti-FoxP1 (Rabbit polyclonal) | abcam | Abcam Cat# ab16645, RRID:AB_732428 | IB, 1:1000 |
| Antibody | anti-pCREB (Rabbit monoclonal) | Cell Signaling | Cell Signaling Technology Cat# 9198, RRID:AB_2561044 | IB, 1:1000 |
| Cell line (M. musculus) | P19 | ATCC | ATCC Cat# CRL-1825, RRID:CVCL_2153 | embryonic carcinoma cells |
| Cell line (Homo-sapiens) | HEK293T | ATCC | ATCC Cat# CRL-3216, RRID:CVCL_0063 | |
| Cell line (M. musculus) | A172L ESC | PMID: 22343290, 22039605 | ||
| Chemical compound, drug | iAKT1/2 | Sigma Aldrich | A6730 | 10 μM |
| Chemical compound, drug | SAG | Calbiochem | MER-566660 | 0.25 μM |
| Chemical compound, drug | Lipofectamine 2000 | Invitrogen | Cat. 52887 | |
| Chemical compound, drug | Superfect | Qiagen | Cat. 301307 | |
| Chemical compound, drug | SuperScript III First-Strand Synthesis System | Invitrogen | Cat. 18080085 | |
| Chemical compound, drug | SYBR-Green kit | Enzynomics | RT501S | |
| Software, algorithm | GPS 3.0 | PMID: 15980451 | http://gps.biocuckoo.cn/ |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.46683.032





