FcγRIIB-I232T polymorphic change allosterically suppresses ligand binding
Figures
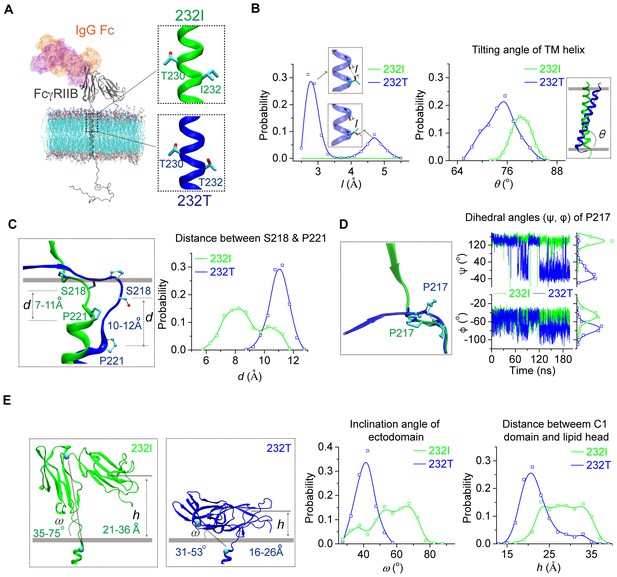
MD simulations reveal different conformations of 232I and 232T.
(A) The modeled structures of almost full-length FcγRIIB (232I and 232T, residues A46-I310, shown in gray cartoon) are complexed with IgG Fc (using the complex structure of IgG Fc and FcγRIIB ectodomain, PDB ID: 3WJJ as a reference, shown in color shaded surface) and imbedded in an asymmetric lipid bilayer (lines with atoms colored by element type: P, tan; O, red; N, blue; C, cyan). The helical structures in the vicinity of residue 232 for 232I (green) and 232T (blue) are shown in the insets. (B) Probability distributions of the distance between T232 Oγ atom and its nearest backbone O atom from residue V228 (left), and of the tilting angles between TM helix and lipid bilayer (right). The inclination of TM for 232T can be observed clearly. Blue dashed line in the upper inset of the left panel indicates H-bond. (C) The representative snapshot comparison of 232I and 232T at the stalk and TM region by superposing the lipid bilayers (left), and the length distribution of S218-P221 backbone in normal direction of lipid bilayer (right). (D) Conformational comparison of I212-S220 regions by aligning residues S218 to S220 (left), and the time courses of the dihedral angles (ψ, φ) of residue P217 (right). (E) Representative snapshots of 232I and 232T with the inclination angles and C1(Ig-like C2-Type one domain)/bilayer distances, probability distributions of the inclination angle between FcγRIIB ectodomain and lipid bilayer (left), and the distances between C1 domain and lipid bilayer (right).
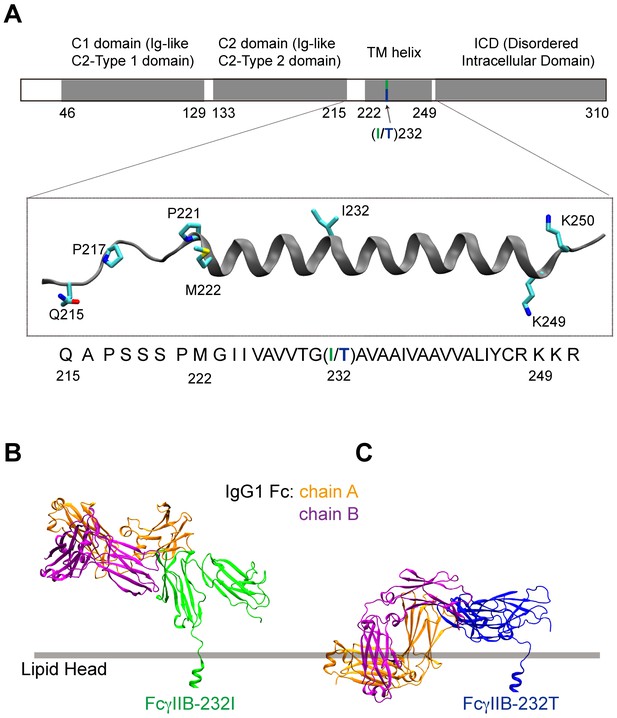
I232T polymorphic change of FcγRIIB induces the ectodomain recumbent and may impair its binding to Fc portion of antibodies.
(A) The modeled structure of the transmembrane region is highlighted. Residue 232 is the only difference between 232I and 232T. (B) The ectodomain of 232I (green) stands straight and is free for Fc binding. (C) For FcγRIIB-I232T polymorphic change, although the Fc binding site is still accessible, it occurs the clashes between the Fc and membrane. Therefore, the binding ability of 232T (blue) with Fc may be significantly reduced. The ectodomain in the FcγRIIB/Fc complex structure (PDB code: 3WJJ) is superimposed to that observed by the MD simulation.
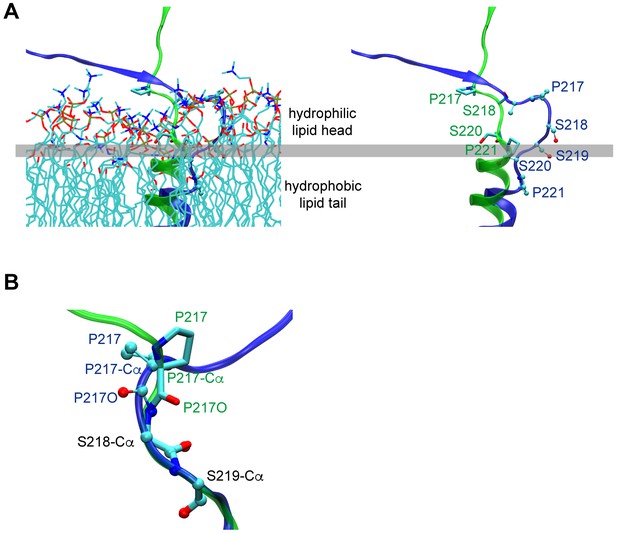
Conformation differences of ecto-TM linker and its vicinity between 232I (green) and 232T (blue) are obtained by MD simulations.
(A) The conformation of the ecto-TM linker in lipid bilayer (left) or lipids removed (right, the residues number in this region is indicated). (B) Conformation differences of I212-S220 region were showed through aligning residues S218 to S220.
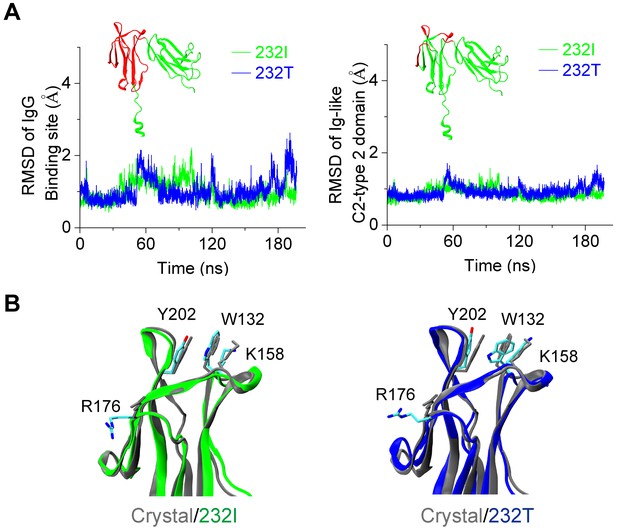
IgG-binding sites in both 232I and 232T do not undergo significant structural change.
(A) RMSD time courses of protein backbone atoms in Ig-like C2-type two domain (left panel, colored red on the crystal structure shown in the inset) and IgG binding site (composed mainly with the residues W132, K158, R176 and Y202; right panel and colored red on the crystal structure shown in the inset) for 232I (green) or 232T (blue). No significant change in the domain and the binding site can be observed. (B) Structural comparison between crystal structure (gray) and representative snapshots of MD simulations in the 232I (green) or 232T(blue) system.
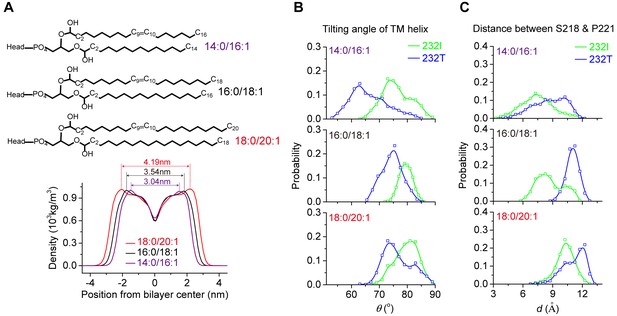
The length of fatty acid chain and membrane thickness do not alter the effect of I232T polymorphism on the tilting of TM domain.
(A) Schematic of 14:0/16:1, 16:0/18:1 and 18:0/20:1 lipids (upper), and the membrane thickness comparisons with different lengths of lipid fatty acid chain (bottom). MD simulation results of TM helix tilting angle (B), and S218-P221 prolongation (C) in lipid membranes with different-length lipids and membrane thickness.
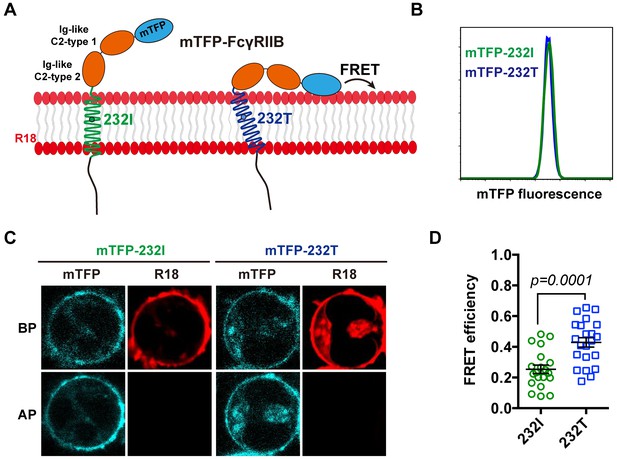
The 232T ectodomain prefers to a more recumbent orientation on the plasma membrane.
(A) Schematic of mTFP-R18 FRET experimental setup to measure the FRET signals between the ectodomain of 232I (green) or 232T (blue) (N-terminal of ectodomain fused with mTFP as FRET donor, cyan) and the plasma membrane (stained with R18 dye as FRET acceptor, red). (B) Comparison of mTFP fluorescence intensities of A20II1.6 B cell lines expressing either mTFP-232I (green) or mTFP-232T (blue) constructs by FACS analysis. (C) Representative images of de-quenching FRET assay. R18-labeled mTFP-232I or mTFP-232T cell images were acquired in both channels before or after R18 photo-bleaching (BP or AP). (D) FRET efficiency comparison of mTFP-232I (green circle) and mTFP-232T (blue square) cells (~20 cells, respectively) with a p-value indicated. Error bars represent SEM.
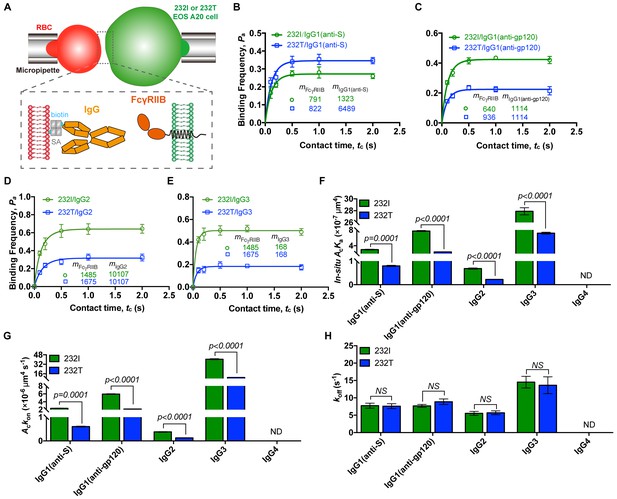
232T exhibits significantly reduced 2D IgG binding affinity and on-rate in comparison with 232I.
(A) Schematics of experimental setup for single-cell in situ 2D kinetic measurement. Two opposing micropipettes aspirated a human red blood cell coated with a monoclonal IgG (red) and an A20II1.6 B cell expressing either 232I or 232T (green) to operate contact-retraction cycles manipulation, respectively. (B–H) Plots of adhesion frequency Pa versus contact time tc of FcγRIIB (either 232I or 232T) binding with human IgG1 antibody (anti-S, B, or anti-gp120, (C)/IgG2 (D)/IgG3 (E), corresponding in situ 2D effective binding affinity AcKa (F), on-rate Ackon (G) and off-rate koff (H) are compared, respectively. mFcγRIIB and mIgG are surface molecular densities of respective proteins. Error bars represent SEM. ND, not detectable. NS, not significantly different.
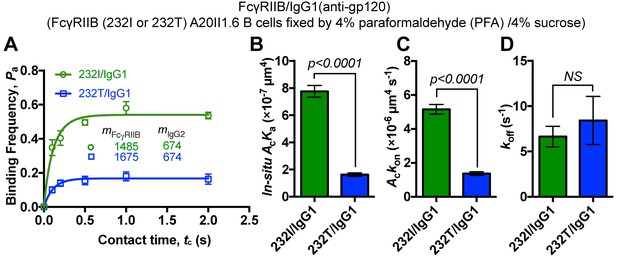
232T exhibits significantly reduced 2D affinity and on-rate of binding to IgG1 (anti-gp120) in comparison with 232I, although FcγRIIB (either 232I or 232T) A20II1.6 B cells are fixed by 4% paraformaldehyde (PFA) plus 4% sucrose.
(A) Adhesion curves of FcγRIIB (either 232I or 232T) and human IgG1 (anti-gp120) according to probabilistic kinetic model. (B–D) Comparisons of in situ 2D effective binding affinity (AcKa) (B), on-rate (Ackon) (C) and off-rate (koff) (D) between 232I and 232T binding to IgG1 when cells are fixed. Error bars represent SEM. NS, not significantly different.
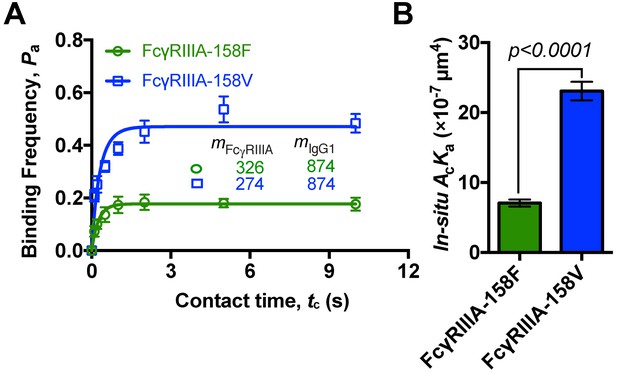
FcγRIIIA-158V shows a higher 2D affinity than FcγRIIIA-158F in binding with IgG1.
(A) Adhesion curves of FcγRIIIA (158F or 158V) and human IgG1 antibody (anti-S) according to probabilistic kinetic model. (B) From the adhesion curves, in situ 2D effective binding affinities (AcKa) were calculated. Error bars represent SEM.
Tables
Association analysis between homozygous FcγRIIB-I232T genotype and SLE in subphenotype-control cohorts, adjusting for age and sex
https://doi.org/10.7554/eLife.46689.003| Genotype frequency | Subphenotype vs. controls | |||
|---|---|---|---|---|
| Tt+tc (%)/CC (%) | OR | 95% | P value | |
| Controls | 376+286 (96.2)/26 (3.8) | |||
| Disease Onset, age < 37 | 207+150 (92.7)/28 (7.3) | 2.739 | 1.456–5.152 | 0.002 |
| Disease Onset, age >= 37 | 171+133 (93.3)/22 (6.7) | 1.657 | 0.872–3.150 | 0.123 |
| Arthritis = 1 | 203+166 (92.5)/30 (7.5) | 2.074 | 1.206–3.565 | 0.008 |
| Arthritis = 0 | 132+87 (94.8)/12 (5.2) | 1.41 | 0.698–2.847 | 0.338 |
| Hematological involvement = 1 | 242+185 (93.4)/30 (6.6) | 1.797 | 1.048–3.084 | 0.033 |
| Hematological involvement = 0 | 90+75 (94.8)/9 (5.2) | 1.369 | 0.627–2.988 | 0.431 |
| Anemia = 1 | 126+94 (91.7)/20 (8.3) | 2.323 | 1.270–4.249 | 0.006 |
| Anemia = 0 | 153+125 (94.2)/17 (5.8) | 1.552 | 0.828–2.907 | 0.17 |
| Leukopenia = 1 | 140+114 (91.7)/23 (8.3) | 2.294 | 1.284–4.099 | 0.005 |
| Leukopenia = 0 | 136+107 (94.9)/13 (5.1) | 1.345 | 0.680–2.663 | 0.394 |
| dsDNA = 1 | 198+151 (92.1)/30 (7.9) | 2.224 | 1.293–3.826 | 0.004 |
| dsDNA = 0 | 129+94 (94.5)/13 (5.5) | 1.48 | 0.747–2.932 | 0.261 |
| ANA = 1 | 308+241 (93.4)/39 (6.6) | 1.82 | 1.094–3.030 | 0.021 |
| ANA = 0 | 25+16 (93.2)/3 (6.8) | 1.893 | 0.549–6.526 | 0.312 |
| Total Ig = 1 | 131+101 (92.8)/18 (7.2) | 1.969 | 1.060–3.66 | 0.032 |
| Total Ig = 0 | 120+88 (93.7)/14 (6.3) | 1.706 | 0.874–3.328 | 0.118 |
| Complement Decrease = 1 | 227+162 (92.4)/32 (7.6) | 2.1 | 1.233–3.578 | 0.006 |
| Complement Decrease = 0 | 58+54 (95.7)/5 (4.3) | 1.124 | 0.423–2.991 | 0.814 |
| Hematuria = 1 | 95+65 (90.9)/16 (9.1) | 2.634 | 1.370–5.063 | 0.004 |
| Hematuria = 0 | 180+140 (94.7)/18 (5.3) | 1.423 | 0.768–2.635 | 0.262 |
| Leucocyturia = 1 | 64+57 (91.0)/12 (9.0) | 2.541 | 1.246–5.178 | 0.010 |
| Leucocyturia = 0 | 196+143 (93.9)/22 (6.1) | 1.651 | 0.922–2.957 | 0.092 |
| Serositis = 1 | 66+45 (92.5)/9 (7.5) | 2.108 | 0.961–4.627 | 0.063 |
| Serositis = 0 | 225+174 (94.1)/25 (5.9) | 1.615 | 0.919–2.838 | 0.096 |
| SLEDAI >= 12 | 20+25 (88.2)/6 (11.8) | 3.327 | 1.273–8.696 | 0.014 |
| SLEDAI < 12 | 116+93 (95.9)/9 (4.1) | 1.072 | 0.493–2.328 | 0.861 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Mus musculus) | A20II1.6 B cell | ATCC | a gift from S.K. Pierce (National Institute of Allergy and Infectious Diseases, Bethesda, MD) | |
| Cell line (Mus musculus) | 232I A20II1.6 B cell line | (Xu et al., 2016) | ||
| Cell line (Mus musculus) | 232T A20II1.6 B cell line | (Xu et al., 2016) | ||
| Cell line (Mus musculus) | mTFP-232I A20II1.6 B cell line | This paper | Stable mTFP-232I expressing A20II1.6 B cell lines were acquired by lentivirus infection. | |
| Cell line (Mus musculus) | mTFP-232T A20II1.6 B cell line | This paper | Stable mTFP-232T expressing A20II1.6 B cell lines were acquired by lentivirus infection. | |
| Antibody | Anti-gp120 IgG1, human monoclonal | a kind gift from Dr. Y. Shi, The Institute of Microbiology of the Chinese Academy of Sciences | Dosage: 20 μg | |
| Antibody | Anti-S IgG1, human monoclonal | a kind gift from Dr. L. Zhang and X. Wang, Tsinghua University | Dosage: 20 μg | |
| Antibody | IgG2, human monoclonal | a kind gift from Dr. H. Wang, Hisun, China | RANKL human IgG2 monoclonal antibody (HS629) | Dosage: 20 μg |
| Antibody | IgG3, human monoclonal | InvivoGen | Catalog#bgal-mab3; RRID: AB_2810285 | recombinant Anti-β-Gal-hIgG3 was produced in CHO cells; dosage: 20 μg |
| Antibody | IgG4, human monoclonal | a kind gift from Dr. Y. Shi, The Institute of Microbiology of the Chinese Academy of Sciences | Dosage: 20 μg | |
| Recombinant DNA reagent | PHAGE-mTFP-232I (plasmid) | This paper | mTFP was fused to N-terminal of FcγRIIB-232I in a pHAGE backbone | |
| Recombinant DNA reagent | PHAGE-mTFP-232T (plasmid) | This paper | mTFP was fused to N-terminal of FcγRIIB-232T in a pHAGE backbone | |
| Commercial assay or kit | TIANamp Blood DNA Midi Kit | TIANGEN Biotech, China | Catalog#DP332-01 | TaqMan probe C: 5’-VIC-CGCTACAGCAGTCCCAGT-NFQ-3’, TaqMan Probe T: 5’-FAM-CGCTACAGCAATCCCAGT-NFQ-3’ |
| Commercial assay or kit | TaqMan Genotyping Assays | Life Technology | Catalog#4351376 | |
| Commercial assay or kit | ClonExpress MultiS One Step Cloning Kit | Vazyme, China | Catalog#C113 | |
| Chemical compound, drug | Octadecyl rhodamine B (R18) | Invitrogen | Catalog#O246 | |
| Chemical compound, drug | Biotin-PEG-SGA | JenKem Technology, China | Catalog#ZZ324P050 | |
| Chemical compound, drug | Streptavidin | Sangon Biotech, China | Catalog#C600432 | |
| Chemical compound, drug | EZ-Link Sulfo-NHS-LC-Biotin kits | Thermo Fisher Scientific | Catalog#21435 | |
| Software, algorithm | VMD (Visual Molecular Dynamics) | University of Illinois at Urbana-Champaign | Visual Molecular Dynamics, RRID:SCR_001820 | http://www.ks.uiuc.edu/Research/vmd/ |
| Software, algorithm | NAMD | University of Illinois at Urbana-Champaign | NAMD, RRID:SCR_014894 | http://www.ks.uiuc.edu/Research/namd/ |
| Software, algorithm | CHARMM Force Field | Alex Mackerell lab at School of Pharmacy, University of Maryland | http://mackerell.umaryland.edu/charmm_ff.shtml | |
| Software, algorithm | GraphPad Prism | GraphPad | GraphPad Prism, RRID:SCR_002798 | https://www.graphpad.com/ |
Additional files
-
Supplementary file 1
Association analysis of rs1050501 with SLE (adjusted for sex and age).
- https://doi.org/10.7554/eLife.46689.013
-
Supplementary file 2
Demographic characteristic of SLE cohort.
- https://doi.org/10.7554/eLife.46689.014
-
Transparent reporting form
- https://doi.org/10.7554/eLife.46689.015





