Extensive ribosome and RF2 rearrangements during translation termination
Figures
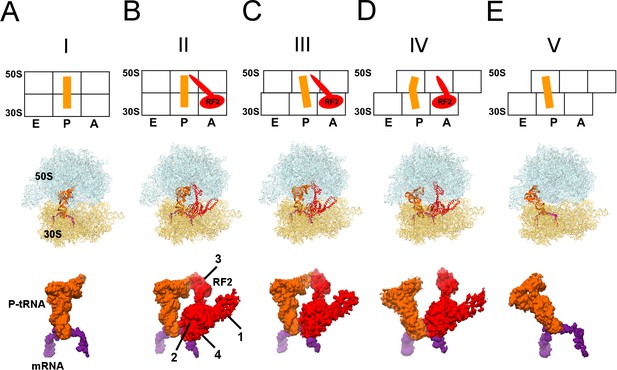
Cryo-EM Structures I to V.
Panels A-E show for each structure: upper row: a schematic of the conformations of the 70S ribosome, tRNA and RF2, with ribosomal subunits (50S and 30S) and A, P and E sites labeled; middle row: structures with RF2 shown in red, tRNAfMet in orange, 30S subunit in yellow, 50S subunit in cyan and mRNA in purple; lower row: cryo-EM density for mRNA, tRNA and RF2, colored as in the middle row. Domains of RF2 are labeled in panel B, illustrating the codon-recognition superdomain (domains 2 and 4) and catalytic domain (3).
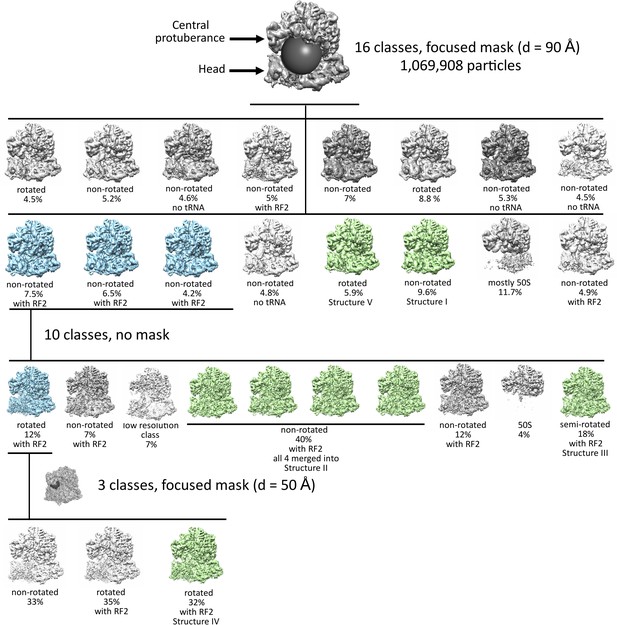
Maximum-likelihood classification strategies to obtain the final maps and state occupancies (shown as percentages of particles for each classification).
Cyan classes were used for further classifications, green classes were used for obtaining the final maps. The views show the 50S (top) and 30S (bottom) subunits and the positions of spherical masks.
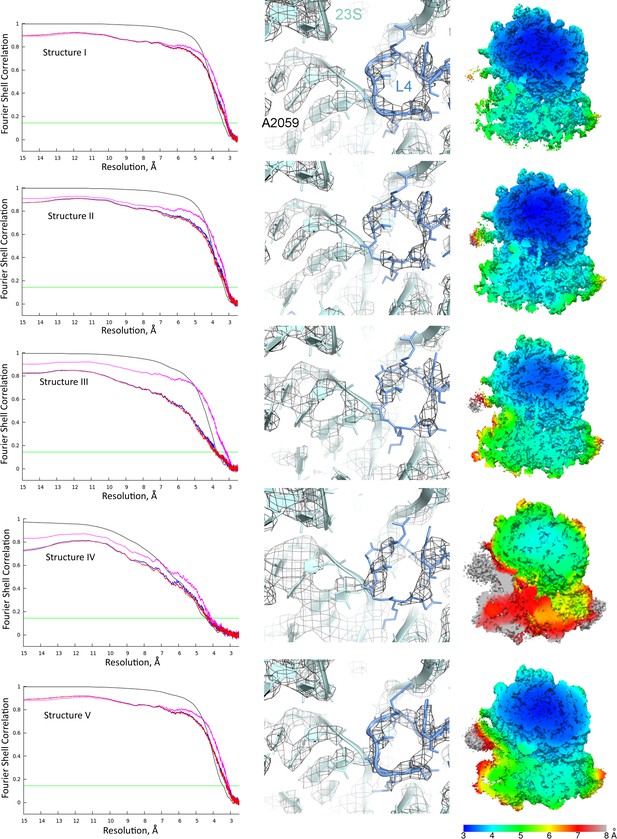
Global and local resolution of Structures I-V.
Left column: Fourier shell correlation (FSC) between even- and odd-particle half maps (black) show that map resolutions range from 3.3 to 4.4 Å for Structures I-V (at FSC = 0.143, green line); FSC between final models and final maps (magenta), self (blue) and cross-validation half-maps (red) masked FSC are also shown. Middle column: Examples of local map resolution for rRNA and protein in the vicinity of the peptidyl transferase center are shown for each structure (23S rRNA and protein L4 are labeled for reference). Right column: Local resolutions in cryo-EM densities for Structures I-V, calculated using Blocres. Slab views show the ribosome interior in the vicinity of the long helix α7 of domain 3 of RF2 (the helix is visible at the centers of Structures II and III).
A representative micrograph of the 70S•RF2 sample showing ribosome particles.
https://doi.org/10.7554/eLife.46850.005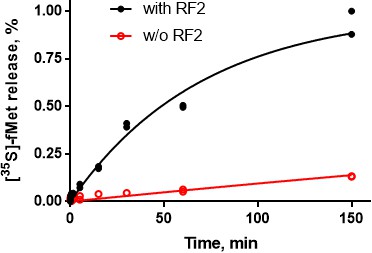
RF2 mediates formyl-methionine release from 70S ribosomes.
The time course shows RF2-catalyzed [35S]-fMet release from 70S•[35S]-fMet-tRNAfMet•mRNA(UGA) (black curve and data points), in contrast to slow spontaneous release in the absence of RF2 (red). The reactions were performed using buffer conditions and concentrations identical to those used for cryo-EM grids preparation (see Materials and methods); N = 2 for all data points.
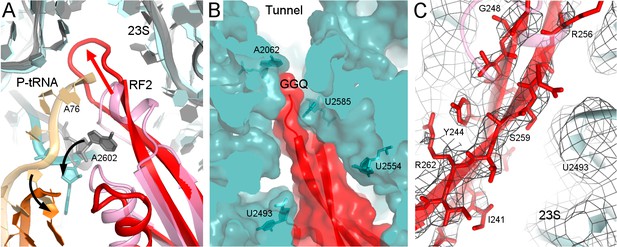
Rearrangement of the catalytic domain of RF2 upon tRNA displacement from the PTC.
(A) Rearrangements of RF2 in Structure II (RF2 in red, P-tRNA in orange and 23S in cyan) in comparison with the canonical 70S•RF2 conformation (X-ray structure: Korostelev et al., 2008; RF2 in pink, P-tRNA in light orange, 23S in gray). Arrows show rearrangements for RF2 (red), tRNA acceptor arm and 23S nucleotide A2602 (black). The superposition was achieved by structural alignment of 23S rRNA. (B) The GGQ region of RF2 forms a long β-hairpin that reaches into the constriction of the peptide tunnel (at A2062) and thus plugs the tunnel. Nucleotides of 23S rRNA and the GGQ motif are labeled. (C) Cryo-EM density for the β-hairpin formed by the GGQ region. Residues of RF2 and 23S rRNA (also shown in panel B for reference) are labeled.
Rearrangements of PTC nucleotides (cyan) upon formation of the β-hairpin structure by the catalytic region of RF2 (red; Structure II is shown).
Nucleotides in the crystal structure with an α-helical conformation of the GGQ motif of RF2 (not shown; see Figure 2A) are shown in gray (Korostelev et al., 2008). (A) Cryo-EM density (gray mesh, shown at σ = 3.0) for G2505 and U2506 (shown as sticks). (B) Cryo-EM density (gray mesh, shown at σ = 2.5) for A2602 (shown as sticks).
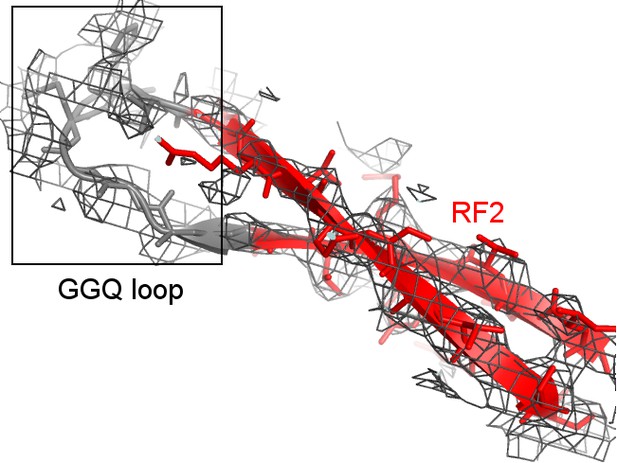
Cryo-EM density (mesh) shows that the tip of the GGQ loop of RF2 is poorly ordered (gray model) when the catalytic domain adopts the β-hairpin conformation (red; Structure II is shown).
https://doi.org/10.7554/eLife.46850.011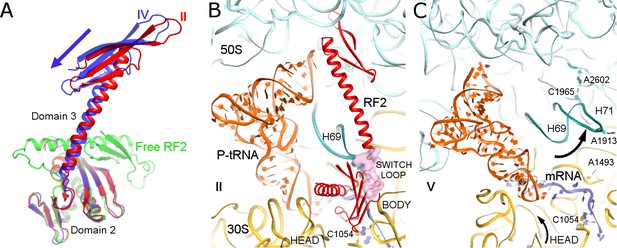
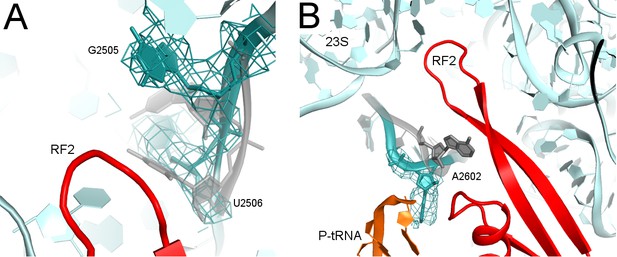
Differences in conformations of RF2 and helix 69 of 23S rRNA in Structures II, IV and V.
(A) RF2 in Structures II (red) and IV (blue) in comparison with the crystal structure of isolated (free) RF2 (green, PDB 1GQE [Vestergaard et al., 2001]). The arrow shows the direction of domain three repositioning from Structure II to IV to free RF2. Structures were aligned by superposition of domain 2. (B) Interactions of H69 (teal) with the switch loop (pink surface) of RF2 in Structure II (RF2 in red, P-tRNA in orange, 50S in cyan and 30S in yellow). Domains 2 and 3 of RF2, head and body domains of the 30S subunit and nucleotides A1913 (at H69) and C1054 (head) are labeled. (C) In Structure V, dissociation of H69 from the decoding center and packing on H71 next to A2602 disassembles intersubunit bridge B2a. The structure is colored as in panel B. View in panels B and C is rotated by ~180° relative to panel A.
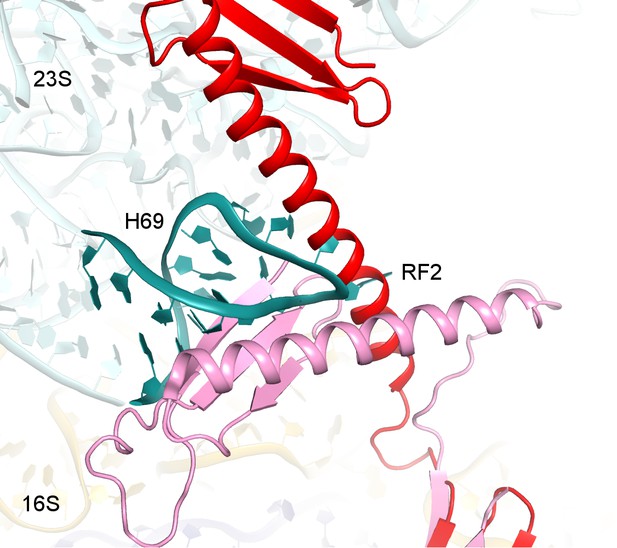
Structure alignments show that the disengaged H69 in Structure V (teal) is incompatible with the extended (red, Structure II) and compact (pink, PDB 1GQE [Vestergaard et al., 2001]) conformations of RF2 due to steric hindrance.
Structures II and V were superimposed via 16S rRNA. Codon-recognition domain 2 of compact RF2 was superimposed with domain 2 of RF2 in Structure II.
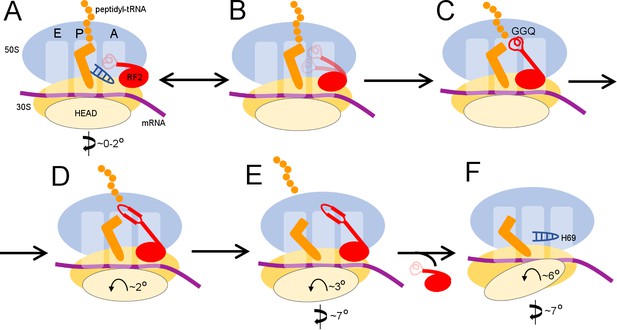
Scheme of the termination mechanism.
https://doi.org/10.7554/eLife.46850.014Videos
An animation showing transitions between the structures of the 70S ribosome upon interaction with RF2.
https://doi.org/10.7554/eLife.46850.015Tables
Cryo-EM structures obtained from a single sample of the 70S•RF2 complex.
https://doi.org/10.7554/eLife.46850.007| I | II | III | IV | V | |
|---|---|---|---|---|---|
| tRNA and RF2 occupancy | P-tRNA | P-tRNA, RF2 | P-like (L5) tRNA, RF2 | P/E-like (L1) tRNA, RF2 | P/E tRNA |
| 30S (body) rotation | 1.6° | 1.6° | 4.9° | 7.4° | 6.8° |
| 30S head rotation (swivel) | 2.0° | 1.4° | 1.7° | 2.8° | 6.0° |
| Resolution (FSC = 0.143) | 3.3 Å | 3.3 Å | 3.7 Å | 4.4 Å | 3.4 Å |
Data collection and refinement statistics for cryo-EM Structures I-V.
https://doi.org/10.7554/eLife.46850.008| STRUCTURE | I | II | III | IV | V |
|---|---|---|---|---|---|
| PDB ID EMD code | 6OFX 20048 | 6OG7 20052 | 6OGF 20056 | 6OGG 20057 | 6OGI 20058 |
| Data collection | |||||
| EM equipment | FEI Titan Krios | ||||
| Voltage (kV) | 300 | ||||
| Detector | K2 summit | ||||
| Pixel size (Å) | 1.33 | ||||
| Electron dose (e-/Å2) | 29.4 | ||||
| Defocus range (μm) | – 0.5 – 1.8 | ||||
| Reconstruction | |||||
| Software | Frealign v9.11 | ||||
| Number of particles in final map | 102,723 | 62,029 | 28,549 | 5,881 | 63,383 |
| Final resolution (Å) | 3.3 | 3.3 | 3.7 | 4.4 | 3.4 |
| Average sharpening B factor (Å2) | -30 | -26 | -17 | -4 | -30 |
| Structure Refinement | |||||
| Model Fitting | Chimera/Pymol | ||||
| Refinement | |||||
| Software | RSRef/Phenix | ||||
| Correlation Coefficient,cc_mask * | 0.83 | 0.84 | 0.84 | 0.76 | 0.82 |
| Real-space R-factor † | 0.25 | 0.23 | 0.22 | 0.25 | 0.25 |
| Validation (proteins) | |||||
| MolProbity Score ‡ | 2.22 | 2.34 | 2.42 | 2.26 | 2.24 |
| Clash score, all atoms ‡ | 17.4 | 17.4 | 16.5 | 15.1 | 16.3 |
| Poor rotamers (%) ‡ | 0.4 | 0.7 | 0.7 | 0.6 | 0.5 |
| Favored/allowed rotamers (%) ‡ | 99.6 | 99.3 | 99.3 | 99.4 | 99.5 |
| Ramachandran-plot statistics | |||||
| Outlier (%) ‡ | 0 | 0.6 | 0.8 | 0.0 | 0.4 |
| Favored (%) ‡ | 92.2 | 88.1 | 82.3 | 89.0 | 90.7 |
| R.m.s. deviations †,§ | |||||
| Bond length (Å) | 0.008 | 0.006 | 0.006 | 0.006 | 0.009 |
| Bond angle (°) | 1.064 | 0.892 | 0.922 | 0.848 | 1.113 |
| Validation (RNA) | |||||
| Good sugar puckers (%) ‡ | 99.7 | 99.8 | 99.8 | 99.8 | 99.8 |
| Good backbone conformation (%) ‡ | 82.9 | 84.6 | 85.0 | 85.0 | 82.0 |
-
*from Phenix
†from RSRef
-
§root-mean-square deviations
# RNA backbone suites that fall into recognized rotamer conformations defined by MolProbity
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | Escherichia coli MRE600 | (Cammack and Wade, 1965) | ATCC 29417 | |
| Strain, strain background (Escherichia coli) | Escherichia coli BLR (DE3) | Novagen | ||
| Recombinant protein | C-terminally His-tagged K-12 E. coli RF2 | (Demo et al., 2017) | ||
| Recombinant DNA reagent | E. coli RF2 plasmid (vector pET24b) | (Demo et al., 2017) | ||
| Chemical compound, drug | E. coli tRNAfMet | Chemical Block | ||
| Chemical compound, drug | [35S]-methionine | Perkin Elmer | NEG709A500UC | |
| Chemical compound, drug | Biodegradable Scintillation Cocktail | Econo-Safe | ||
| Sequence-based reagent | RNA GGCAAGGAGGUAAAAAUGUGAAAAAAA | IDT | ||
| Software, algorithm | PHENIX | (Adams et al., 2002) | https://www.phenix-online.org/ | |
| Software, algorithm | CNS | (Brunger, 2007) | http://cns-online.org/v1.2/ | |
| Software, algorithm | PyMOL | (DeLano, 2002) | https://pymol.org/2/ | |
| Software, algorithm | Chimera | (Pettersen et al., 2004) | https://www.cgl.ucsf.edu/chimera/ | |
| Software, algorithm | Bsoft | (Heymann and Belnap, 2007) | https://lsbr.niams.nih.gov/bsoft/ | |
| Software, algorithm | Frealign | (Lyumkis et al., 2013) | http://grigoriefflab. janelia.org/frealign | |
| Software, algorithm | cisTEM | (Grant et al., 2018) | https://cistem.org/ | |
| Software, algorithm | MolProbity | (Chen et al., 2010) | http://molprobity.biochem.duke.edu/ | |
| Software, algorithm | RSRef | (Chapman, 1995; Korostelev et al., 2002) | http://www.sb.fsu.edu/~rsref/ | |
| Software, algorithm | SerialEM | (Mastronarde, 2005) | http://bio3d.colorado.edu/SerialEM/ | |
| Other | Holey-carbon grids QUANTIFOIL R 2/1, Cu 200 | Quantifoil Micro Tools | ||
| Other | Vitrobot MK4 | FEI | ||
| Other | Titan Krios microscope | FEI | ||
| Other | K2 Summit camera | Gatan |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.46850.016





