Subtle selectivity in a pheromone sensor triumvirate desynchronizes competence and predation in a human gut commensal
Figures
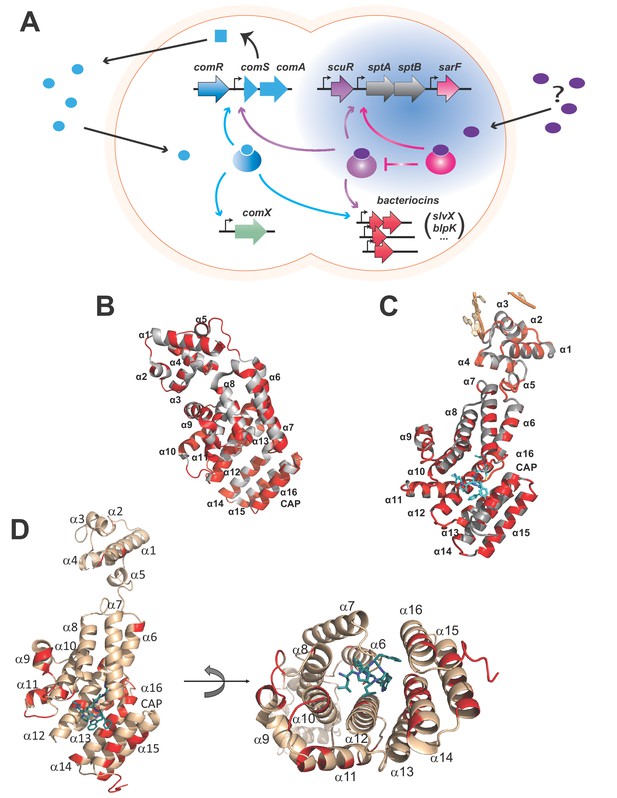
Rgg-based decision for competence-predation activation in S. salivarius.
(A) Scheme of genomic organization and transcriptional dependencies (color-coded arrows) between competence activation (comX) and bacteriocins production (blpK, slvX, …) in S. salivarius. Promoters are depicted with broken arrows. Regulators (large gradient-colored ellipses) and the ComS pheromone (uniform blue shapes) are colored according to their encoding genes. The ComS precursor is produced (curled plain arrow) as an intracellular precursor (blue square) before secretion, maturation and import as an active pheromone (blue ellipses). The newly described two-Rgg system (Figure 1—figure supplement 1) is highlighted in blue and the T arrow pinpoints the inhibitory role of SarF on ScuR. Their cognate pheromone of unknown origin is purple-colored. (B) Crystal structure of the monomeric apo form of S. thermophilus ComR (PDB ID 5JUF). The protein is shown as cartoon colored in gray. The HTH and TPR domains are labeled, as well as the linker region. Residues which are not conserved in ScuR are highlighted in red. The numbering of the α-helices is indicated to gain clarity. (C) Crystal structure of S. thermophilus ComR in complex with sComS (LPYFAGCL) (PDB ID 5JUB). Only one ComR subunit of the dimeric ComR•sComS•DNA complex is shown in gray with ScuR substitution sites in red as in (B). The bound peptide is highlighted in blue sticks. Part of the bound DNA is shown in orange and labeled, as the HTH and TPR domains. (D) Orthogonal views of the ScuR-sBI7 model. The protein is shown as cartoon colored in beige except for residues substituted in SarF, which are highlighted in red as in Figure 8C. The bound peptide is shown in blue sticks with the conserved tryptophan (W). The numbering of α-helices is indicated to gain clarity. In the right panel, the HTH domain is hidden in the back of the figure. The model was obtained by homology modeling using the i-TASSER server and the ComR•sComS•DNA complex (PDB ID 5JUB) as template.
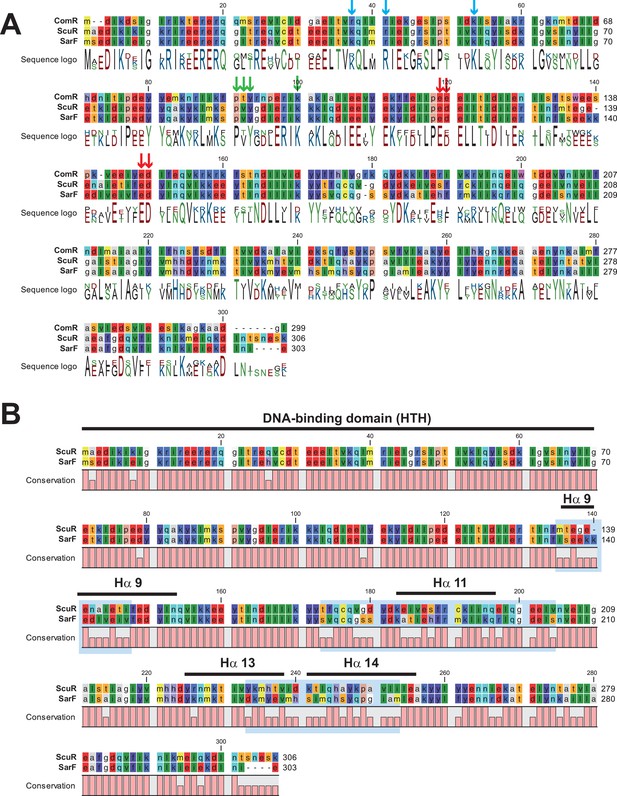
Sequence alignments of ComR paralogs.
(A) Sequence alignment of ComR, ScuR and SarF from S. salivarius. Residues are color-coded according to the Rasmol color scheme. The consensus sequence is shown underneath with the following nomenclature: positively-charged residues in blue, negatively-charged residues in red, polar residues in green and hydrophobic residues in black. Stained arrows pinpoint residues essential in DNA binding (blue), HTH-TPR sequestration (red), and peptide binding (green) according to the ComR•sComS•DNA structural analysis (Talagas et al., 2016). (B) Primary sequence conservation between the highly similar ScuR and SarF regulators. Residues are color-coded according to the Rasmol color scheme. Remarkable ComR apo secondary structure elements of apo ComR are indicated at the top of the alignment. The conservation between ScuR and SarF is summarized underneath. Light blue boxes highlight the most divergent parts between paralogs. (A and B) The formatting of the multiple alignment has been generated using CLC Main Workbench 7 (https://www.qiagenbioinformatics.com/).
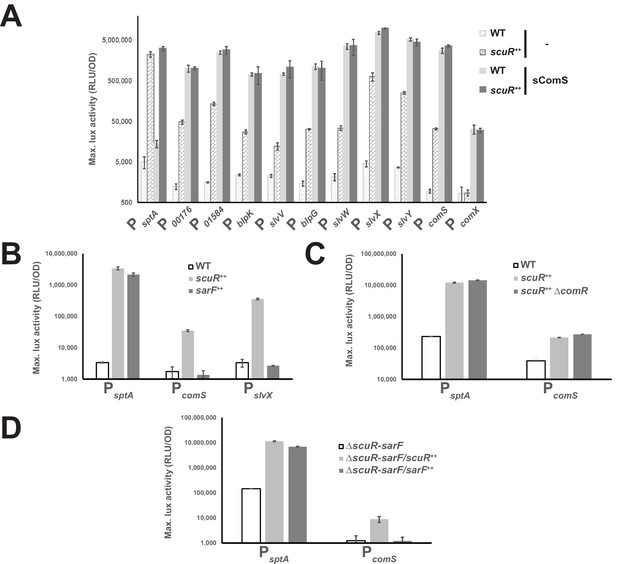
Competence-predation desynchronization in S. salivarius.
(A, B, C and D) Maximum luciferase activity/OD600 ratio (RLU/OD; logarithmic scale) of various promoters involved in competence or bacteriocin production fused to a luxAB reporter system in WT or overexpressing backgrounds (Figure 2—figure supplement 1). (A) Promoter activation of genes upon sComS addition (full bars) vs mock condition (striped bars) in WT (light gray bars) or scuR overexpression mutant (scuR++; dark gray bars). (B) Activity of sptA, comS and slvX promoters in WT strain, and scuR (scuR++) or sarF (sarF++) overexpression mutants. (C) Activity of sptA and comS promoters in WT and scuR ++ mutant deleted or not of comR gene. (D) Activity of sptA and comS promoters in a ΔscuR-sarF background when scuR or sarF are overexpressed. Experimental values represent averages (with standard error of the mean, SEM) of at least three independent biological replicates.
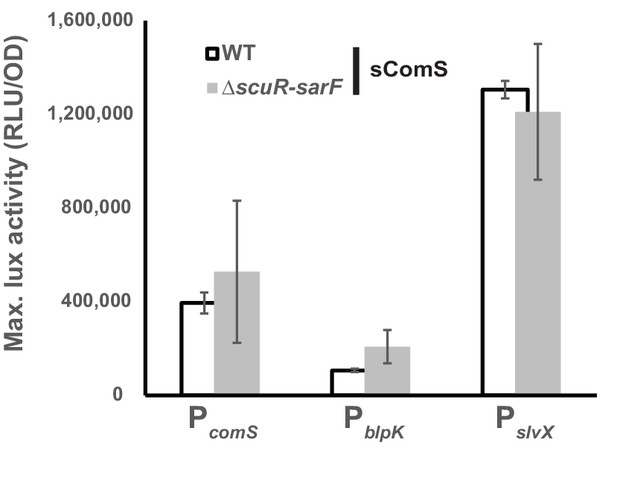
The ComS-driven activation is ScuR/SarF-independent.
Maximum luciferase activity/OD600 ratio (RLU/OD) of comS, blpK and slvX promoters fused to a luxAB reporter system in WT (open bars) or ΔscuR-sarF (gray bars) strain activated with sComS. Experimental values represent the averages (with standard error of the mean, SEM) of at least three independent biological replicates.
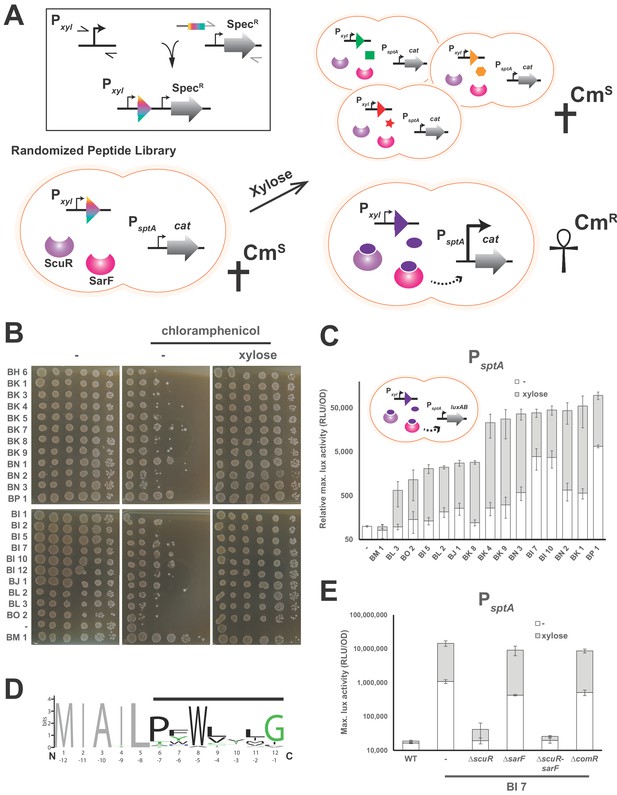
Identification of ScuR/SarF activating peptide.
(A) Cartoon portraying the rational strategy for the peptide randomization-based screen. In the box on the top-left: a degenerated primer (rainbow box) was used to amplify two kinds of xylose-inducible promoters, Pxyl1 (strong and leaky) and Pxyl2 (mild and tight). The 5’ end of this primer hybridizes on a second amplicon encompassing a spectinomycin resistance cassette (SpecR). Both PCR products were grafted with an overlapping PCR. Next, a library of randomized small genes (rainbow arrow head) under inducible promoter control (Pxyl) is transformed into a reporter strain in which the chloramphenicol resistance gene (cat) is translationally fused to sptA promoter. In absence of xylose or upon xylose induction of irrelevant peptides (green square, orange hexagon and red star), sptA promoter remains OFF and does not initiate cat transcription, causing cell sensitivity (CmS) on chloramphenicol-supplemented media. The xylose-driven intracellular production of a cognate peptide (purple ellipse) promotes chloramphenicol resistance (CmR), through PsptA activation by ScuR/SarF (dashed arrow). (B) Viability test of WT and all non-redundant mutants expressing intracellularly activating peptides in the PsptA-cat reporter background. The BM1 clone was used as an irrelevant peptide (negative control). Overnight precultured cells were diluted (OD600 of 0.05) to inoculate fresh M17G medium and grown 3 hr (OD600 of 0.5). Before plating on medium supplemented or not with chloramphenicol (2 mg.ml−1) and/or xylose (1% top panel; 0.1% bottom panel), the culture was sampled and serially diluted (10:10) in M17G. (C) Activity of sptA promoter in WT strains and various mutants intracellularly expressing activating peptides (cartoon) in medium supplemented with xylose (0.1% or 1%; gray bars) vs mock conditions (open bars). The BM1 clone is an irrelevant peptide (negative control). Magnitude is expressed in percentage compared to the WT PsptA-luxAB reporter strain (Relative maximal luciferase activity). Experimental values represent the averages (with standard error of the mean, SEM) of at least three independent biological replicates. (D) Weighted consensus sequence for 22 activating peptides identified in the randomization-based screen (Figure 3—figure supplement 1). Randomized residues are highlighted with a horizontal black bar while the non-variable amino acids are gray-coloured. The Bits represent the relative frequency of residues. Information content is plotted as a function of residues position and depicted from the N-terminus (1 to 12) or the C-terminus (−1 to −12). The sequence logo image was generated using the WebLogo application (http://weblogo.berkeley.edu/logo.cgi). (E) Promoter activity of the sptA gene in response to the BI7 encoded peptide in various scuR, sarF or comR mutant backgrounds. Media were supplemented with 0.1% xylose (open bars) or water (gray bars). Experimental values represent averages (with standard error of the mean, SEM) of at least three independent biological replicates.
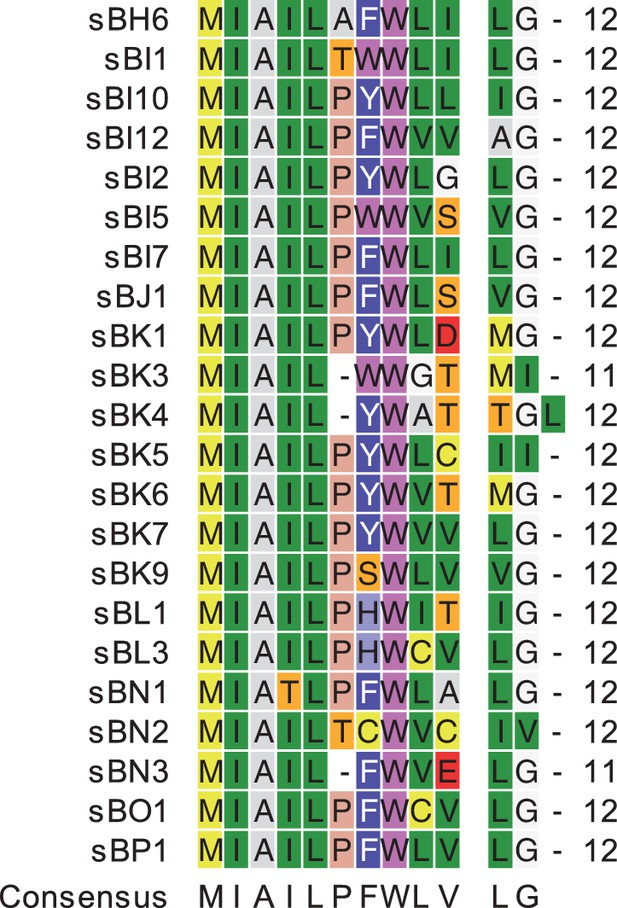
Sequence conservation of ScuR/SarF activating peptides.
Sequence alignment of the 22 peptides activating ScuR or SarF. Residues are color-coded according to the Rasmol color scheme. The consensus sequence is shown underneath with the following nomenclature. The formatting of the multiple alignment has been generated using CLC Main Workbench 7 (https://www.qiagenbioinformatics.com/).
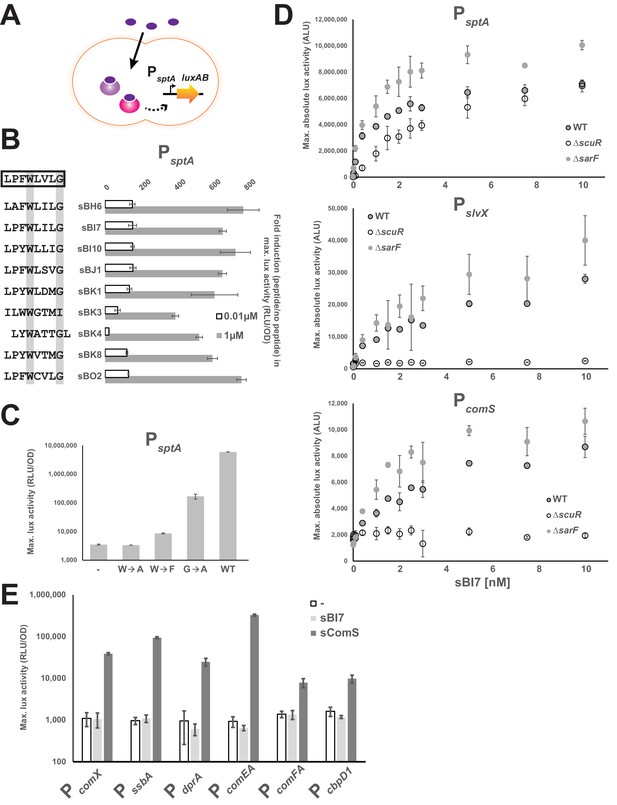
The ScuR/SarF system responds to exogenous peptides.
(A) Cartoon depicting the ScuR/SarF-mediated activation of PsptA upon addition of exogenous synthetic peptides. (B) Fold increase in maximal PsptA activity upon addition of representative synthetic peptide (0.01 or 1 µM) vs mock conditions. Peptide sequences are associated to peptide names and compared to the consensus motif (open box). The highly conserved W and G residues are highlighted with gray boxes. (C) Maximal activity of PsptA exposed to WT and mutant sBI7 peptides (1 nM) (Figure 4—figure supplement 1). (D) Dose response dot plot of sptA, slvX and comS promoter activity upon sBI7 induction at various concentrations in nM (maximal absolute luciferase activity). Promoters were tested in WT strain and ΔscuR or ΔsarF mutants (Figure 4—figure supplement 2). (E) Maximal activity of PcomX and ComX-dependent promoters exposed to the sComS (light gray) or sBI7 (dark gray) peptide (1 µM) in comparison to basal activity (open box) (Figure 4—figure supplement 3). (B, C, D, and E) Experimental values represent averages (with standard error of the mean, SEM) of at least three independent replicates. Some standard errors are too small to be visualized in (B), (D), and (E).
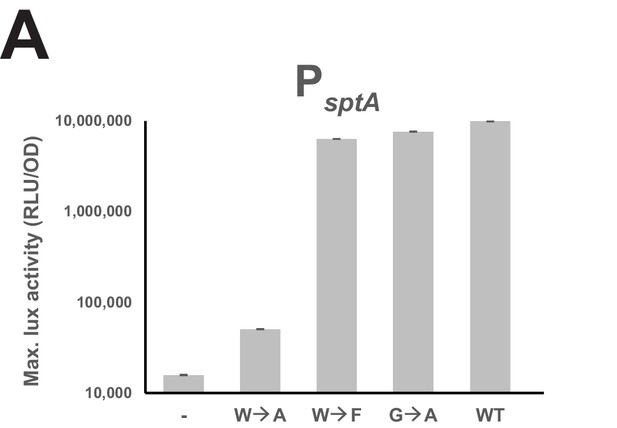
Amino acid requirements for the sBI7-mediated effect.
Maximal luciferase activity of PsptA (RLU/OD; logarithmic scale) exposed to sBI7 (WT) and mutant peptides (1 µM). Experimental values represent the averages (with standard error of the mean, SEM) of at least three independent biological replicates.
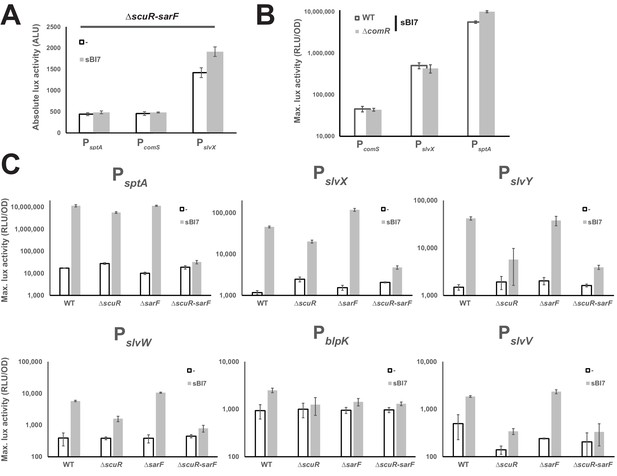
Loss of ScuR/SarF, but not ComR, annihilates the sBI7-mediated effect.
(A) Activity of sptA, comS and slvX promoters (absolute maximal luciferase activity; ALU) in ΔscuR-sarF challenged with sBI7 synthetic peptide (gray bars) vs mock conditions (open bars). (B) Maximum luciferase activity/OD600 ratio (RLU/OD; logarithmic scale) of comS, slvX and sptA promoters fused to a luxAB reporter system in WT (open bars) or ΔcomR (gray bars) strain activated with the sBI7 synthetic peptide. (C) Maximal activity of sptA and salivaricin promoters exposed to the peptide sBI7 (1 µM) in WT strain and ΔscuR or ΔsarF mutants. (A, B and C) Experimental values represent the averages (with standard error of the mean, SEM) of at least three independent biological replicates.
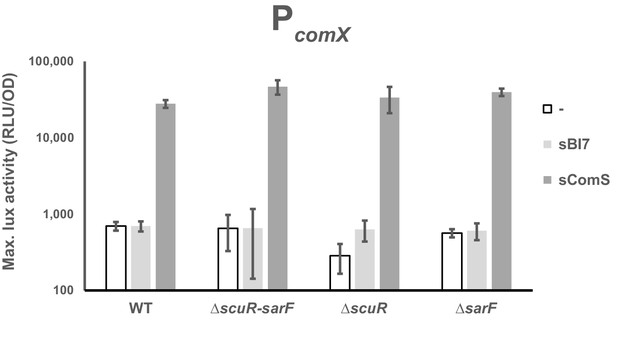
sBI7 has no effect on the comX promoter activation.
Maximal activity of PcomX exposed to the sComS or sBI7 peptide (1 µM) in WT strain and ΔscuR or ΔsarF mutants. Experimental values represent the averages (with standard error of the mean, SEM) of at least three independent biological replicates.
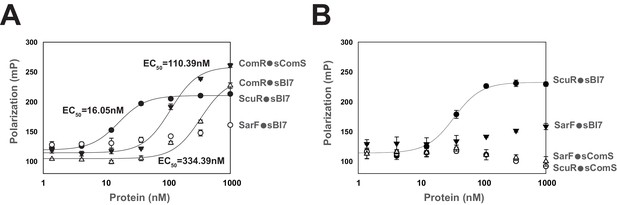
Peptide binding specificity toward ComR paralogs.
(A and B) Fluorescence polarization of synthetic peptide-regulator pairs. Effective concentration of peptide was 10 nM. Hill equation was used to fit sigmoid curves on profiles that reach saturation, namely the ComR•sComS, ComR•sBI7, and ScuR•sBI7 couples, and calculate an EC50 affinity factor. Experiments (A) and (B) were performed independently. (A) Titration of ComR, ScuR and SarF with a fixed concentration of their cognate or non-cognate peptide. (B) Titration of ScuR (circle) and SarF (triangles) with a fixed concentration of their cognate (sBI7; black symbols) or non-cognate (sComS; open symbols) peptides. Experimental values represent averages (with standard error of the mean, SEM) of at least three independent replicates. Some standard errors are too small to be visualized.
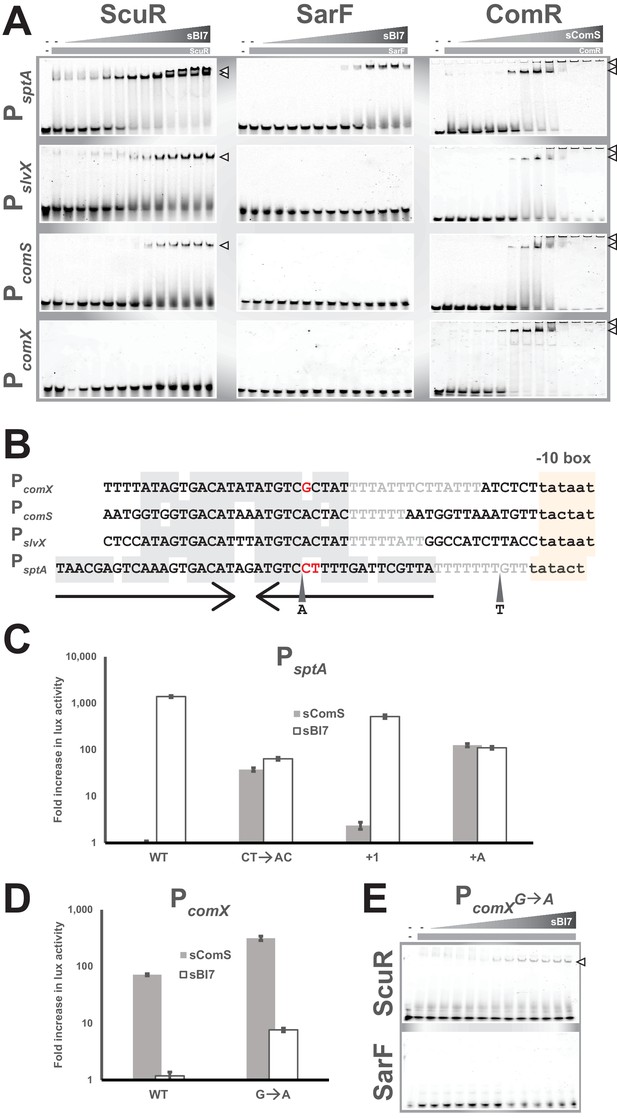
Singularities in promoter recognition of ComR paralogs.
(A) Mobility shift assays of comX, comS, slvX and sptA promoter probes conducted with purified ComR paralogs and decreasing concentrations of their cognate peptide (gray triangles; 2:2 dilutions from 20 µM). Probes are 30 bp (or 40 bp for PsptA), were Cy3-conjugated and used at 40 ng. Protein concentration remained constant (gray boxes; 4 µM). Open triangles show ternary complexes (peptide-regulator-DNA) (Figure 6—figure supplement 1). (B) Nucleotide alignment of comX, comS, slvX and sptA promoters. The (pseudo-)palindromic stretches (converging arrows) and the sigma-bound DNA sequence (−10 boxes) are highlighted in gray or yellow, respectively. The characteristic T-rich region is gray-font. Red-marked nucleotides highlight the potential mismatches in the pseudo-palindromic structure of PcomX and PsptA that were substituted to restore a genuine dyad symmetry sequence (see Figures 6C and 4D). A and T represent the position and nature of single nucleotide insertion in the sptA promoter (see Figure 6C) (Figure 6—figure supplement 1). (C and D) Fold increase in maximal luciferase activity of WT and mutated promoters of sptA (C) or comX (D) exposed to sBI7 or sComS (1 µM). Nucleotides substitutions and insertions are shown in Figure 6B. Experimental values represent the averages (with standard error of the mean, SEM) of at least three independent biological replicates. (E) Mobility shift assays of mutated comX promoter probes conducted with a single concentration of ScuR or SarF (gray boxes; 4 µM) and decreasing concentrations of sBI7 peptide (gray triangles; 2:2 dilutions from 20 µM). Open triangles showcase ternary complexes (peptide-regulator-DNA).
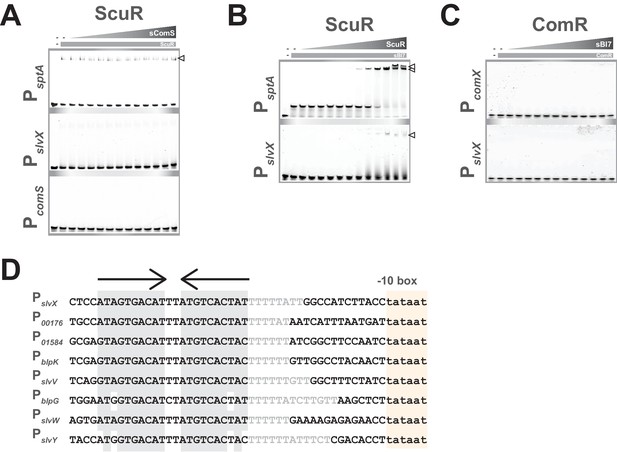
Promoter and peptide specificities toward ScuR and SarF.
(A) Mobility shift assays of comS, slvX and sptA promoter probes conducted with purified ScuR and decreasing concentrations of the non-cognate sComS peptide (gray triangles; 2:2 dilutions from 20 µM). Probes are 30 bp (or 40 bp for PsptA), were Cy3-conjugated and used at 40 ng. Protein concentration remains constant (gray boxes; 4 µM). Open triangle showcases binary complexes (ScuR-DNA). (B) Mobility shift assays of slvX and sptA promoter probes conducted with sBI7 peptide and decreasing concentrations of purified ScuR (gray triangles; 2:2 dilutions from 8 µM). Probes are 30 bp (PslvX) or 40 bp (PsptA), were Cy3-conjugated and used at 40 ng. Peptide concentration remains constant (gray boxes; 1 µM). Open triangles showcase ternary complexes (sBI7-ScuR-DNA). (C) Mobility shift assays of comX and slvX promoter probes conducted with purified ComR and decreasing concentrations of the non-cognate sBI7 peptide (gray triangles; 2:2 dilutions from 20 µM). Probes are 30 bp, were Cy3-conjugated and used at 40 ng. Protein concentration remains constant (gray boxes; 4 µM). (D) Nucleotide alignment of bacteriocin-related gene promoters. The pseudo-palindromic stretches (converging arrows) and the sigma-bound DNA sequence (−10 boxes) are highlighted in gray or yellow, respectively. The characteristic T-rich region is gray font.
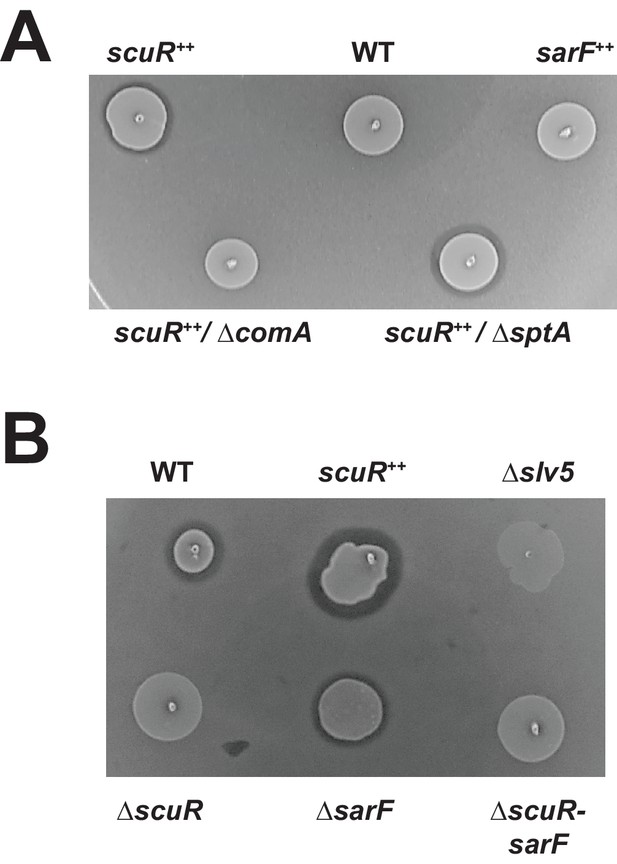
Activated ScuR drives bacteriocin production.
(A and B) Bacteriocin inhibition assay of S. salivarius WT and mutant derivatives. The indicator strain (L. lactis) was embedded in the top soft agar layer, while sBI7 was added into the bottom agar layer as required. Producer strains were spotted on top of the two agar layers. (A) Killing properties of scuR or sarF overexpression mutants compared WT without sBI7 induction. (B) Effect of sBI7 addition (1 µM) on WT strain and various scuR/sarF mutants. scuR ++ and bacteriocin null (Δslv5) mutants were used as positive and negative controls, respectively (Figure 7—figure supplement 1).
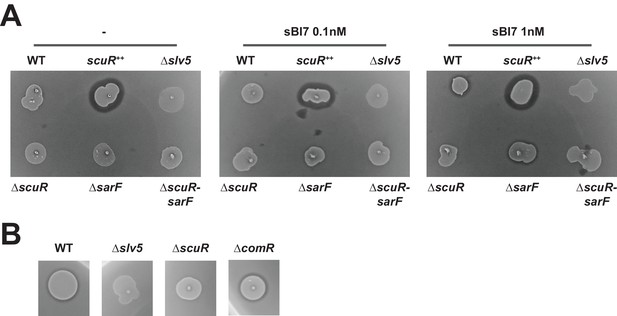
Bacteriocin production through ScuR activation with sBI7.
(A and B) Bacteriocin inhibition assay of S. salivarius WT and scuR/sarF or comR mutant derivatives. The indicator strain (L. lactis) was embedded in the top soft agar layer and producer strains were spotted on top of the two agar layers., (A) sBI7 was supplemented or not into the bottom agar layer as stated. scuR ++ and bacteriocin null (Δslv5) mutants were used as positive and negative control, respectively. (B) the sBI7 pheromone was added at a final concentration of 1 µM. All spots are originated from the same plates but reorganized for optimal visualization.
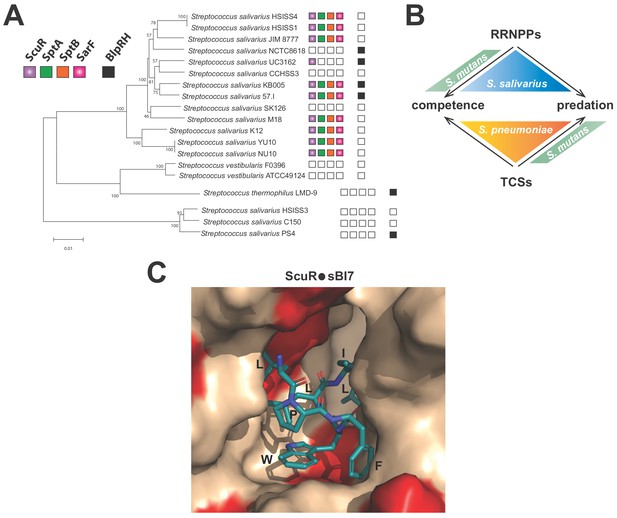
Distribution and activation of ComR paralogs for predation control in S.
salivarius (A) Conservation of ScuR-encoding locus, and BlpRH system across S. salivarius (Supplementary file 5). Functional BlpRH pair (black), ScuR (purple), SptA (green), SptB (orange), and SarF (pink) were screened for homologs in various S. salivarius strains. The phylogenetic tree (100 bootstrap replicates) was adapted from Yu et al. (2015). An empty box means that no functional ortholog was found in the strain genome. Scale bar: 0.01 substitutions per site. (B) Figurative illustration of RRNPPs vs two-component systems (TCSs) drift toward competence and predation regulation in model streptococci (S. pneumoniae, S. mutans and S. salivarius). Arrows symbolize the species-specific control of quorum sensing regulators on developmental processes. (C) Close view of the ScuR peptide-binding pocket. The model of the peptide-bound form of ScuR was obtained by homology modeling using the i-TASSER server and the ComR•sComS•DNA complex (PDB ID 5JUB) as template. It is shown as surface colored in beige except for residues substituted in SarF, wich are highlighted in red. The bound sBI7 peptide is shown in blue sticks and its residues LPFWLIL are labeled. The C-terminal glycine, deeply buried in the pocket, is not visible in this view. The figure was prepared using the graphic software PyMol.
Tables
Competence development (transformation frequencya) in S. salivarius HSISS4 derivatives
https://doi.org/10.7554/eLife.47139.012| Strains | No peptide | sComS | sBI7 |
|---|---|---|---|
| Wild-type | ND | 1.1 (±0.08) E-03 | ND |
| ΔscuR | ND | 0.7 (±0.2) E-03 | ND |
| ΔsarF | ND | 1.5 (±0.5) E-03 | ND |
| P32-scuR | ND | NA | NA |
| P32-sarF | ND | NA | NA |
-
acalculated as the ratio of transformants (chloramphenicol-resistant CFU) to the total CFU count per 0.1 ug of linear DNA. Transformation frequencies are expressed as the arithemtic mean of three independent experiments. Geometric means ± standard deviations are provided. ND: not detected (<1.0 E-08), NA: not applicable.
Additional files
-
Supplementary file 1
Bacterial strains, plasmids and oligonucleotides list, and DNA engineering.
- https://doi.org/10.7554/eLife.47139.019
-
Supplementary file 2
RNAseq data on WT, ΔscuR and ΔsarF strains.
- https://doi.org/10.7554/eLife.47139.020
-
Supplementary file 3
RNAseq data on WT, scuR ++and sarF++ strains.
- https://doi.org/10.7554/eLife.47139.021
-
Supplementary file 4
Random peptide list.
- https://doi.org/10.7554/eLife.47139.022
-
Supplementary file 5
Conservation of ScuR-SarF system in streptococci.
- https://doi.org/10.7554/eLife.47139.023
-
Transparent reporting form
- https://doi.org/10.7554/eLife.47139.024





