In vivo Firre and Dxz4 deletion elucidates roles for autosomal gene regulation
Figures
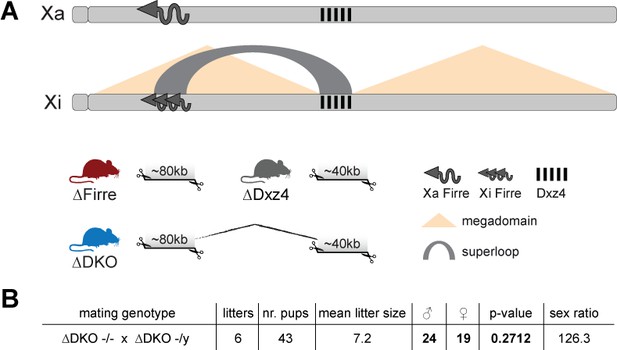
Mice carrying a single or double deletion of Firre and Dxz4 are viable and fertile and show the expected litter sizes and sex ratios.
(A) Schematic representation of the active (Xa) and inactive X (Xi) chromosomes. The deleted loci of the SKO and DKO mouse strains are indicated (Firre (red), Dxz4 (gray) and DKO (blue)). The Firre locus escapes random XCI resulting in a full-length transcript from Xa chromosome and multiple short isoforms from Xi. (B) Sex type results from homozygote intercrosses. The p-value was calculated based on binomial distribution using R (binom.test(x = 19, n = 43, p=0.5, alternative = ‘less’, conf.level = 0.95)) and the sex ratio by using the following formula: number of males/number of females x 100.
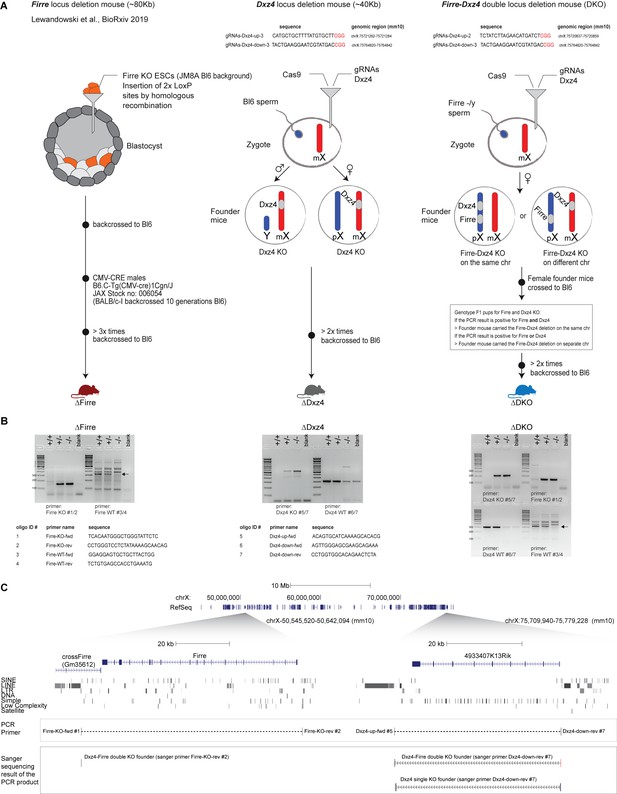
Generation of mice that carry a single or double deletion of Firre and Dxz4.
(A) Schematic overview of the three genome-editing strategies, more details in the Materials and methods section. (B) Genotype strategy and required primers to identify the KO and WT allele for Firre and Dxz4 (C) UCSC genome browser showing the Firre and Dxz4 region. The PCR product of the KO bands was Sanger sequenced, and the resulting sequence was aligned to the UCSC genome browser to confirm the deletion of these loci.
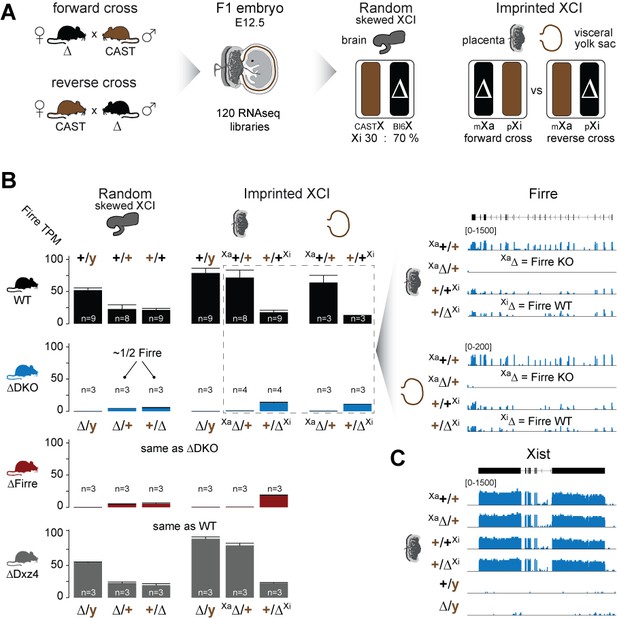
Firre is exclusively expressed from the maternal X chromosome in extra-embryonic tissues.
(A) Allele-specific RNA-seq approach to test the functional impact of the maternal and paternal deletion on random and imprinted XCI using the Allelome.PRO pipeline (Andergassen et al., 2015). While the brain undergoes random/skewed XCI (Bl6 X chromosome inactive in 70% of cells), the placenta and visceral yolk sac undergo imprinted XCI (paternal X chromosome 100% inactive). Allele-specific analysis of the placenta and visceral yolk sac allows to distinguish the effects of maternal inheritance of the deletion (Xa, forward cross) versus paternal inheritance of the deletion (Xi, reverse cross). (B) Firre expression (mean and SD) in female and male brains, placentas and visceral yolk sacs for WT and KO mouse strain. Notably, Firre is approximately 4 times higher expressed from the Bl6 allele compared to the CAST allele, as observed by comparing the expression levels between the forward cross (Xa Bl6) and reverse cross (Xa CAST) in the placenta and visceral yolk sac. (C) Xist expression abundance in placenta for the Firre-Dxz4 double KO strain.
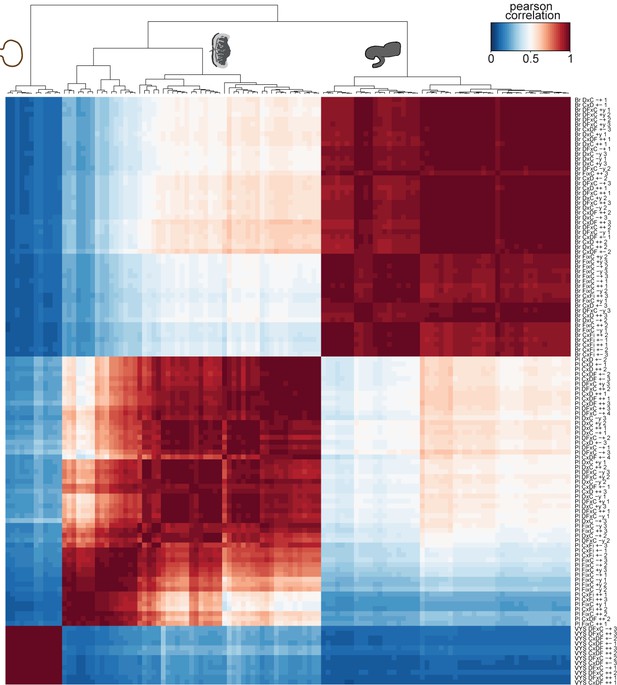
RNA-seq quality control from F1 brains, placentas, and visceral yolk sacs.
Heatmap showing unsupervised clustering of a Pearson correlation matrix (120 brain, placenta, and visceral yolk sac samples) from expression data (TPM), confirming the expected developmental relationship.
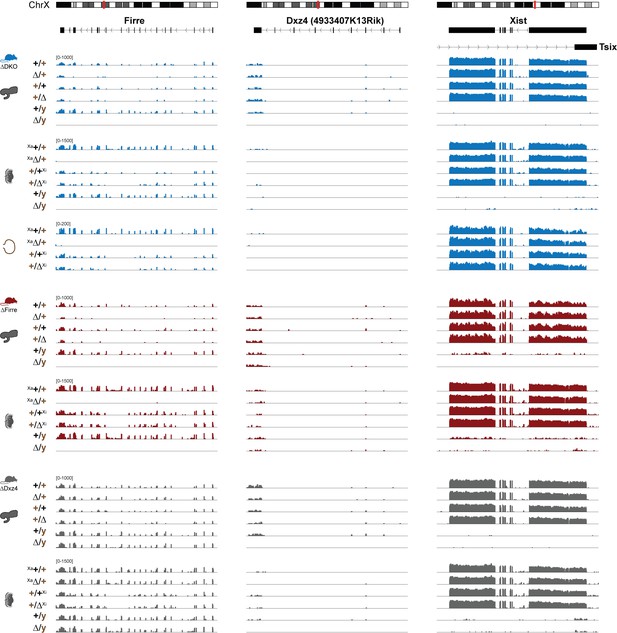
Firre, Dxz4 and Xist expression across tissue, sex and genotype.
Firre, Dxz4 (4933407K13Rik) and Xist expression abundance in brain, visceral yolk sac and placenta, collected from the forward (WT and maternal inheritance of the deletion) and reverse cross (WT and paternal inheritance of the deletion) for all three strains. Only one of the replicates is shown.
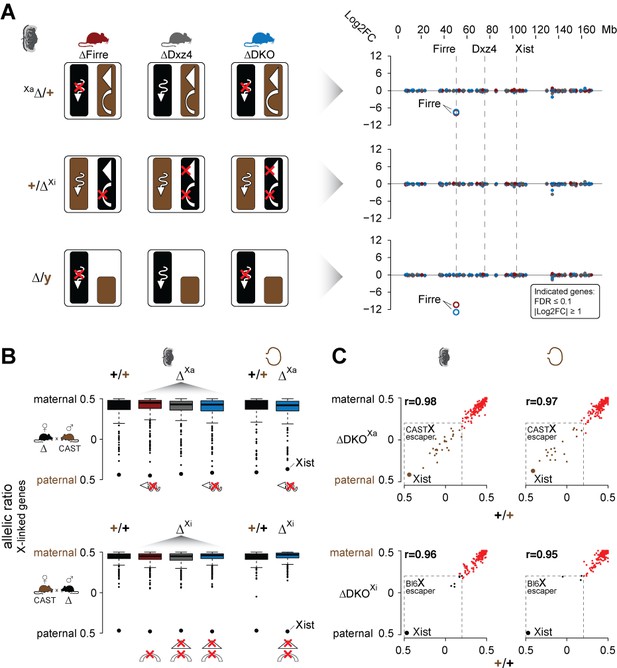
Mice carrying a single or double deletion of Firre and Dxz4 undergo normal random and imprinted X chromosome inactivation (XCI).
(A) Schematic overview showing the effect in the placenta of the deletion in females on Xa (top) or Xi (middle), and in males (bottom) for every KO strain (left). Log2FC across the X chromosome between wildtype and KO strains (right). Firre is the only differentially expressed gene on the X chromosome (DEseq2: FDR ≤ 0.1, |log2FC| ≥ 1). (B) Boxplot showing the allelic ratios for X-linked genes in the placenta and visceral yolk sac in WT and in the KO strains, for the forward cross (deletions on the maternal X = Xa) and reverse cross (deletions on the paternal X = Xi). (C) Scatter plot showing the allelic ratios for X-linked genes in the placenta and visceral yolk sac between WT and DKO on Xa and Xi. Pearson correlation coefficient r. Maternal ratios (red) and strain-specific escaping from the CAST (brown) and Bl6 (black) X chromosome are indicated (dashed line, escaper cutoff: allelic ratio < 0.2).
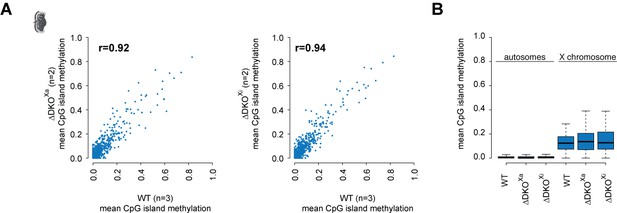
Placentas lacking Firre and Dxz4 on Xaor Xi show a similar methylation levels as in wildtype.
(A) Scatter plot illustrating the relationship between mean CpG islands methylation as measured by reduced representation bisulfite sequencing (RRBS) for wildtype placenta (x axis, n = 3) versus DKO deletion on Xa or Xi (y axis, n = 2). (B) Boxplot showing CpG islands methylation levels on autosomes and on the X chromosome between wildtype and DKO deletion on Xa and Xi.
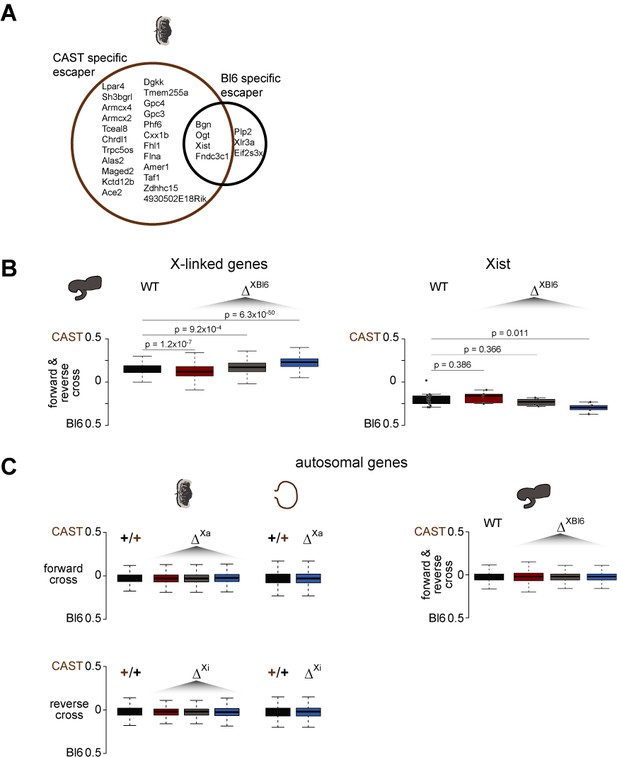
Allelic ratio analysis from F1 brains, placentas and visceral yolk sacs.
(A) Venn diagram showing common and strain specific escaper genes in the placenta. (B) Boxplot showing the allelic ratio for WT and each KO strain of X-linked genes (left) and Xist (right) for the brain, an organ that undergoes random/skewed XCI (Bl6 X chromosome in 70% of cells inactive). The allelic ratio of Xist is significantly skewed towards inactivation of the Bl6 X chromosome in the DKO (t-test, FDR-adjusted p-value=0.0108). (C) Boxplot showing the allelic ratio on autosomes for WT and the three KO strains in the brain, placenta, and visceral yolk sac.
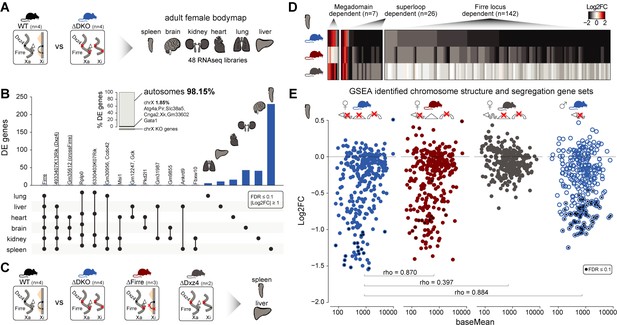
Homozygous deletion of Firre and Dxz4 loci results in organ-specific expression changes on autosomes.
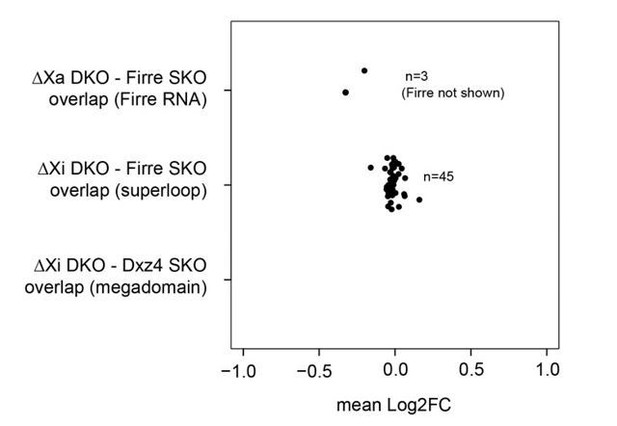
(A) Cartoon illustrating the structural differences of the X chromosome between wildtype and DKO female mice (left), and the organs collected from 4 WT and 4 DKO six-weeks old adult females to generate the transcriptomic bodymap (right). (B) Overview of the differentially expressed genes in the female bodymap and their overlap across the six organs (DEseq2: FDR ≤ 0.1, |log2FC| ≥ 1). The bar plot shows the proportion of differentially expressed genes between autosomes and the X chromosome. Genes that are directly affected by the deletion are underlined (Firre, Dxz4 transcript (4933407K13Rik) and crossFirre (Gm35612, antisense to Firre). (C) Cartoon illustrating the structural differences of the X chromosome between WT and each of the KO strains (left), and the SKO organs collected from six-weeks old adult females (Firre = 3, Dxz4 = 2) (right). (D) Heatmap showing the fold changes of megadomain, superloop and Firre locus dependent gene sets in the spleen. (E) MA plot for all KO strains of the genes extracted from the top five gene sets (GSEA analysis) identified in the DKO and Firre SKO spleen (more details in the Materials and methods section and Supplementary file 1 sheet I). The black dot indicates significant differentially expressed genes (DEseq2: FDR ≤ 0.1).
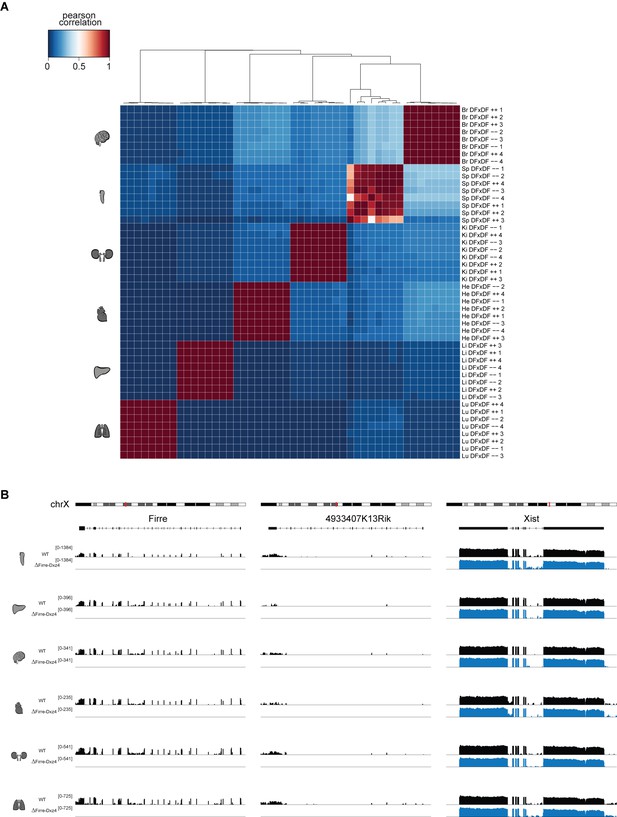
Analyzing adult female organs carrying a homozygous deletion of Firre and Dxz4.
(A) Heatmap showing unsupervised clustering of a Pearson correlation matrix (48 samples, DKO bodymap) from expression data (TPM), confirming the expected developmental relationship. The DKO bodymap includes spleen, brain, kidney, heart, lung and liver from 6 weeks old females (4 WT and 4 DKO replicates). (B) Firre, Dxz4 (4933407K13Rik) and Xist expression abundance in the bodymap organs. One of the replicates is shown.
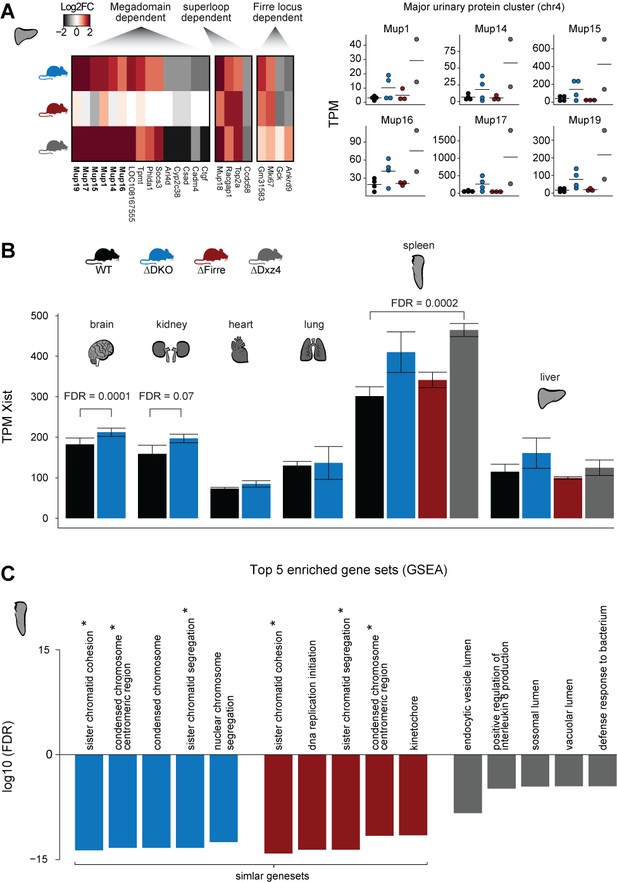
Analyzing adult female organs carrying a homozygous deletion of Firre and/or Dxz4.
(A) Heatmap showing the log2FC of megadomain, superloop and Firre locus dependent gene sets in the liver (left). The major urinary protein gene cluster, which was previously found to show high interindividual expression variation (Thoß et al., 2015) shows megadomain-dependent upregulation (right). (B) Bar plot showing the Xist expression abundance in the bodymap organs. FDR values obtained by DEseq2 analysis. (C) Top five enriched dysregulated gene sets in the spleen of the DKO and of the Firre and Dxz4 SKO (p-value corrected for multiple testing using the p.adjust function in R according to the Benjamini and Hochberg method). The GSEA analysis was performed on DEseq2 test statistics with all GO gene sets (c5.all.v6.2.symbols). Asterix indicates the overlapping gene sets.
Additional files
-
Supplementary file 1
RNA and reduced representation bisulfite sequencing analysis.
(A) Log2FC (lfcMLE) and adjusted p-values (padj) from DEseq2 differential expression analysis, computed by comparing female WT placentas of the forward cross (n = 8) with each of the deletion strains (deletion on Xa, n = 3–4) and female WT placentas of the reverse cross (n = 9) with each of the deletion strains (deletion on Xi, n = 3–4). (B) Log2FC (lfcMLE) and adjusted p-values (padj) from DEseq2 differential expression analysis, computed by comparing male WT placentas of the forward cross (n = 9) with each of the KO strains (n = 3). (C) Methylation levels as measured by reduced representation bisulfite sequencing (RRBS) for WT placenta and DKO deletion on Xa or Xi. (D) Imprinted ratios of X-linked placenta genes for WT and each of the deletion strains (deletions on Xa or Xi, 0.5 = 100% maternal, −0.5 = 100% paternal). The allelic ratios for each replicate per genotype was combined by using the median. (E) Imprinted ratios of X-linked visceral yolk sac genes for WT and DKO (deletions on Xa or Xi, 0.5 = 100% maternal, −0.5 = 100% paternal). The allelic ratios for each replicate per genotype was combined by using the median. (F) Differentially expressed genes (DEseq2 differential expression analysis) in all tissues (bodymap) of DKO female and liver and spleen of SKO female. (G-H) Log2FC of megadomain, superloop and Firre locus dependent gene sets in the liver and spleen. (I) Top enriched gene sets identified in the DKO and Firre SKO spleen. (J) Information of every analyzed sample in this study.
- https://doi.org/10.7554/eLife.47214.013
-
Transparent reporting form
- https://doi.org/10.7554/eLife.47214.014