tec-1 kinase negatively regulates regenerative neurogenesis in planarians
Figures
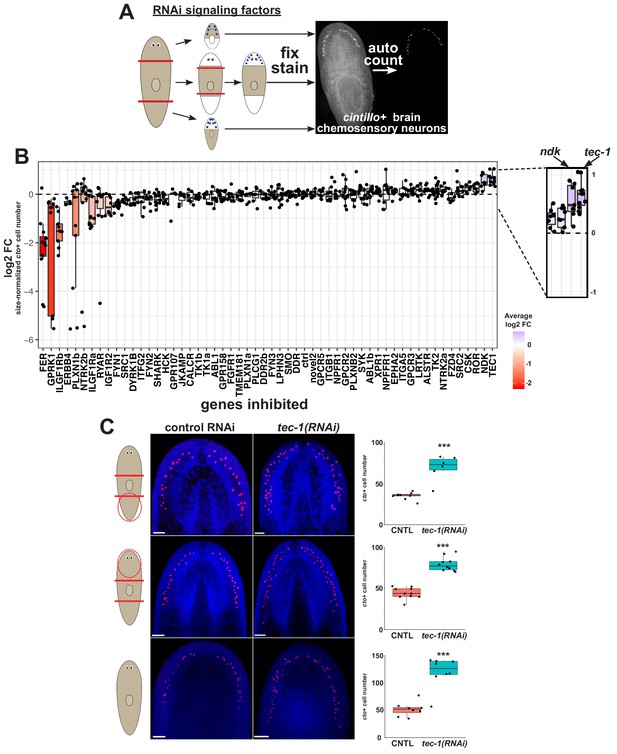
An RNAi screen identifies regulators of neuronal regeneration.
(A) Design of RNAi screen. Animals were fed over several weeks, challenged to regenerate after head and tail amputation, and then fixed and stained for cto expression using Hoechst as a counterstain to detect total animal tissue (heads and tail fragments fixed at d23, while and regenerating trunks were fed dsRNA at d10 then amputated again the following day and then fixed 12 days later). Numbers of cto+ cells were enumerated using CellProfiler and normalized to animal size as measured by Hoechst area in CellProfiler. (B) Log2-transformed fold changes of area-normalized cto-cell number were determined by comparison to similar fragments treated with control dsRNA, combined across head, trunk and tail fragment types for each dsRNA treatment and plotted in ascending order of average log2-fold change. Dots represent log2-fold change for each regenerating fragment, with boxplot shading representing average log2-fold change compared to controls. Knockdown of tec-1 increased cto+ cell number more than nou-darake positive control on average. Dotted line indicates log2FC for control RNAi conditions. (C) To measure tec-1 RNAi’s effect on head regeneration and injury-induced remodeling, animals were fed dsRNA for 2 weeks before decapitation and allowed to regenerate for 16 days. To measure the effects of tec-1 inhibition in homeostasis, animals were fed dsRNA for 60 days and fixed without injury. An increase in cto+ cell number was detected in all contexts. ***p<0.001 by two-tailed t-test. Scale bars: 100 μm.
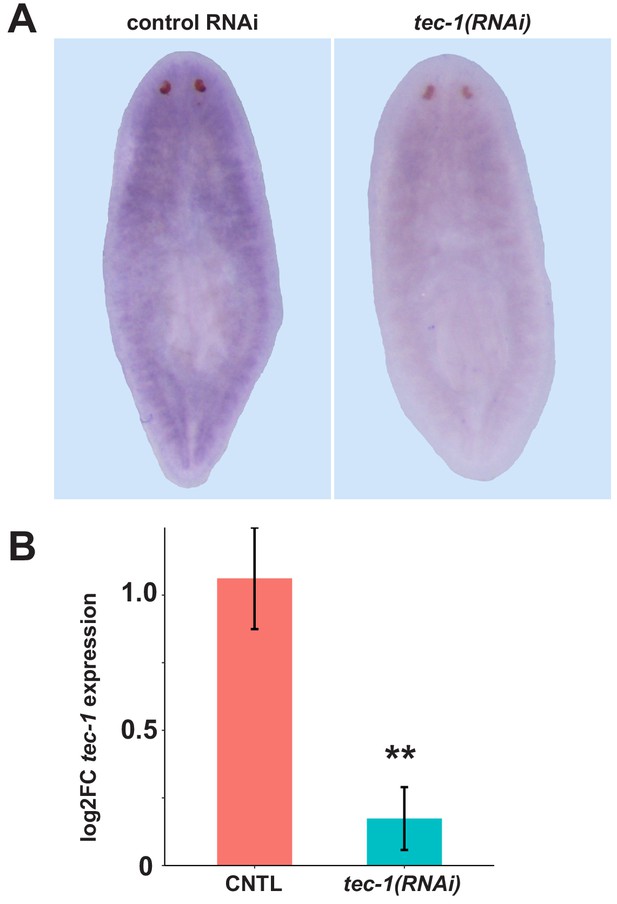
tec-1 mRNA is depleted by dsRNA feeding.
(A) Animals were fed tec-1 or control dsRNA for two weeks, fixed 3 days later, stained with a tec-1 riboprobe, and visualized by colorimetric in situ hybridization. tec-1 in situ hybridization signal was decreased by tec-1 dsRNA in 5/5 animals investigated. (B) Animals were fed dsRNA for 2 weeks and RNA was collected after an additional 17 days. tec-1 mRNA levels were assayed by qRT-PCR and analyzed by ΔΔCt to determine fold change. tec-1 mRNA levels are approximately 5-fold lower in tec-1(RNAi) animals. Significance determined by two-tailed t-tests (n.s., p>0.01). Error bars = s.d.

tec-1 and tec-2 encode Tec family kinases.
(A) Predicted protein sequences for dd_Smed_v6_4818_0_1 (tec-1) and dd_Smed_v6_4313_0_1 (tec-2) were aligned to non-receptor tyrosine kinase sequences from well-annotated proteomes. The clade containing known Tec kinases is marked in blue. SMED-TEC-1 and SMED-TEC-2 consistently cluster together as TFKs and did not consistently cluster with any of the five mammalian TFKs. (B) Human Tec protein domain structure compared to TEC-1 and TEC-2 protein domain structure, as predicted by SMART [107, 108]. Species codes: mm = Mus musculus, hs = Homo sapiens, dm = Drosophila melanogaster, dr = Danio rerio, ce = Caenorhabditis elegans).
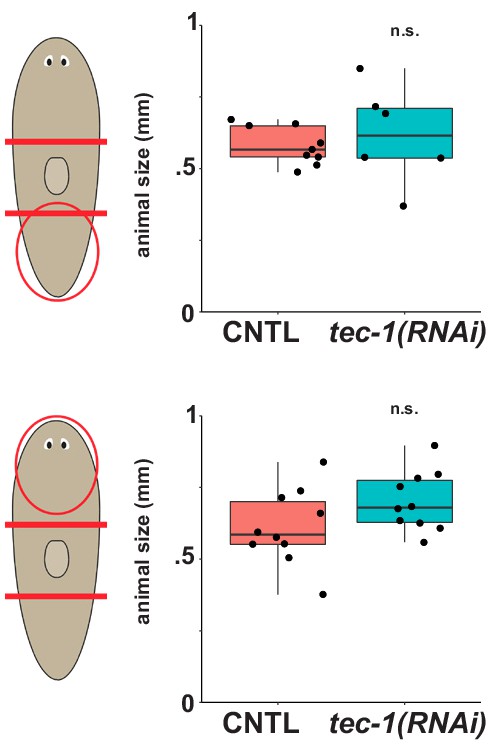
tec-1 RNAi does not alter body size.
Data from regenerating animals in Figure 1C. The area of the animals was measured using Hoechst nuclear stain. Despite the increase in cto+ cell numbers from Figure 1C, animal size is not significantly altered after tec-1 inhibition in the context of both regenerating heads (top) and remodeling heads (bottom) (n.s., p>0.05 by two-tailed t-test).
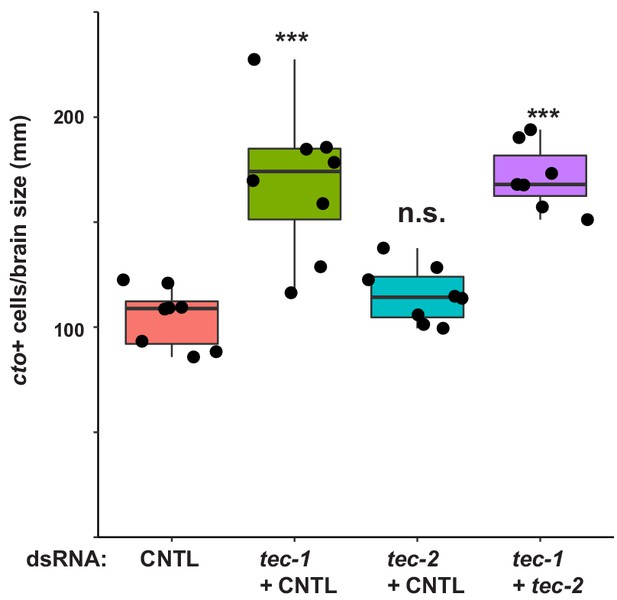
tec-2 does not act with tec-1.
Animals were fed dsRNA to inhibit tec-1, tec-2 or both genes simultaneously as indicated, amputated to remove heads and tails, and allowed to regenerate for 15 days. Regenerating trunk fragments were fixed and stained for cto expression, and relative cto+ cell abundance plotted by normalizing cto+ cell number to animal size measured by the square root of animal area determined by Hoechst staining. tec-2 RNAi did not increase cto+ cell number, nor did it enhance tec-1 RNAi (***, p<0.001, n.s., p>0.05 by two-tailed t-test).
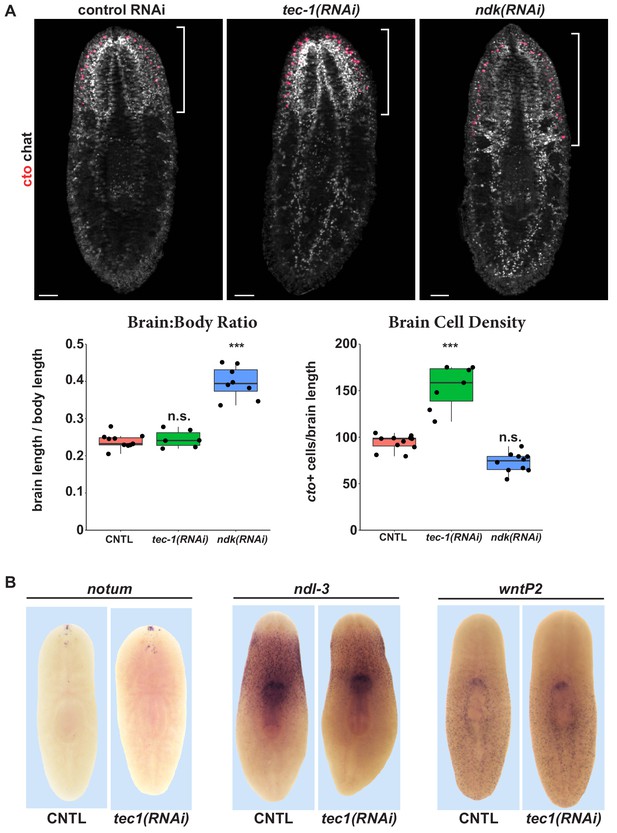
tec-1 inhibition increases chemosensory neuron abundance but not head regionalization.
(A) Animals were fed dsRNA for 2 weeks, amputated to remove heads and tails, and allowed to regenerate for 16 days. FISH was performed on trunk fragments to simultaneously measure cto+ chemosensory neurons and chat+ cholinergic neurons that allow visualization of the brain. Effects on the brain:body size and density of cto+ cells normalized to brain size were quantified. ndk RNAi resulted in an increase in brain:body size but not in numbers of cto+ cells normalized to brain size. By contrast, tec-1 RNAi increased density of cto+ cells within the brain without increasing the proportion of the body axis occupied by the brain. (B) tec-1 RNAi did not strongly affect the expression of anterior-posterior positional control genes notum, ndl-3, and wntP-2 as measured by WISH (images representative for 27/27 animals probed). Scale bars: 100 μm.
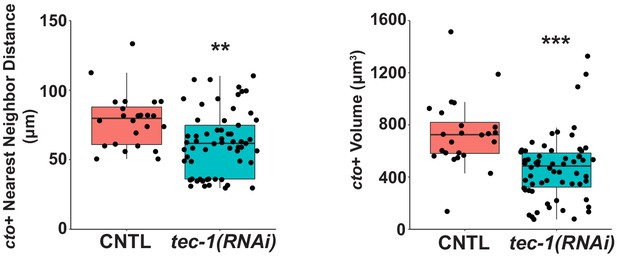
tec-1 inhibition reduces the nearest-neighbor distance and volume of cto+ neurons.
Day 16 regenerating tails were stained for cto. Confocal image stacks were analyzed using Fiji to determine cell volume and nearest-neighbor distances (**, p<0.01; ***, p<0.001).
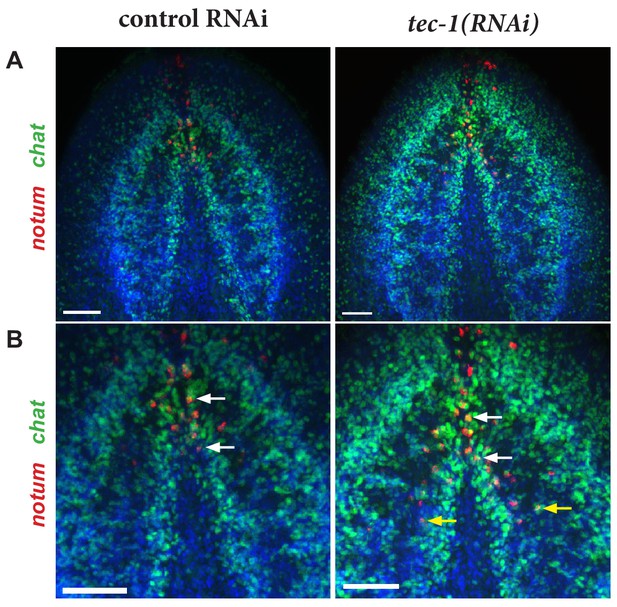
Excess notum+ neurons in the brain after tec-1 inhibition.
(A) Day 15 regenerating head fragments were stained with WISH for notum and chat. (B) The region of the brain commissure containing notum+chat+ neurons from (A). Example brain commissure notum+chat+ cells are marked with white arrows. Examples of excess brain commissure notum+chat+ cells in tec-1 RNAi animals are marked with yellow arrows. Images show Hoechst-stained maximum projections. Scale bars: 100 μm.
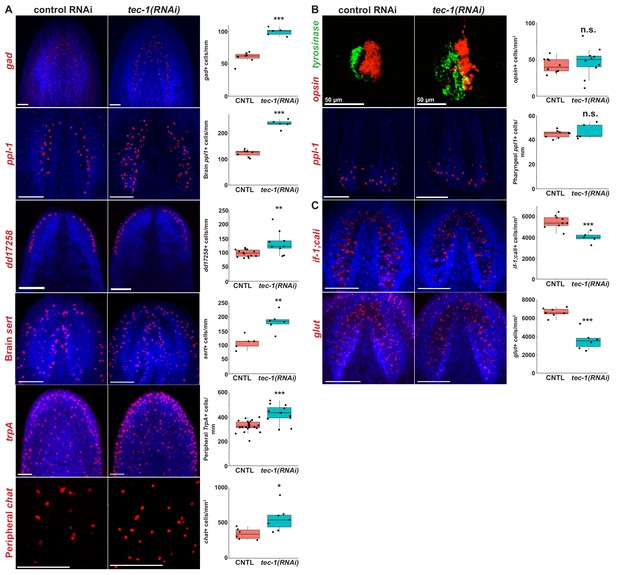
tec-1 inhibition increases abundance of several CNS and PNS neurons.
(A-C) Animals were fed dsRNA for two weeks, amputated to remove heads and tails, and allowed to regenerate for 15 days. Regenerating head fragments were fixed and stained for gad expression, regenerating tail fragments were stained for ppl-1, trpA, opsin/tyrosinase, glut, or if-1/cali expression, and regenerating trunks were stained for chat or dd17258 expression. Cell types amenable to total animal enumeration were quantified by normalizing cell number to body size by dividing by the square root of whole animal area (gad+, brain and pharynx ppl1+, brain sert+, dd17258+, opsin+ cells). Abundances of cell types too numerous for whole-body counting were quantified by manually scoring cell numbers in a region of interest and normalizing to the area of that region (trpA+ cells were scored in an anterolateral region, peripheral chat+ neurons scored in a postpharyngeal ventromedial region, if-1+;cali, and glut+ cells scored within the brain defined by Hoechst staining). (A) tec-1 RNAi animals had increased numbers of gad+ neurons, ppl-1+ neurons within the brain, dd17258+ neurons, sert+ brain neurons, trpA+ peripheral neurons, and chat+ peripheral neurons. (B) tec-1(RNAi) animals regenerated disorganized photoreceptors but had no alteration in opsin+ cell numbers. Likewise, tec-1 inhibition did not alter numbers of pharyngeal ppl-1+ neurons. (C) tec-1 knockdown decreased the density of glut+ and pooled if-1/cali+ glial cells in the brain. Significance determined by two-tailed t-tests (*, p<0.05; ***, p<0.001; n.s. p>0.05). Images show Hoechst-stained maximum projections except for maximum-projected pharyngeal ppl-1+ cells shown for clarity overlayed with a single slice of Hoechst-labeled pharynx tissue. Scale bars: 100 μm unless otherwise noted.
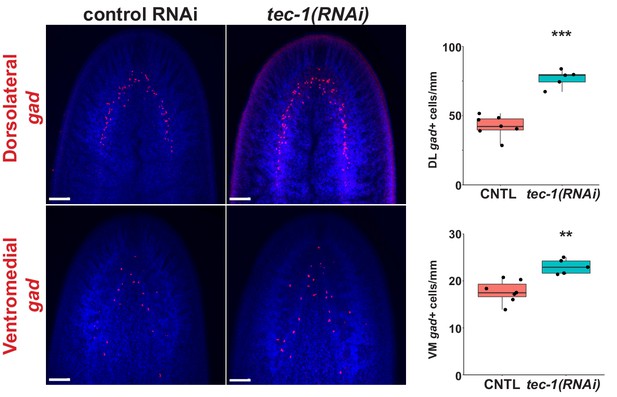
Two GABAergic neuron populations are regulated by tec-1.
Images and quantification of dorso-lateral (top) and ventro-medial populations (bottom) gad+ neurons in animals from Figure 3A (**p<0.01, ***p<0.001 by two-tailed t-test). Scale bars: 100 μm.
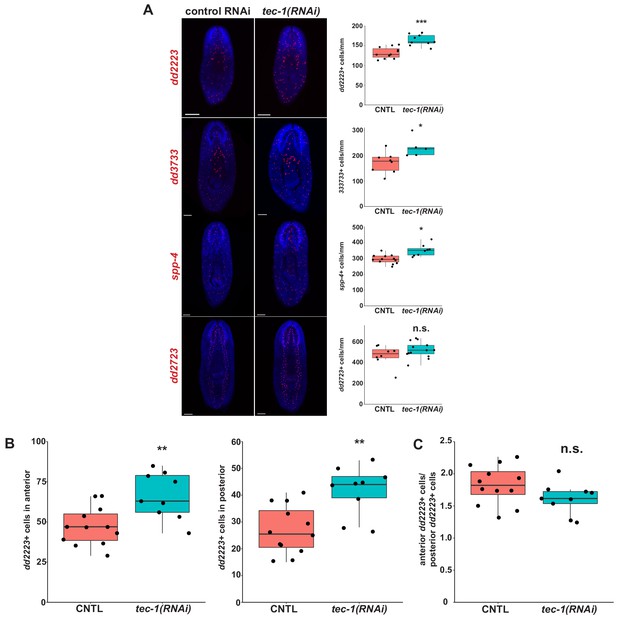
Effects of tec-1 inhibition on neurons expressed across the A-P axis.
Animals were fed dsRNA for two weeks, amputated to remove heads and tails, and allowed to regenerate for 15 days. (A) Regenerating trunks were stained for markers of neuronal subpopulations that are expressed throughout the planarian body and quantified for changes in tec-1 RNAi (left). These genes were chosen for their body-wide expression, countability, and specficity to putatitve cell types identified by Fincher et al (right). (B and C) dd2223+ cells from (A) were scored manually for their position along the body in ImageJ, binned into either the anterior half or posterior half of the body, and counted. (B) Cell numbers in the anterior and posterior halves of the tec-1(RNAi) animals are significantly higher than those of CNTL animals. (C) The ratio of anterior dd2223+ cells to posterior dd2223+ cells was not altered in tec-1(RNAi) animals, suggesting tec-1 can act equally across the entire body axis (*, p<0.05; ***, p<0.001; n.s. p>0.05). Significance determined by two-tailed t-tests (*, p<0.05; n.s., p>0.05). Scale bars: 100 μm.
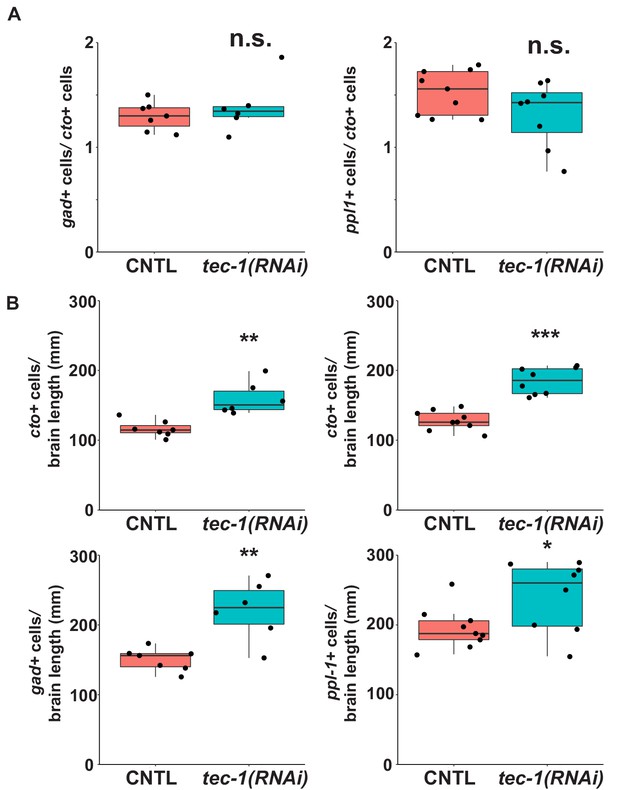
Additional analysis of neuron density in tec-1 RNAi.
Day 15 regenerating trunk fragments were fixed and stained for cto and either gad or ppl-1expression. (A) The ratio of cells expressing each marker was determined for each animal, and the difference between tec-1(RNAi) and CNTL animals was not found to be statistically significant for either pair of markers. (B) The animals stained for cto and gad (left) and the animals stained for cto and ppl-1 (right) had increased neuronal cell density. Significance determined by two-tailed t-tests (*, p<0.05;**, p<0.01; ***, p<0.001, n.s., p>0.05).
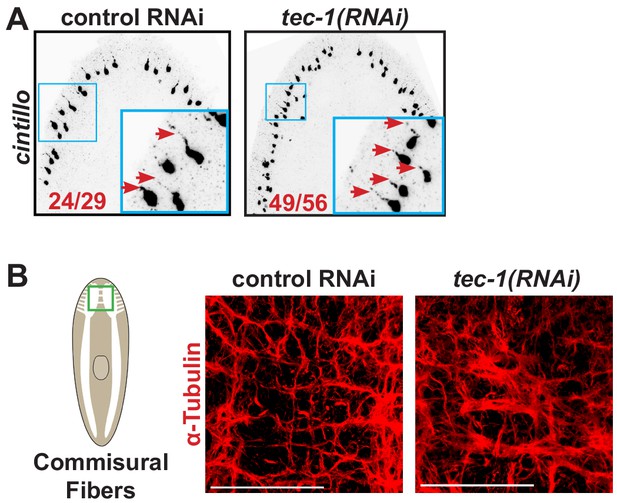
Tec-1(RNAi) neurons form projections.
(A) FISH to detect cintillo identified projections associated with cto+ cell bodies (red arrows). Such processes could be identified in the majority cells from control and tec-1(RNAi) animals (red numbers). (B) α-tubulin staining to detect commissural fibers joining the cephalic ganglia. Fibers appeared thicker in tec-1(RNAi) animals. Scale bars: 100 μm.
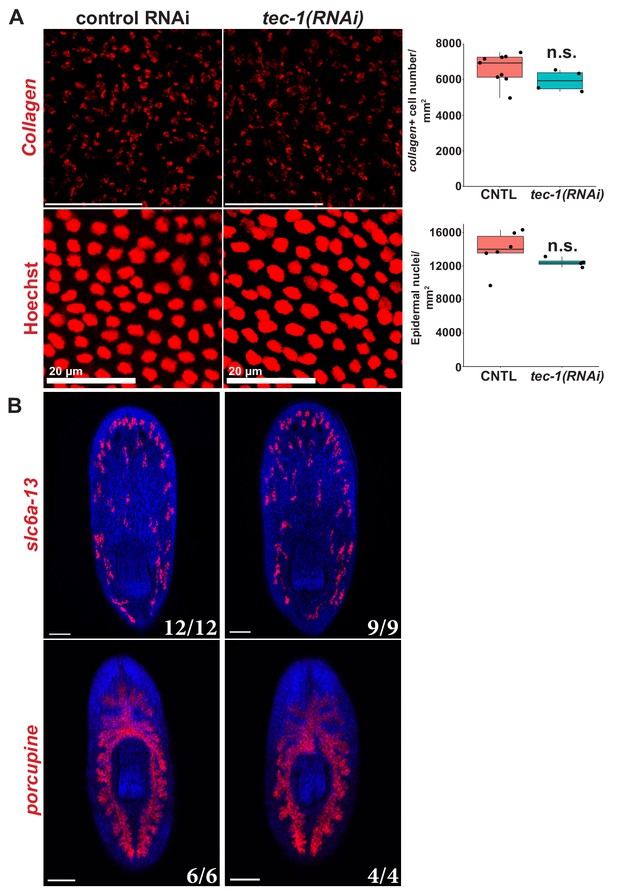
Non-neural cell types not affected by tec-1 RNAi.
(A) Day 15 regenerating trunk fragments were stained by FISH used to measure the density of body-wall muscle and epidermis by quantifying the numbers of collagen+ muscle cells and Hoechst+ epidermal nuclei scored in medial postpharyngeal and pre-pharyngeal domains, respectively, and normalized to the size of the imaged field. (B) Day 15 regenerating head or tail fragments were stained with FISH for slc6a-13, marking excretory cells, or porcupine, marking gut tissue, respectively. Images show Hoechst-stained maximum projections. (n.s., p>0.05 by two-tailed t-test). Scale bars: 100 μm except where otherwise noted.
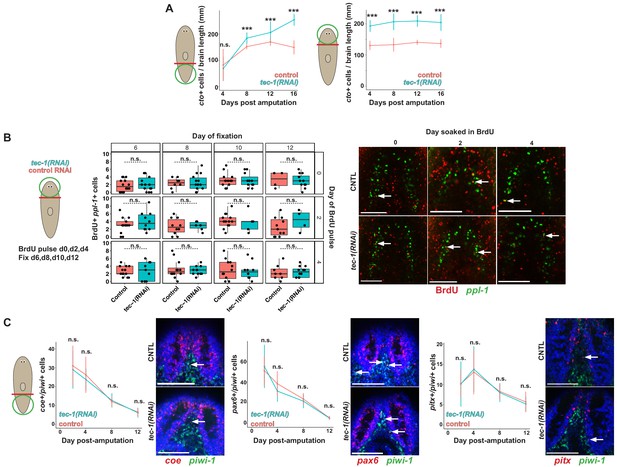
tec-1 inhibition does not increase rates of neuron differentiation.
All animals were fed dsRNA for 2 weeks before surgeries as indicated by cartoons. (A) Time courses of cto+ cell number in regeneration of a new head (left) or remodeling of a pre-existing head (right). Each data point represents sample size of between 4 and 12 animals. (B) Animals were transtioned into high-salt water one week before surgery, and either whole animals (day 0) or head fragments (days 2 and 4) were soaked in BrdU for 4 hr. Heads fragments were fixed at days 6, 8, 10, and 12 post-amputation. Co-staining of control versus tec-1(RNAi) animals for ppl-1 mRNA and BrdU showed no detectable increase in numbers of BrdU+ppl-1+ cells at any time point (left). Single confocal slices show staining of BrdU and ppl-1 in day 12 animals (right). White arrows show colocalization. (C) Regenerating tail fragments were fixed at 2, 4, 8, or 12 days post injury and stained with FISH to detect piwi-1 and coe or pax6 transcription factors, which label broad neuronal progenitors, or pitx transcription factor, labeling progenitors of serotonergic neurons. Single confocal slices show staining of transcription factors, piwi-1, and Hoechst counterstain in day 12 animals. No significant differences in numbers of neural progenitor cells were detected (***, p<0.001, n.s. p>0.05 by two-tailed t-test, error bars represent standard deviation). Scale bars: 100 μm.
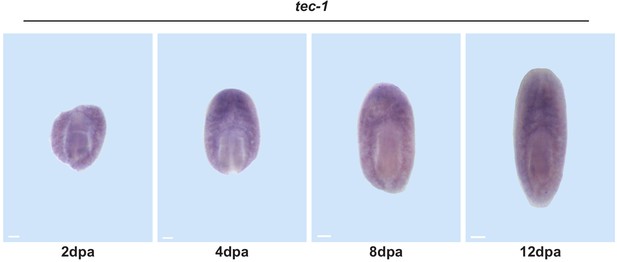
tec-1 is expressed during regeneration.
Trunk fragments were fixed 2, 4, 8, and 12 days post amputation and stained for tec-1 expression. tec-1 is expressed throughout the animal, including the anterior, throughout regeneration. Scale bars: 100 μm.
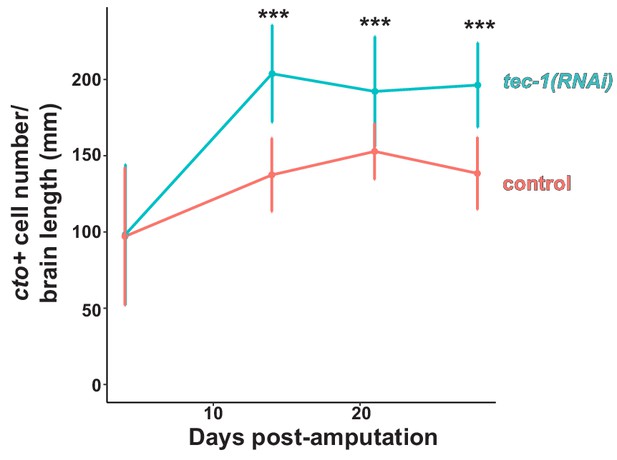
tec-1 RNAi phenotype reaches a steady state by 2 weeks of head regeneration.
Trunk fragments were fixed 4, 14, 21, and 28 days post amputation and stained for cto expression. The increase in cto+ cell number after tec-1 inhibition was sustained for several weeks, suggesting cto+ neurons reach steady-state levels after two weeks of regeneration under these conditions. Significance determined by two-tailed t-tests (***, p<0.001, error bars represent standard deviation).
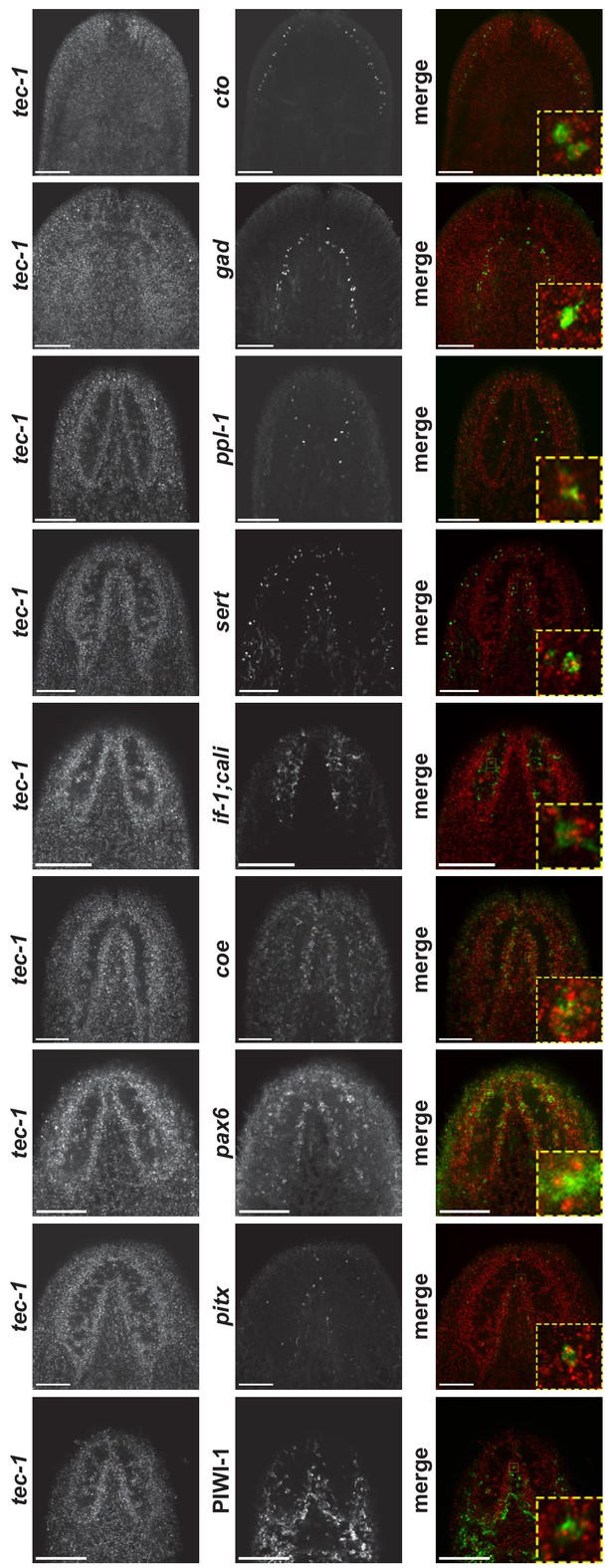
tec-1 is broadly expressed in neural cells and progenitors.
Animals were stained with tec-1 riboprobe and the indicated neural or progenitor marker (left and center) using in situ hybridizations or immunostainings (for PIWI-1 protein). Channels are merged (right) and a magnified image demonstrating colocalization is shown (right inset). Images show a single confocal slice. Scale bars: 100 μm.
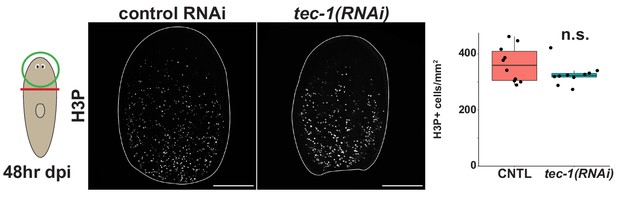
tec-1 RNAi does not affect mitotic activity 48 hr after injury.
Animals were fed dsRNA for 2 weeks before amputating and fixing at 48 hr to examine mitotic activity at the time of a proliferative burst in early regeneration. Regenerating head fragments were immunostained to detect phosopho-ser10-Histone3, and no significant difference in numbers of H3P+ cells were detected (n.s. p>0.05 by two-tailed t-test). Scale bars: 100 μm.
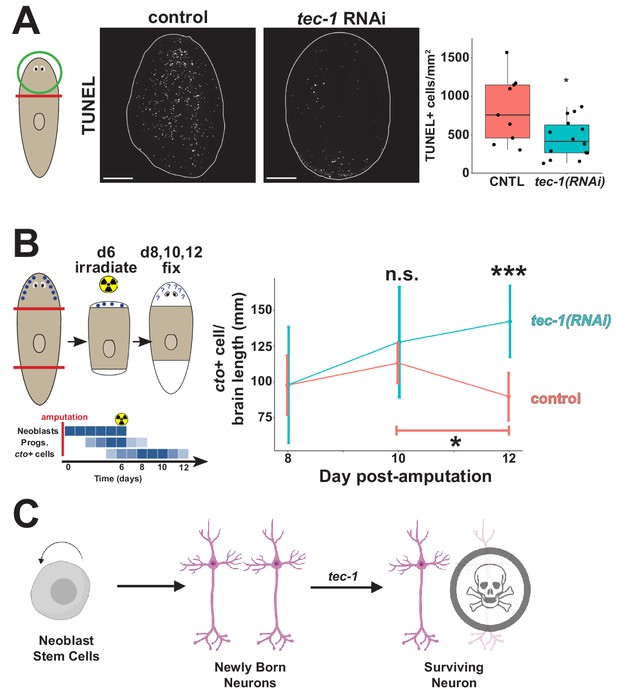
tec-1 promotes cell death and limits survival of new neurons.
Animals were fed dsRNA for 2 weeks before amputation to measure numbers of dying cells and persistence of regenerated chemosensory neurons after lethal irradiation. (A) Head fragments were TUNEL stained 72 hr after injury and numbers of TUNEL+ cells normalized to fragment area quantified to find that tec-1 inhibition resulted in diminished numbers of dying cells. (B) Amputated trunk fragments were allowed to regenerate for 6 days, treated with 6000 rads of X-rays to eliminate neoblasts and subsequent neuron differentiation, then fixed at the indicated times post-injury, and numbers of cto+ cells detected and counted by FISH and normalized to brain length determined by Hoechst staining. Schematic shows predicted effects of lethal irradiation on population abundances (blue shading) for cells involved in producing new cto+ neurons in head regeneration: neoblasts, neural progenitors, and differentiated CNS neurons after irradiation during head regeneration over the course of days. In control animals, the density of cto+ neurons (red) decreased between four and six days after irradiation (bracket). By contrast, tec-1(RNAi) animals had normal numbers of newly formed cintillo+ cells, cto+ number is not significantly greater than controls at d10 post amputation, but is significantly greater than controls two days later. Each data point represents sample size of between 4 and 9 animals. (C) Model of normal neuronal production, where tec-1 acts to cull excess neurons in homeostasis and regeneration (*p<0.05, **p<0.01, ***p<0.001, n.s. p>0.05 by two-tailed t-test, error bars represent standard deviation). Scale bars: 100 μm.
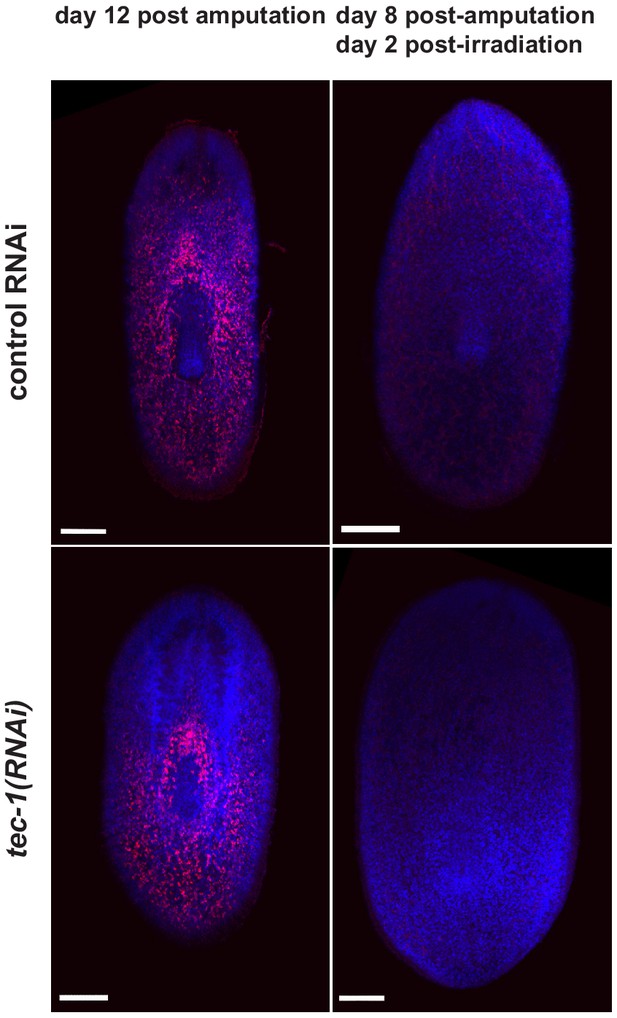
X-ray irradiation ablates neoblasts in tec-1(RNAi) animals.
Trunk fragments were irradiated on day 6 post-amputation and fixed 48 hours later or allowed to regenerate until 12 days post-amputation without irradiation. Animals were stained for piwi-1 and counterstained with Hoechst. Neoblasts were completely ablated in control and tec-1(RNAi) animals, indicating that increased neuronal cell numbers after irradiation are not caused by resistance to irradiation. Scale bars: 100 μm.
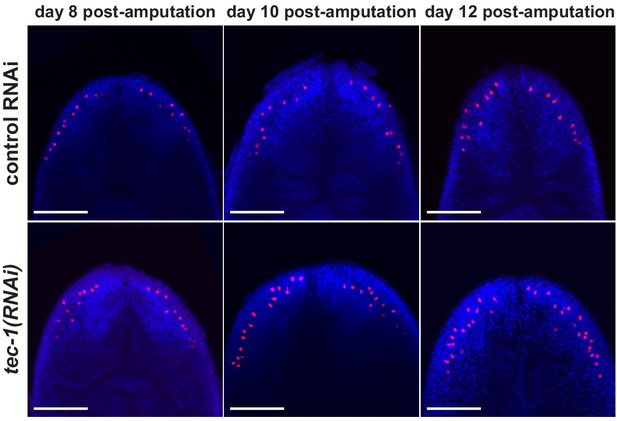
cto expression after irradiation in regenerating animals.
FISH images of regenerating trunk fragments 8-12 days after amputation (left to right) from Figure 5B. All animals were irradiated 6 days post amupation. Scale bars: 100 μm.
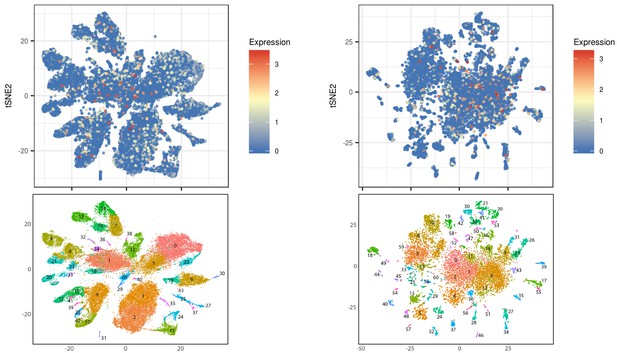
tec-1 is broadly expressed.
Single-cell RNA sequencing and tSNE subcluster analysis from Fincher et al. (2018), comparing tec-1’s expression in all cells and neurons (top) to clusters determined by tSNE (bottom). tec-1 is broadly expressed in numerous different cell types (left). tec-1 is expressed in many different neural subclusters (right) but is enriched in subcluster 6, previously identified as containing neural progenitors (adjusted p-value of 3.11e-23).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Schmidtea mediterranea) | Tec-1 | Planmine | dd_Smed_v6_4818_0_1 | |
| Antibody | Polyclonal rabbit anti-digoxigenin-POD, Fab fragments | Sigma/Roche | # 11207733910 RRID: AB_514500 | 1:2000 dilution |
| Antibody | Polyclonal rabbit anti-fluorescein-POD, Fab fragments | Sigma/Roche | #11426346910 RRID: AB_840257 | 1:2000 dilution |
| Antibody | Polyclonal rabbit digoxigenin-AP | Sigma/Roche | #11093274910 | 1:4000 dilution |
| Antibody | Rat polyclonal Anti-BrdU | Abcam | 6326 | 1:1000 dilution |
| Antibody | Rabbit monoclonal anti-phospho-ser10 Histone H3 | Cell Signaling | D2C8 | 1:3000 dilution |
| Antibody | Mouse monoclonal anti-TUBULIN-ALPHA AB-2 | Thermo/Fisher | MS581P1 | 1:1000 dilution |
| Commercial assay or kit | TUNEL labeling kit | Thermo/Fisher | EP0162 | N/A |
| Antibody | Goat polyclonal anti-rat HRP | Jackson ImmunoResearch | 112-036-072 | 1:1000 dilution |
Additional files
-
Supplementary file 1
Information about genes investigated in RNAi screen.
Table describes ddv6 contig identifier from Planmine, provisional name, and blastx annotation information obtained from Planmine, as well as primers used for cloning cDNA for each gene.
- https://cdn.elifesciences.org/articles/47293/elife-47293-supp1-v1.xlsx
-
Supplementary file 2
RNAi screen data.
Table describes the measurement of cto cell abundance after inhibition of each gene. Genes are described by their ddv6 contig identifier from Planmine and with a provisional name. Data from analysis of head, trunks, and tail fragments were pooled to obtain an average value of cto cell number normalized to animal area and standard deviations calculated. Log2-normalized values of (average cto cells/area) are additionally presented. Unadjusted and Benjamini-Hochberg-adjusted p-values are shown from t-tests to compare cto/area measurements between each indicated RNAi condition and control RNAi treatment (C. elegans unc-22, ‘ctrl’).
- https://cdn.elifesciences.org/articles/47293/elife-47293-supp2-v1.xlsx
-
Supplementary file 3
Data used for plotting figure graphs.
Each subpanel or plot is indicated by the name of the corresponding tab in the file. Data of cell counts or normalized cell counts across specimens is indicated.
- https://cdn.elifesciences.org/articles/47293/elife-47293-supp3-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/47293/elife-47293-transrepform-v1.docx





