Sox1a mediates the ability of the parapineal to impart habenular left-right asymmetry
Figures
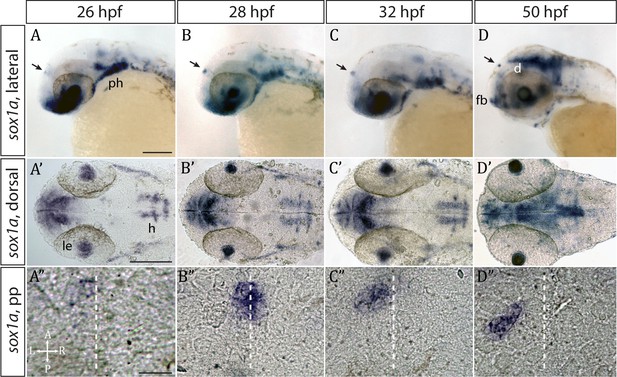
sox1a is expressed in the parapineal from the onset of its formation.
(A–D’) Lateral (A–D) and dorsal (A’–D’) views of zebrafish embryos showing sox1a mRNA expression at stages indicated. In addition to the parapineal (indicated by black arrows in A-D), sox1a is also expressed in the lens vesicle (le), hindbrain (h) and pharyngeal arches (ph). At 50 hpf, sox1a is also detected in the ventral diencephalon (d) and the anterior forebrain (fb). Scale bars 100 µm. (A”–D”) Dorsal views of the epithalamus showing sox1a mRNA expression in the migrating parapineal at stages shown above. Dashed line indicates the midline. Scale bars 25 µm.
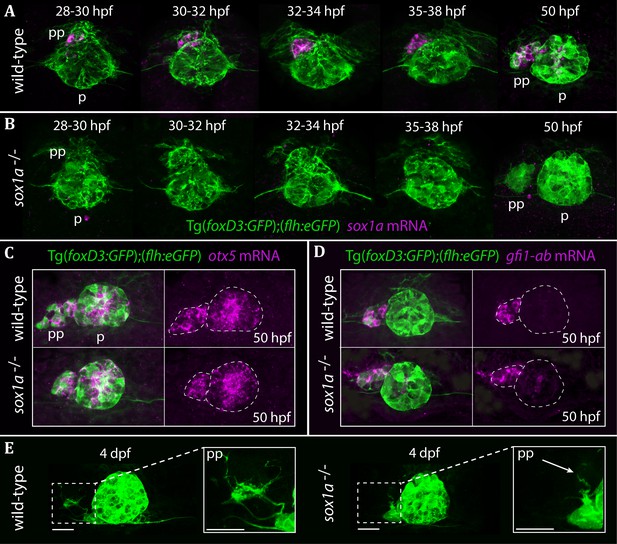
The parapineal is specified and migrates normally in sox1a-/- mutants.
All images show dorsal views of the epithalami of wild-type or sox1a mutant embryos with expression of Tg(foxD3:GFP)zf104 and Tg(flh:eGFP)U711 transgenes (green) in the pineal (p) and the parapineal (pp). mRNA expression of genes indicated is shown in magenta. (A–B) Time-course of parapineal migration in Tg(foxD3:GFP);(flh:eGFP) (A) and sox1a-/- Tg(foxD3:GFP);(flh:eGFP) (B) embryos. Note the absence of sox1a mRNA in the parapineal cells of the sox1a-/- mutants. (C–D) otx5 (pineal and parapineal) and gfi1ab (parapineal) mRNA expression in wild-type and sox1a-/- mutant embryos at 50 hpf. (E) Efferent parapineal projections to the left dHb in wild-type and sox1a mutant embryos at 4 dpf. Note the stunted projection arising from the sox1a-/- parapineal (arrow). Scale bars 25 µm.
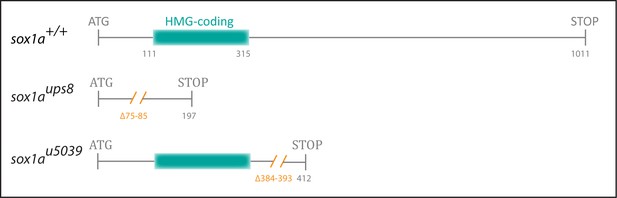
Generation of mutant sox1a alleles.
An 11 bp (sox1aups8) and a 10 bp (sox1au5039) deletion (orange) was introduced into the single 1011 bp exon of sox1a, leading to a premature stop codon at positions 197 and 412, respectively. In sox1aups8, the deletion leads to disruption of the sequence coding for the HMG DNA binding domain (blue), whereas in sox1au5039, the HMG domain sequence is intact.
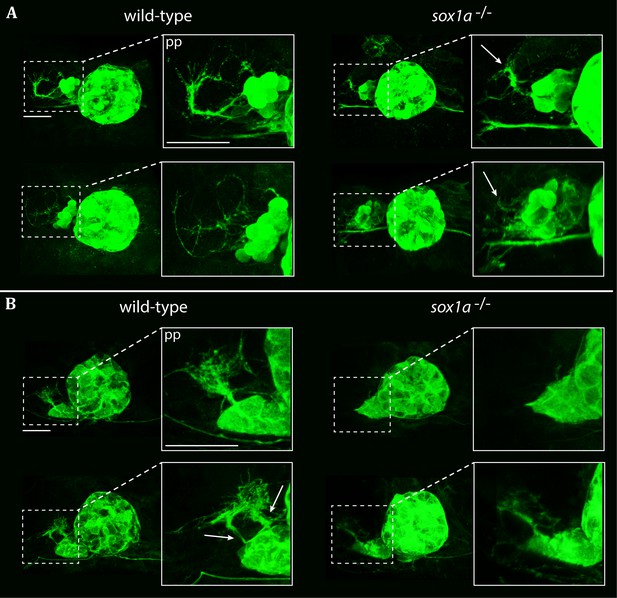
Parapineal projections show reduced growth in sox1a-/- mutants.
Pineal complexes in Tg(foxD3:GFP);(flh:eGFP) and sox1a-/- Tg(foxD3:GFP);(flh:eGFP) embryos. Scale bars 25 µm. pp – parapineal. (A) Parapineal projections at 50 hpf in two typical examples of sox1a-/- mutants and wild-type siblings. In wild-type siblings, parapineal projections exhibit extensive outgrowth in 75% of the embryos (n = 37). Parapineal projections in sox1a-/- mutants show reduced outgrowth in all embryos (white arrows) (n = 30). (B) Parapineal projections at 4 hpf in two typical examples of sox1a-/- mutants and wild-type siblings. In wild-type larvae, parapineal projections show extensive branching in all larvae (n = 31). Two distinct stemming points can often be observed at 4 dpf (white arrows). In sox1a-/- mutants, parapineal projections are either stunted (85% of the larvae) or missing (15% of the larvae) (n = 41).
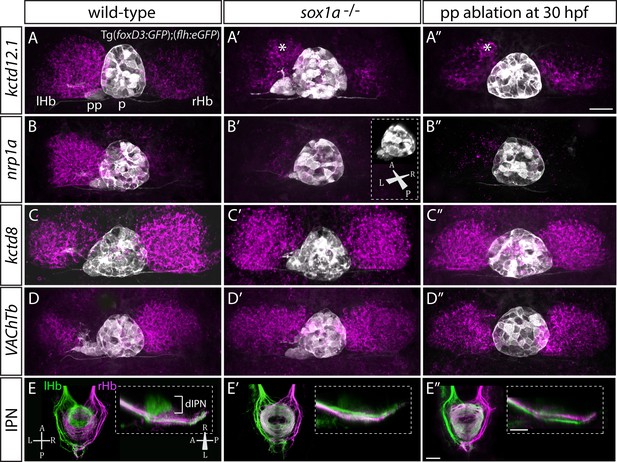
sox1a-/- mutants show a double-right habenular phenotype.
(A–D”) Dorsal views of the epithalami of wild-type (A–D), sox1a mutant (A’–D’) and parapineal-ablated (A’’–D’’) larvae at 4 dpf showing expression of Tg(foxD3:GFP)zf104 and Tg(flh:eGFP)U711 transgenes (grey) in the pineal (p) and the parapineal (pp). Habenular mRNA expression of genes indicated on the left is shown in magenta (lHb – left habenula, rHb – right habenula, asterisk – residual asymmetry). Scale bar 25 µm. (E–E”) Dorsal (left images) and lateral (right images) views of the midbrain interpeduncular nucleus (IPN) labelled by anterograde tracing of axons from left dorsal habenula (lHb, green) and right dorsal habenula (rHb, magenta) at 4 dpf. Note the loss of dorsal IPN (dIPN) innervation by the left habenula in the sox1a-/- mutant (E’) and parapineal-ablated larva (E”). Scale bars 25 µm.
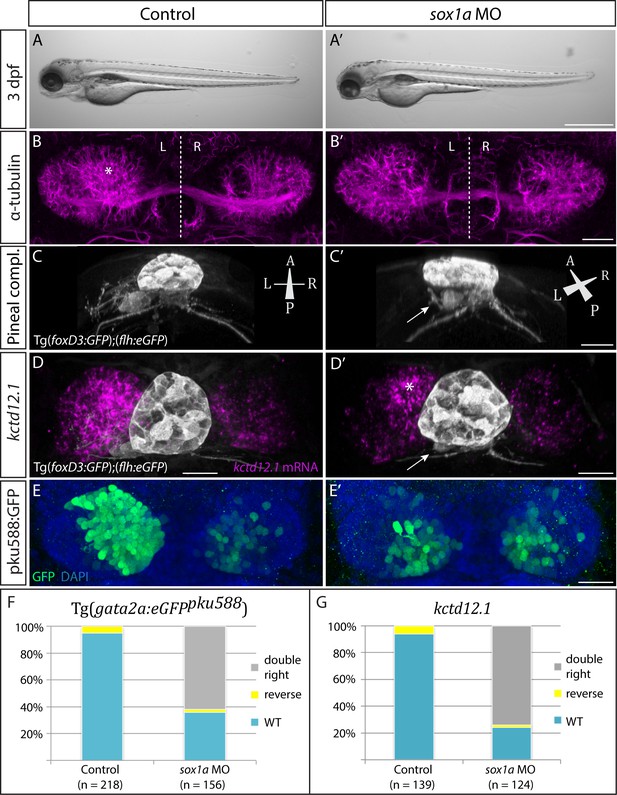
sox1a morphants show a double-right habenular phenotype comparable to sox1a mutants.
(A–A’) sox1a morpholino (MO)-injected embryo showing normal overall morphology at 3 dpf. Scale bar 500 µm. (B–B’) Dense left dHb neuropil domain (asterisk) is reduced in sox1a morphants at 4 dpf (acetylated α-tubulin immunostaining). Scale bar 25 µm. (C–C’) MO injections into Tg(foxD3:GFP);(flh:eGFP) transgenic fish reveal that the parapineal migrates to its normal position next to the pineal stalk by 4 dpf in the sox1a morphant, but has shortened axonal projections (arrow in C’). Scale bar 25 µm. (D–D’) Left habenula kctd12.1 mRNA expression at 4 dpf is decreased in the sox1a morphant despite normal parapineal migration (arrow), with only a small residual asymmetric domain (asterisk). Scale bar 25 µm. (E–E’) Numbers of GFP-positive neurons in Tg(gata2a:eGFPpku588) line labelling the left-dominant lateral domains of the dHb (located medially in larval zebrafish) is reduced in the 4 dpf sox1a morphant. Scale bar 25 µm. (F–G) Percentages of larvae with double-right, wild-type and reversed habenular phenotypes in controls and upon sox1a MO injections. Scoring is based on Tg(gata2a:eGFPpku588) GFP expression (E–E’, F) and kctd12.1 mRNA expression (D–D’, G) at 4 dpf.
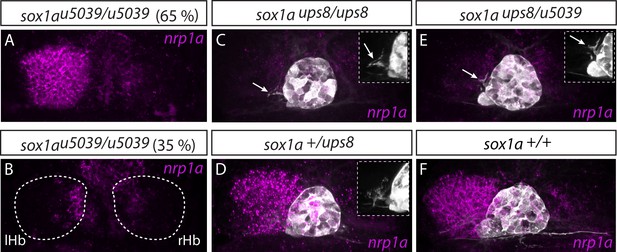
Two sox1a mutant alleles do not complement each other.
(A–B) The sox1au5039 allele shows incomplete penetrance with ~65% (n = 117/181) of homozygous mutants having a double-right dHb as confirmed by nrp1a mRNA in situ hybridisation at 4 dpf. lHb– left habenula, rHb – right habenula. (C–D,F) sox1aups8 allele exhibits complete penetrance with all homozygous mutants having a double-right dHb, as confirmed by nrp1a mRNA in situ hybridisation at 4 dpf in Tg(foxD3:GFP);(flh:eGFP) background with GFP expressed in the pineal complex (C and Figure 3). sox1a+/ups8 heterozygotes (n = 45) (D) have a phenotype identical to wild-type siblings (F). (E) The two mutant sox1a alleles do not complement each other – all trans-heterozygote larvae are double-right for nrp1a mRNA expression (n = 45/45). Similar to sox1aups8, trans-heterozygotes of the two mutant alleles exhibit stalled outgrowth of parapineal axons (white arrows in C and E).
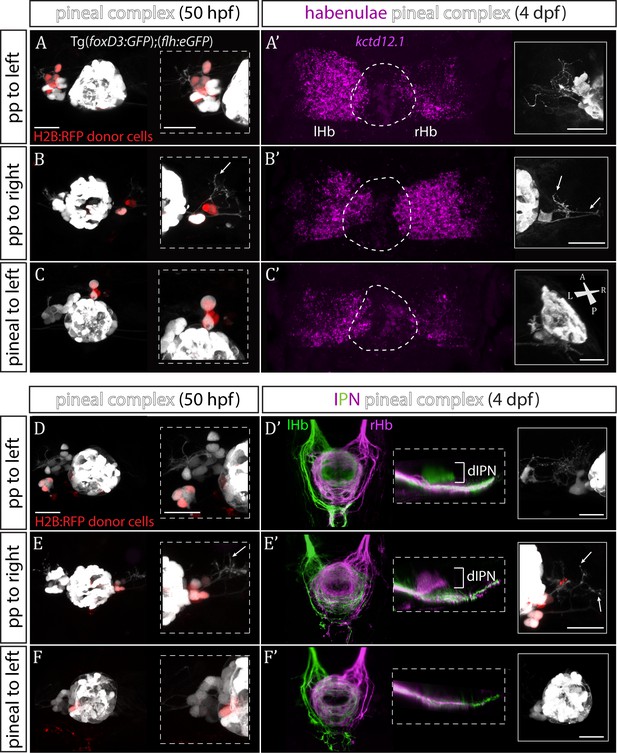
Transplanted wild-type parapineal cells rescue habenular asymmetry in sox1a-/- mutants.
(A–F) Parapineal (pp) cells from an H2B:RFP mRNA-injected Tg(foxD3:GFP);(flh:eGFP) donor embryo transplanted to the left (A, D) or right (B, E) side of a sox1a-/- Tg(foxD3:GFP);(flh:eGFP) recipient at 32 hpf send projections to the habenula at 50 hpf (white arrows in B and E), as shown by live-imaging. Transplanted pineal cells (C, F) do not send projections to the habenula and locate to the midline (C) or reincorporate into the pineal (F) by 50 hpf. Scale bars 25 µm. (A’–F’) By 4 dpf, the habenula adjacent to the transplanted wild-type parapineal cells acquires a left habenula phenotype in sox1a-/- mutants as shown by kctd12.1 mRNA expression (A’,B’) and anterograde labelling of habenula-IPN projections (dorsal and lateral views) (D’,E’). sox1a-/- larvae with pineal cell transplant have double-right habenulae (C’,F’). Solid boxes show the transplanted parapineal cells at 4 dpf, sending out long projections (arrows in B’ and E’) to the adjacent habenula. The whole pineal complex is shown for the pineal cell transplanted larvae (C’,F’). lHb – left habenula, rHb – right habenula, dIPN – dorsal interpeduncular nucleus. Scale bars 25 µm.
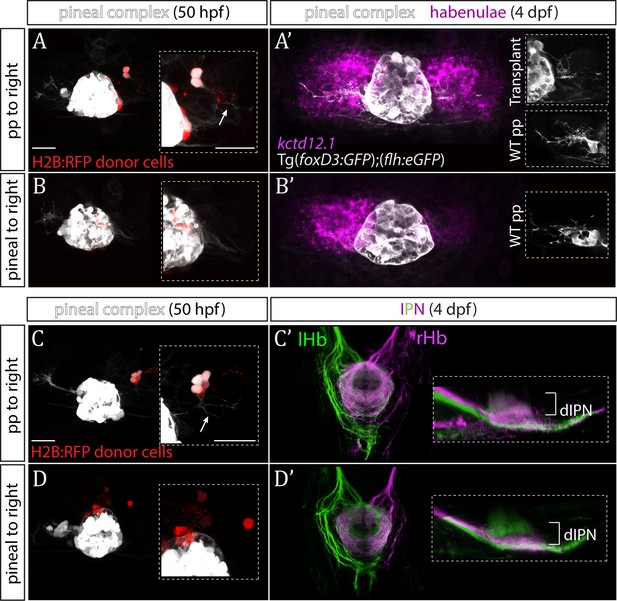
Parapineal cells impose left habenula character on right dorsal habenula neurons.
(A–D) Parapineal cells from an H2B:RFP mRNA-injected Tg(foxD3:GFP);(flh:eGFP) donor embryo transplanted to the right side of a Tg(foxD3:GFP);(flh:eGFP) recipient at 32 hpf send projections to the habenula at 50 hpf (arrows in A and C), whereas transplanted pineal cells (B, D) do not (but reintegrate into the pineal and/or locate to the midline). Scale bars 25 µm. (A’–D’) By 4 dpf, the right habenula adjacent to the transplanted parapineal cells acquires a left habenula phenotype, as shown by increase in kctd12.1 mRNA expression (A’) and anterogradely labelled efferent habenula-IPN projections (dorsal and lateral views) (C’). Note the innervation of dorsal IPN (dIPN) by both, the left and right habenula (lHb, rHb) in parapineal-transplanted larvae. Larvae with pineal cell transplants exhibit a wild-type habenular phenotype at four dpf (B’,D’).
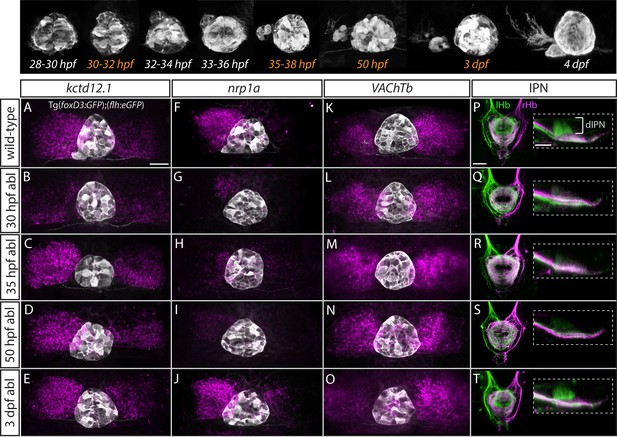
Step-wise regulation of habenular asymmetries by the parapineal.
The top panel shows the time-course of parapineal development in Tg(foxD3:GFP);(flh:eGFP) fish. The time-points selected for parapineal ablations are shown in orange. (A–O) Dorsal views of the epithalami of wild-type and parapineal-ablated larvae at 4 dpf showing expression of (foxD3:GFP)zf104 and (flh:eGFP)U711 transgenes (grey) in the pineal complex. Habenular mRNA expression of kctd12.1, nrp1a and VAChTb is shown in magenta. Parapineal ablations were carried out at time-points indicated on the left. Scale bar 25 µm. (P–T) Dorsal (left images) and lateral (right images) views of the interpeduncular nucleus labelled by anterograde tracing of the axons from the left dorsal habenula (lHb, green) and right dorsal habenula (rHb, magenta) at 4 dpf. Parapineal ablations were carried out at time-points indicated on the left. dIPN – dorsal interpeduncular nucleus. Scale bar 25 µm.
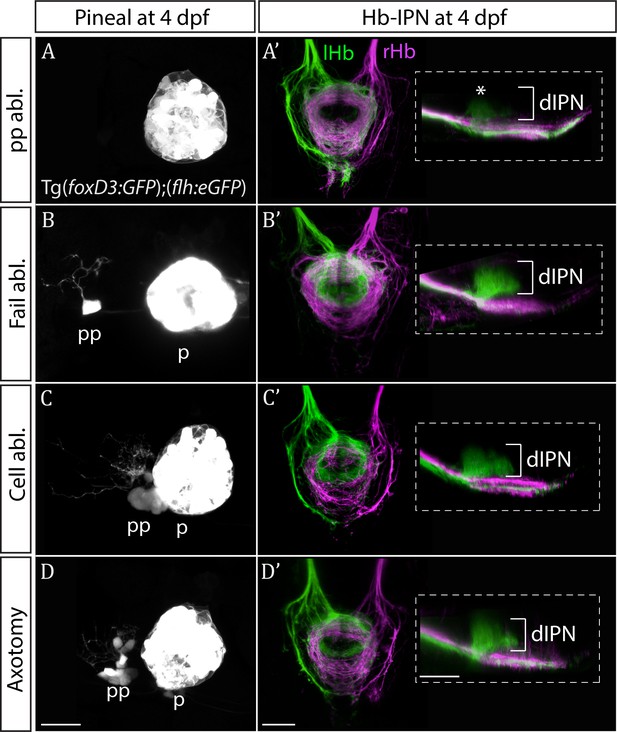
Only ablation of all parapineal cells leads to loss of habenular asymmetries.
(A–D) Live-imaging of the pineal complex at 4 dpf showing expression of Tg(foxD3:GFP)zf104 and Tg(flh:eGFP)U711 transgenes in the pineal (p) and the parapineal (pp). Parapineal ablations were carried out at 50 hpf, parapineal axotomies repeatedly at 50, 60 and 72 hpf. Fail ablation (n = 7) demarks one or two parapineal cells left unablated (B), cell ablation (n = 4) is ablation of 2–3 parapineal cells (C) and axotomies (n = 5) were carried out by severing the parapineal axon bundle approximately 10 µm from the cell bodies, which often also disrupted the compact structure of the parapineal (D). (A’–D’) Dorsal and lateral views of the interpeduncular nucleus labelled by anterograde tracing of the axons from the left dorsal habenula (lHb, green) and right dorsal habenula (rHb, magenta) at 4 dpf. Note that innervation of dorsal interpeduncular nucleus (dIPN) from the left habenula is lost only upon ablation of all parapineal cells. The asterisk indicates a small tuft of neurites in the dorsal anterior IPN that are present upon complete parapineal ablation (Bianco et al., 2008) and can also be detected in sox1a mutants (Figure 3E’). All scale bars 25 µm.
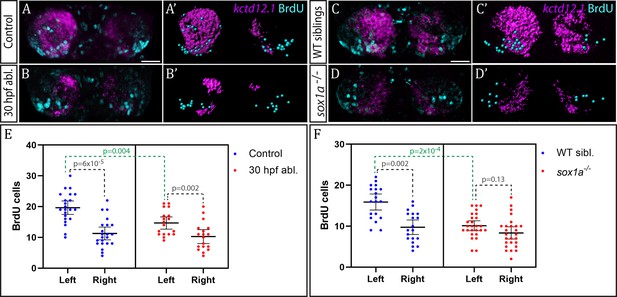
Neurogenesis is reduced in the left habenula upon early parapineal ablation and in sox1a-/- mutants.
(A–D’) Confocal images (A–D) and reconstructions (A’–D’) of kctd12.1 mRNA in situ hybridisation (magenta) and BrdU immunohistochemistry (cyan) in in the dHb of control (A, A’) and parapineal-ablated larvae (B, B’), as well as sox1a-/- mutants (D, D’) and wild-type siblings (C, C’) at 4 dpf. Ablations were carried out at 30 hpf and BrdU pulse given at 32 hpf. Scale bar 25 µm. (E–F) Plots showing the number of BrdU-positive cells born at 32 hpf in the left and right dHb. (E) Control (n = 22) and parapineal-ablated (n = 18) embryos. (F) sox1a-/- mutants (n = 26) and wild-type siblings (n = 19). Cells were counted as shown in A’-D’, mean and 95% CI are shown. The reduction in left dHb BrdU-positive cells upon 30 hpf ablation (p=0.004, Wilcoxon-Mann-Whitney test) and in sox1a-/- mutants (p=2×10−4) is indicated in green. Counts for left and right habenula of each individual embryo are given in Figure 6—source data 1.
-
Figure 6—source data 1
BrdU-positive cell counts in wild-type, parapineal-ablated and sox1a-/- embryos.
- https://doi.org/10.7554/eLife.47376.014
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Danio rerio) | sox1a | NA | Ensembl ENSDARG00000069866.5 | Line maintained at S Wilson lab |
| Strain, strain background (Danio rerio, AB/TL) | sox1au5039 | this paper | Line maintained at S Wilson lab | |
| Strain, strain background (Danio rerio, AB/TL) | sox1aups8 | this paper | Line maintained at S Wilson lab | |
| Strain, strain background (Danio rerio, AB/TL) | Tg(foxD3:GFP)zf104 | PMID: 12062041 | Line maintained at S Wilson lab | |
| Strain, strain background (Danio rerio, AB/TL) | Tg(flh:eGFP)U711 | PMID: 12895418 | Line maintained at S Wilson lab | |
| Strain, strain background (Danio rerio, AB/TL) | Et(gata2a:eGFP)pku588 | PMID: 18164283 | ||
| Antibody | Rabbit polyclonal anti-GFP | Torrey Pines Biolabs | Cat# TP401 | (1:1000) |
| Antibody | Mouse monoclonal anti-ascetylated tubulin | Sigma | Cat# T7451 | (1:1000) |
| Antibody | Mouse monoclonal anti-BrdU | Roche | Cat# 11170376001 | (1:400) |
| Antibody | Goat polyclonal Alexa 488, 568 and 647-conjugated secondary | Molecular Probes | Cat# A32731, A21144, A21126 | (1:250) |
| Other | DAPI stain | Molecular Probes | (1:1000) | |
| Recombinant DNA reagent (plasmid) | Cas9 plasmid | PMID: 23918387 | NA | Provided by W Chen lab |
| Commercial assay or kit | mMESSAGE mMACHINE Kit | Ambion | Cat# AM1344 | |
| Commercial assay or kit | HiScribe T7 High-Yield RNA Synthesis Kit | New England BioLabs | Cat# E2040S | |
| Commercial assay or kit | Precision Melt Supermix | Bio-Rad | Cat# 172–5112 | |
| commercial assay or kit | KASP chemistry for sox1a genotyping | LGC Genomics | NA | Assay designed by manufacturer |
| Software, algorithm | CHOPCHOP | PMID:24861617 | https://chopchop.cbu.uib.no/ |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.47376.015





