Hemimetabolous insects elucidate the origin of sexual development via alternative splicing
Figures
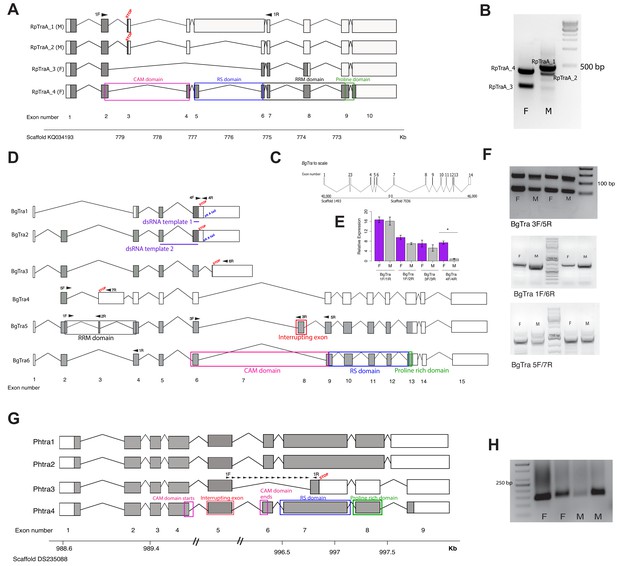
Transformer splicing follows the holometabolous pattern in the kissing bug Rhodnius prolixus, but not in the cockroach Blattella germanica or louse Pediculus humanus.
Schematics showing transformer transcripts isolated from R. prolixus, B. germanica, and P. humanus. Coding sequences are in gray, UTRs are in white. All genes are shown as transcribed from left to right. (A) Four transcripts isolated from RpTraA with 5’ and 3’ RACE and confirmed by RNA-seq. Sex-specificity of each transcript is noted in parentheses at the end of the transcript name. Stop codon in the male-specific 3rd exon is indicated with ‘STOP.’ Predicted Transformer protein domains are shown with colored boxes and labels, using RpTraA_4 as example. (B) PCR on male and female cDNA with RpTraA primers (RpTra_checkF + RpTra_checkR in Supplementary file 7) indicated by black arrows in (A) confirms the sex-specificity of each transcript; all amplicons were verified by Sanger sequencing. (C) BgTra spans two scaffolds in the B. germanica genome. Genome coordinates are shown at the beginning and end of each scaffold. Both scaffolds continue beyond BgTra; scaffold 1493 is 552 kb and scaffold 7036 is 55 kb. (D) Six BgTra transcripts were recovered in PacBio Isoseq data and verified by exon-specific RT-PCR. BgTra1, BgTra2 and BgTra4 were recovered from the female Isoseq dataset; BgTra3, BgTra5, and BgTra6 were recovered from the male Isoseq dataset; however, PCR and qPCR on male and female cDNA showed the presence of BgTra3-6 in both sexes (E, F). Nucleotides that code for important protein domains are color-coded as in panel A, using BgTra5 and BgTra6 as examples. BgTra1 and BgTra2 have alternative polyadenylation sites, marked with blue text. In BgTra1 and BgTra2, exon six extends ~500 bp further 3’ than in all other transcripts. The 3’ end of exon six in BgTra3-6 falls six bp upstream of the stop codon that ends the conceptual translation of BgTra1 and BgTra2. Exon 8 codes for 47 amino acids that interrupt the CAM domain, which spans the end of exon six and the start of exon 9. Purple lines show the locations of dsRNA used for RNAi knock-down. Black arrows indicate locations of primers used for RT-PCR and qPCR. (E) qPCR expression of different BgTra isoforms. The truncated transcripts BgTra1 and BgTra2 are strongly female-biased, while the remaining transcripts are sexually monomorphic. (F) RT-PCR expression of different BgTra isoforms. Top panel: both males and females express transcripts with and without the exon interrupting the CAM domain. The shorter band contains BgTra isoforms that skip exon eight and contain an intact CAM domain; the larger product contains exons 6, 8, and 9. Middle panel: both males and females express exon 7, found only in BgTra3 and producing a truncated transcript. Bottom panel: both males and females express the extended exon three with a premature stop codon found in BgTra4. (G) Four PhTra transcripts isolated from P. humanus by 5’ and 3’ RACE. PhTra3 has a premature stop codon and is found in both sexes, as shown by RT-PCR on male and female cDNA in panel (H). Forward primer for PCR shown in (H) spanned the exon 5/7 junction (marked with black arrows in panel (G)); reverse primer for this PCR is in exon 7 (also noted with a black arrow). All PhTra isoforms contain the ‘interrupting exon’ inside of the CAM domain. Double bars on scaffold in panel (G) represent breaks in scaffold scale.
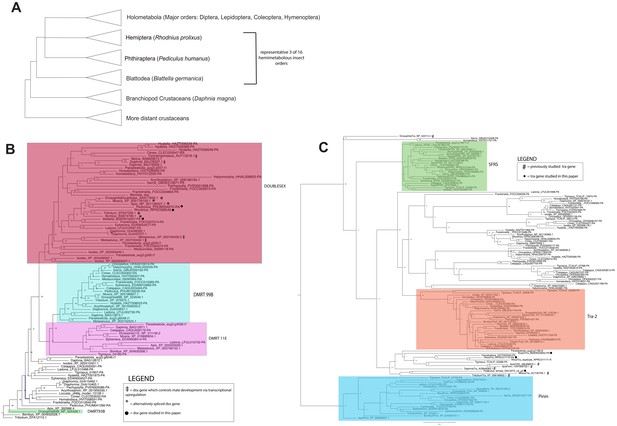
Three trees: a simplified arthropod phylogeny showing species studied, and gene trees for dsx and tra.
(A) A simplified arthropod phylogeny showing the positions of the three hemimetabolous insect orders examined in this study, relative to the monophyletic Holometabola and non-Hexapod crustaceans. (B) Maximum likelihood gene tree of DMRT family proteins in 29 arthropod species (Supplementary file 1) generated using RAxML (Stamatakis, 2014). Node labels indicate bootstrap support values for clades. Clades containing Drosophila DMRT genes are outlined in shaded boxes. doublesex genes that are known to regulate male and female differentiation via alternative spliceforms are indicated with asterisks (*); doublesex genes that promote male differentiation via transcriptional upregulation in males are indicated with a double cross (‡). Genes studied in this paper are indicated with a filled circle (●). For ease of viewing the doublesex clade, the tree is rooted with DMRT93B sequences clade. (B) Transformer proteins do not form a monophyletic group in the gene tree of SR family proteins in 29 arthropod species generated with maximum likelihood (RAxML). Node labels indicate bootstrap values for clades. Previously studied transformer genes are indicated with hashtags (#); genes investigated in this paper are indicated with filled circles.
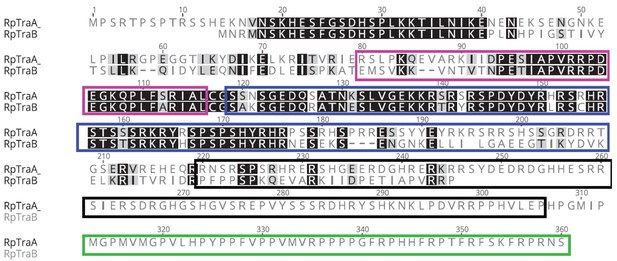
MAFFT protein alignment of RpTraB and the longer female-specific isoform of RpTraA from Rhodnius prolixus.
The CAM domain is boxed in pink, the RS domain in blue, the RRM domain in black, and the proline rich domain in green. R. prolixus shows the first Tra duplication reported outside of Hymenoptera (Geuverink and Beukeboom, 2014).

RRM domains found in select Transformer proteins.
Predicted sequences of Tra proteins from Apis mellifera, Rhodnius prolixus, Blattella germanica, and Tigriopus californicus, with the residues predicted to form an RRM domain highlighted in yellow.
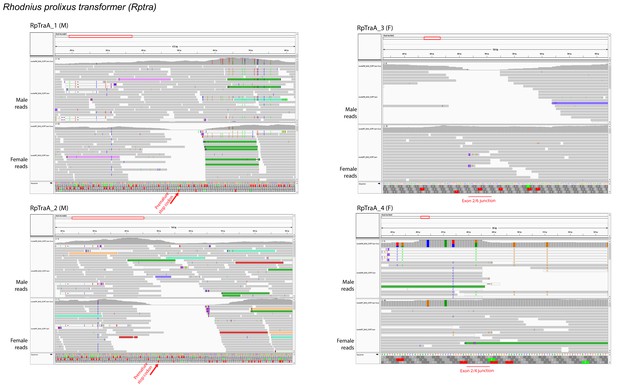
RNA-seq supports the sex-specificity of RpTra isoforms.
Illumina reads from male and female R. prolixus tissue were mapped to RpTra transcripts isolated by RACE using Bowtie (version 2.3.4.1) and then visualized in IGV (v 2.5.3). Left column: male-specific RpTra transcripts; right column: female-specific RpTra transcripts. No female reads map to the male-specific stop codon in male RpTra transcripts, and no male reads map to the female-specific exon junctions of female RpTra transcripts.
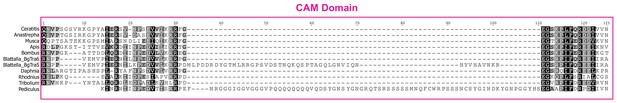
MAFFT protein alignment of the complete CAM domain in nine different insect species and one crustacean.
The cockroach Blattella germanica has three different types of transcripts: some transcripts terminate halfway through the CAM domain (BgTra1, BgTra2), others code for an intact CAM domain (BgTra6, BgTra7, BgTra8), and in some the inclusion of an alternatively spliced exon interrupts the CAM domain with 47 additional residues (BgTra3, BgTra4, BgTra5).
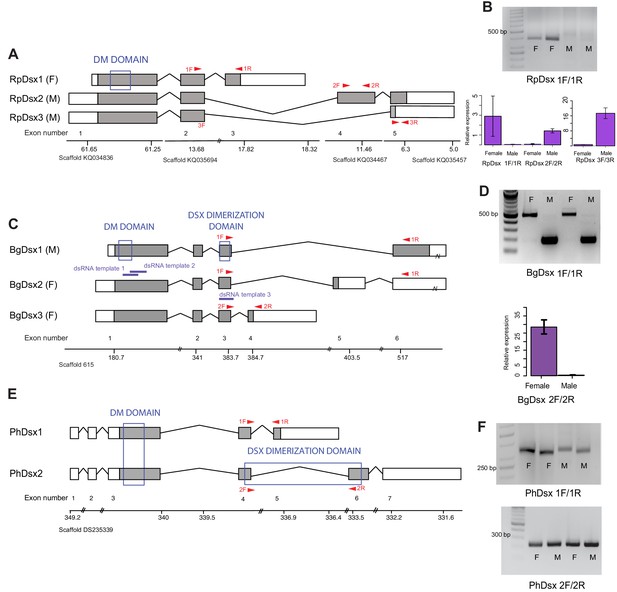
Doublesex shows sex-specific splicing in some but not all hemimetabolous insects.
Schematics showing doublesex transcripts isolated from Rhodnius prolixus, Blattella germanica, and Pediculus humanus. Coding sequences are in gray and UTRs in white. Sex-specificity of each transcript is noted in parentheses at the end of the transcript name. Transcripts are shown mapped to genomic scaffolds; double bars show breaks in scale. Functional domains are labeled and indicated by blue boxes. (A) Three dsx transcripts isolated from R. prolixus span multiple genomic scaffolds. RpDsx1 was isolated from 3’ and 5’ RACE libraries made from female RNA; RpDsx2 and RpDsx3 were cloned from RACE libraries synthesized from male RNA. (B) Top panel: RT-PCR showing female-specificity of RpDsx1, using primers in exons 2 and 3, as indicated with red arrows in (A). Bottom panel: qPCR showing female-specific expression of RpDsx1, and male-specific expression of RpDsx2 and RpDsx3, using primers indicated with red arrows in (A). Forward primer 3F spanned the exon 2/5 junction. (C) Three dsx transcripts isolated by 3’ and 5’ RACE and PacBio Isosequencing from B. germanica. The 3’ UTR of BgDsx1 and BgDsx2 contains a retrotransposon (marked with a break) and cannot be mapped to a single scaffold of the roach genome. Purple lines show the locations of dsRNA used for RNAi. Red arrows show primer locations for RT-PCR and qPCR. (D) Sex-specific expression of BgDsx isoforms. Top panel, PCR on cDNA of adult male and female fat body and reproductive tract with primers in exons 3 and 6. The larger band found exclusively in females contains the alternatively included exon 5, as confirmed by Sanger sequencing. Bottom panel, qPCR for exon 3/4 junction shows amplification only in females. (E) Two dsx transcripts isolated by 3’ and 5’ RACE from P. humanus male and female samples. (F) RT-PCR shows the presence of both PhDsx transcripts in both sexes.
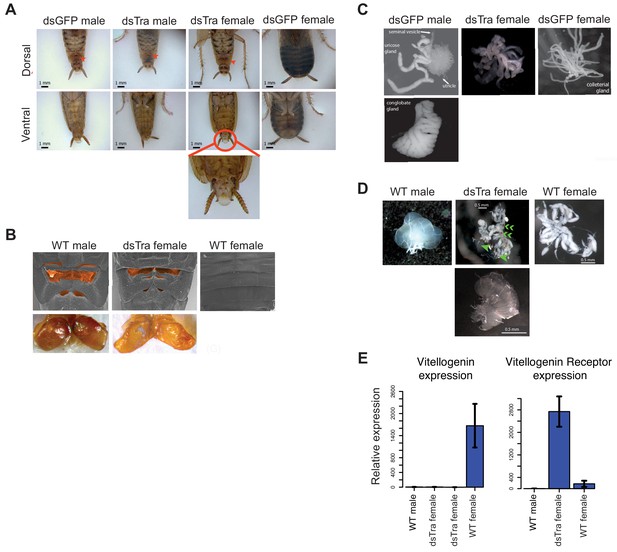
BgTra is necessary for female-specific but not male-specific sexual differentiation in Blattella germanica.
(A) dsBgTra females, injected with dsBgTra in their 4th, 5th, and 6th instars, molt to adults with a masculinized abdomen, in contrast to control animals injected with dsGFP. Top row, dorsal view. Bottom row, ventral view. The tergal gland in wild-type males and dsBgTra females is indicated with red arrows. Red circle outlines abnormal tissue extrusion typical of dsBgTra females; zoomed-in view of this tissue is shown below. (B) Scanning electron microscopy (top row) and light microscopy (bottom row) reveal developed male-like tergal glands (shaded orange in SEMs and dissected for light microscopy) in dsBgTra females compared to wild-type (WT) females. (C) dsBgTra females have malformed, partly masculinized colleterial glands. Left column shows wild-type male accessory glands composed of seminal vesicles, utricles, uricose glands, and a conglobate gland. Right column, wild-type female colleterial gland. Center column, gland dissected from dsBgTra female is located in the same position as the wild-type colleterial gland, but shows thickened tubules reminiscent of the male accessory glands. (D) dsBgTra female gonads are deformed or intermediate between male and female gonads. Left column shows testis of a 5-day-old wild-type male. Middle column, top image shows disorganized gonad tissue in a typical 2-week-old dsBgTra female. Most individuals (n = 12) had underdeveloped ovaries with some undifferentiated tissue, some ovarioles (green arrows), and some sclerotized cuticle (green chevrons) not normally found in the ovary. Middle column, bottom shows that some dsBgTra females (n = 3) have intersex gonads composed of both ovarian and testicular tissue (indicated by green arrow). Right column shows ovaries of a 2-day-old wild-type female. (E) vitellogenin, an egg yolk protein, production is under the control of BgTra. Left: qPCR measuring relative vitellogenin expression in the fat body of males and females treated with dsBgTra. Right: relative upregulation of vitellogenin receptor transcription in the gonads of dsBgTra females, presumably due to the lack of circulating vitellogenin. vitellogenin and vitellogenin receptor gene expression was normalized to actin.
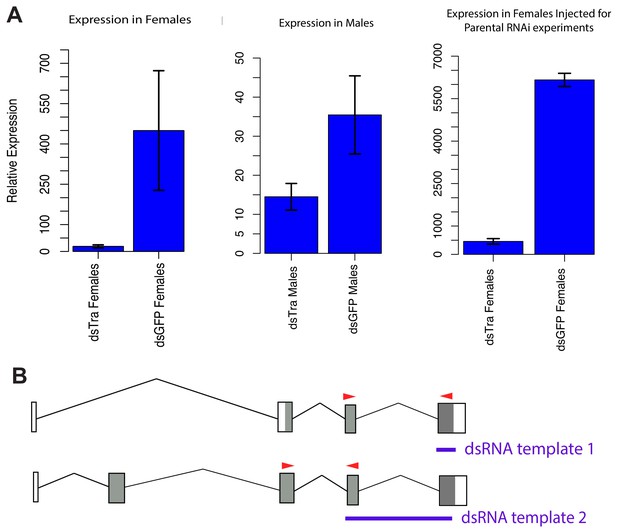
dsBgTra reduces BgTra expression in males and females of Blattella germanica.
(A) Left panel, BgTra expression in the colleterial glands of females injected with dsTra or dsGFP (control) throughout development (n = 2 dsTra and n = 2 dsGFP). Middle panel, BgTra expression in the accessory glands of males injected with dsTra and dsGFP (control) throughout development (n = 3 dsTra and n = 3 dsGFP). Right panel, BgTra in the fat body of females injected with dsTra after terminal molt (n = 3 dsTra and n = 3 dsGFP). This last group of females were used to study the maternal effects of BgTra on embryonic development. All graphs show gene expression measured by qPCR, relative to β-actin. (B) Red arrows show primers used for qPCR, in relation to the dsRNA constructs injected for RNAi (purple). Both the exons targeted by dsTra and the exons used for qPCR are shared by all BgTra isoforms.
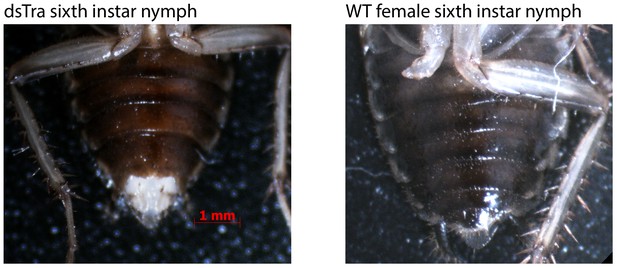
Abnormal morphology in 6th instar dsBgTra female nymphs of Blattella germanica.
Tissue extrusion in dsBgTra females (left) compared with the normal abdomen in wild-type 6th instar females (right).
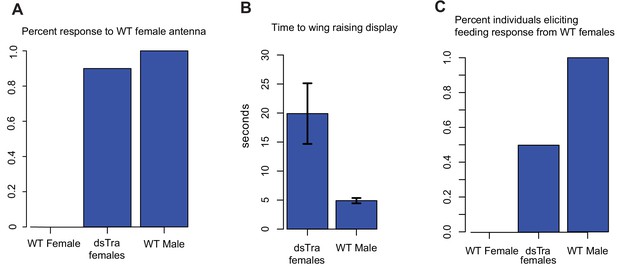
dsBgTra females perform male courtship behavior and elicit courtship responses from wild-type females.
In courtship, wild type male Blattella germanica raise their wings to display their tergal gland. When courtship is successful, wild type females feed upon the secretions of this gland prior to copulation. dsBgTra females (n = 10), wild-type females (n = 10), and wild-type males (n = 10) were exposed to an antenna clipped from a wild-type, 7 day old virgin female, and their response times in seconds were compared. (A) Nine out of ten dsBgTra females performed the stereotypical male wing-raising courtship display in response to a female antenna compared to 0/10 of wild-type females within the one minute of observation time. (B) However, dsBgTra females took longer than wild-type males to respond with the wing-raising display. Wild-type females did not respond to the stimulus throughout the duration of the one-minute observation time. (C) 5/10 dsBgTra females elicited feeding response from wild-type females after raising their wings. Wild-type females, lacking tergal glands, do not elicit a feeding response from other wild-type females.
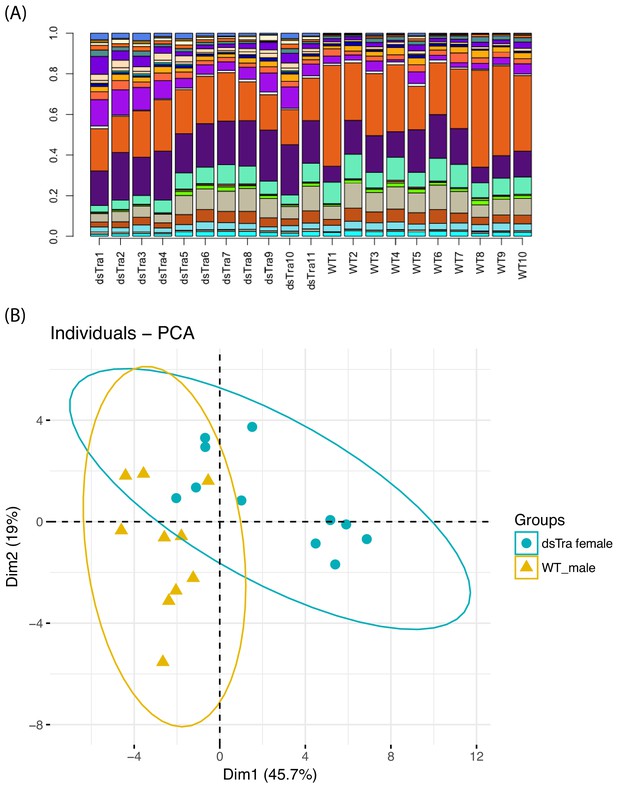
BgTra controls sex-specific oligosaccharide synthesis in tergal glands (A) Relative abundance of oligosaccharides in tergal glands derived from dsBgTra females (dsTra1-dsTra10) and wild-type males (wt1-wt10).
Each bar represents one individual, with different compounds shown in different colors. (B) Principal components analysis on the relative proportions of different oligosaccharides in dsBgTra female and wild-type male tergal glands. These two groups sort into different parts of PCA space, indicating that although all sugars produced by wild type male tergal glands are present in tergal glands from dsBgTra females, they produce these sugars in different relative amounts. Dimension one is primarily driven by the relative abundance of a non-reducing hexose with mass 990.328.
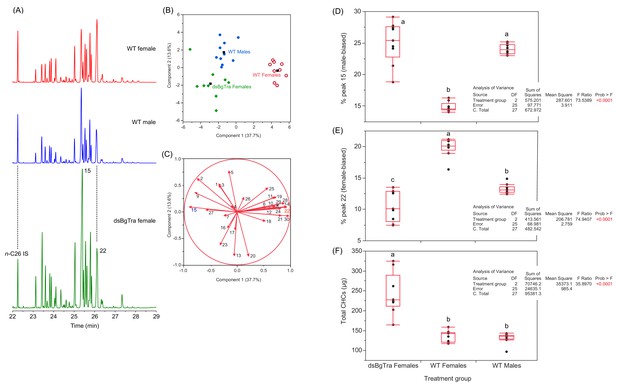
BgTra controls production of sex-specific cuticular hydrocarbons.
Gas chromatograms (A) and PCA (B, C) of the cuticular hydrocarbons of Blattella germanica. The peak numbers correspond to the CHCs in Supplementary file 6 and to Jurenka et al. (1989). In the score plot (B), the solid black squares represent the centroids of their respective groups. Wild-type males in solid blue circles (n = 10), wild-type females in open red circles (n = 9), dsBgTra females in green diamonds (n = 9). In the loadings plot (C), note that peak 15, which is abundant in wild-type males, pulls towards wild-type males and dsBgTra females (to the left), whereas peak 22, which is more abundant in females, pulls towards wild-type females (to the right). (D, E) Statistical analysis of the percentage representation of peak 15 (D) and peak 22 (E) in the total cuticular hydrocarbons of wild-type males, wild-type females and dsBgTra females. Peak 15 (9-, 11-, 13-, and 15-methylnonacosane), which is more common in wild-type males, was more represented in dsBgTra females than in wild-type females. Peak 22 (3,7-, 3,9-, and 3,11-dimethylnonacosane), which serves as precursor to several components of the female contact sex pheromone, was significantly under-represented in dsBgTra females compared to wild-type females. (F) Statistical analysis of the total cuticular hydrocarbons of wild-type males, wild-type females and dsBgTra females. In the box plots, the horizontal line within the box represents the median value and the box represents the 25th to 75th quantiles. Letters within each graph represent the results of Tukey’s HSD, and different letters represent statistically significant differences (p<0.05).
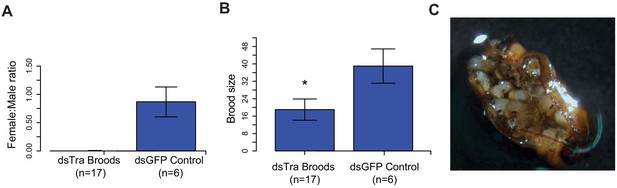
Maternal knockdown of BgTra in Blattella germanica results in all-male broods..
(A) Females injected with dsBgTra (n = 17) three days after emergence and mated to wild type males produced all-male broods, compared to control females injected with dsBgGFP (n = 6) (B) Brood sizes from mothers injected with dsBgTra were significantly smaller than the dsGFP controls (Welch’s t-test, p=0.001). (C) Image of a typical ootheca (egg case) from a dsBgTra mother that failed to hatch. Of 22 mothers injected with dsTra, five egg cases failed to hatch completely. The remaining oothecae had 5–19 dead embryos remaining within it after all males had emerged. Variability in the number of dead embryos can be attributed to surviving offspring eating dead embryos, which was observed by the authors.
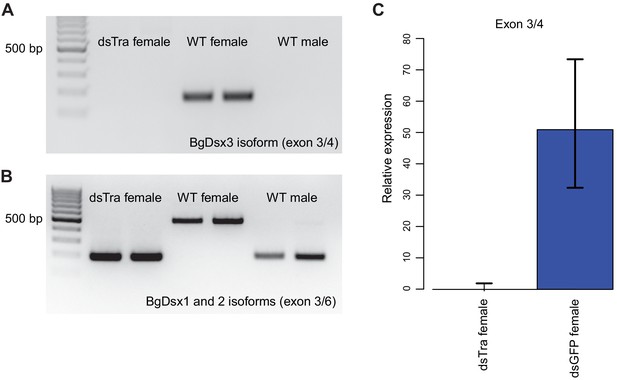
BgTra controls sex-specific splicing of BgDsx.
(A) RT-PCR showing that the female-specific BgDsx exon 3/4 junction (Figure 2B,C) is absent in dsBgTra females (B) RT-PCR showing that the male-specific BgDsx exon 3/5 junction (Figure 2B) is present in dsBgTra females. dsBgTra females do not express any of the female-specific BgDsx isoforms. (C) qPCR shows complete absence of the female-specific BgDsx exon 3/4 junction in dsBgTra females.
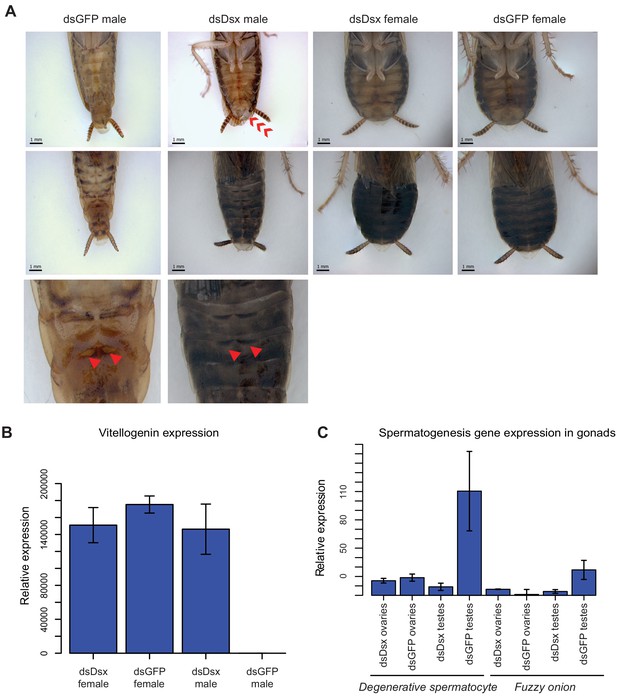
dsBgDsx RNAi in Blattella germanica feminizes males but has no effect in females (A) dsBgDsx males have reduced tergal glands (red arrow), darker female-like pigmentation, and an extrusion of tissue at the distal end of the abdomen (red chevron) compared to dsGFP injected males (see also Figure 7—figure supplement 3).
Females are unaffected by dsBgDsx treatment. Bottom row shows zoomed-in view of dorsal segments containing the tergal gland. The openings from which tergal glands secrete their oligosaccharides and lipids in dsGFP males (red arrows) are greatly reduced in dsBgDsx males. (B) BgDsx represses vitellogenin expression in males, but has no effect in females (n = 3 biological replicates per treatment). (C) BgDsx controls the expression of two spermatogenesis-related genes in males, but has no effect on their expression in females. qPCR showing the expression of two genes important for Drosophila melanogaster spermatogenesis, degenerative spermatocyte and fuzzy onions, relative to actin (n = 3 biological replicates for all columns).
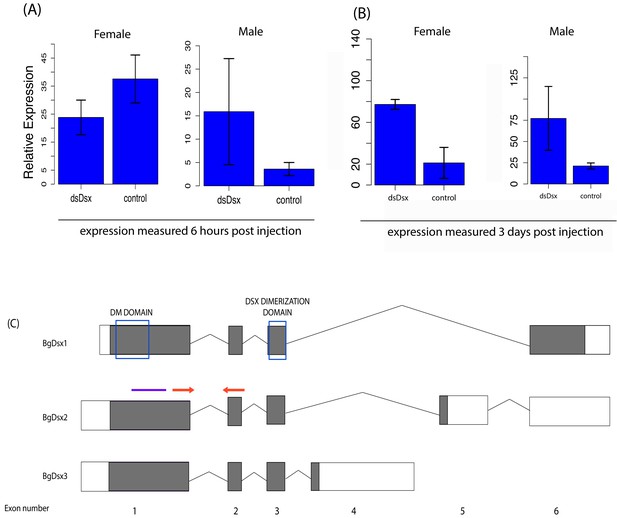
BgDsx RNAi causes an increase in BgDsx transcript abundance in both males and females of Blattella germanica.
BgDsx expression was measured (A) 6 hr and (B) 3 days after injection with dsRNA targeting the BgDsx common region. (C) qPCR primers (red arrows) were located outside the region targeted by dsBgDsx (purple bar). n = 3 females and three males per treatment. Expression levels of BgDsx are normalized to β-actin.
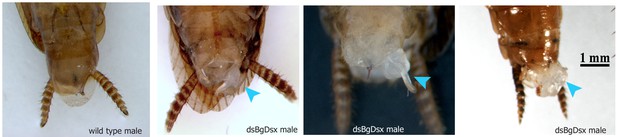
Representative sampling of tissue extrusions found in dsBgDsx males of Blattella germanica (light blue arrowheads).
https://doi.org/10.7554/eLife.47490.019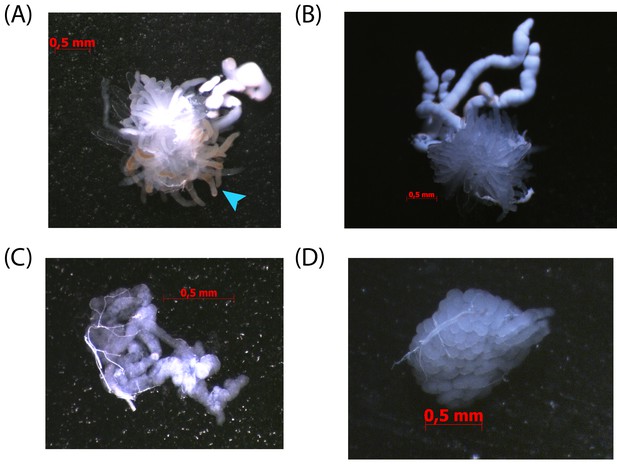
The conglobate glands and utricles of dsBgDsx males of Blattella germanica fail to properly mature.
(A) Utricles and seminal vesicles from five day old dsBgDsx male. Note tissue discoloration of utricles (light blue arrows). (B) Utricles and seminal vesicles of untreated control five day old male. (C) Conglobate gland from five day old dsBgDsx male is malformed compared to control conglobate gland from untreated five day old male (D).
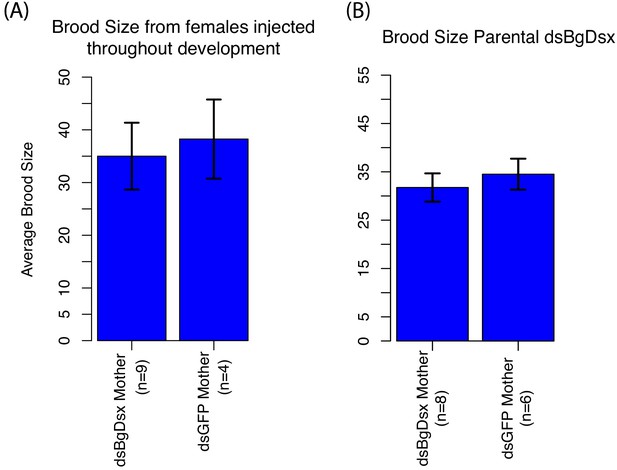
BgDsx has no effect on brood size in females of Blattella germanica that were injected with dsBgDsx either at multiple stages throughout nymphal development (A) or as virgin adult females (B).
https://doi.org/10.7554/eLife.47490.021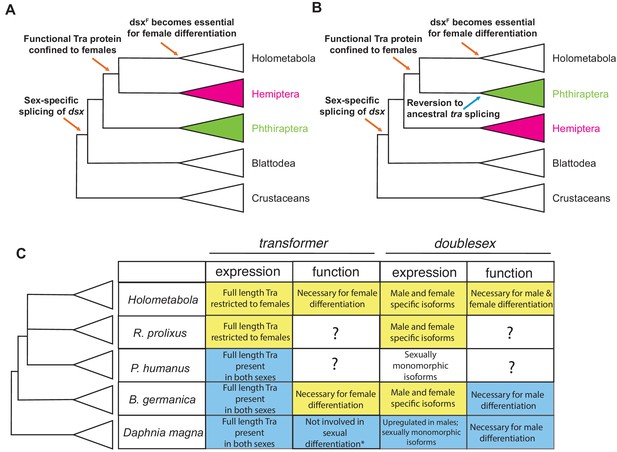
Evolutionary assembly of the insect sexual differentiation pathway.
Uncertainty in the relationships among Hemiptera, Phthiraptera, and Holometabola (Misof et al., 2014; Johnson et al., 2018; Freitas et al., 2018; Li et al., 2015) means that the sequence of events in the evolution of the tra-dsx axis is unclear. If Hemiptera (pink) are the sister group of Holometabola, the key molecular innovations have evolved in a stepwise order (A). If Phthiraptera (green) are the sister of Holometabola instead, the evolution of the tra-dsx axis must have followed a more mosaic pattern, with at least some secondary reversions (B). (C) Summary of tra and dsx expression and function in the three hemimetabolous insect orders in comparison to the canonical holometabolous mechanism and to Daphnia. No effect on sexual differentiation has been reported following tra knock-down in Daphnia (asterisk) (Kato et al., 2011). Hypothesized ancestral states are shown in blue; hypothesized derived states in yellow. Within Holometabola, subsequent events in the evolution of the tra-dsx axis include a secondary loss of tra in Lepidoptera, loss of the CAM domain of Tra in Drosophila, and the evolution of Sex-lethal (Sxl) as the upstream regulator of tra splicing in Drosophila.
Additional files
-
Supplementary file 1
Sources of arthropod gene models used in phylogenetic analyses of the DMRT and SR gene families.
Orders are color-coded as follows: holometabolous insects (orange), hemimetabolous insects (green), non-insect hexapods (red), crustaceans (blue), and chelicerates (purple).
- https://doi.org/10.7554/eLife.47490.023
-
Supplementary file 2
Distances between prospero and doublesex in the genomes of 10 different insect species.
- https://doi.org/10.7554/eLife.47490.024
-
Supplementary file 3
Scaffolds containing prospero and their sizes from nine hemipteran species.
- https://doi.org/10.7554/eLife.47490.025
-
Supplementary file 4
Queries used in BLASTP searches of arthropod gene models for DMRT homologs.
- https://doi.org/10.7554/eLife.47490.026
-
Supplementary file 5
Queries used in BLASTP searches of arthropod gene models for Transformer and SR family proteins.
- https://doi.org/10.7554/eLife.47490.027
-
Supplementary file 6
Estimated percentages of cuticular hydrocarbons in wild-type and dsBgTra-treated Blattella germanica.
Each gas chromatograph (GC) peak is represented as a percentage of the total amount of all 30 hydrocarbons. The peak numbers correspond to the CHCs identified by Jurenka et al. (1989). Peak 15 (9-, 11-, 13-, and 15-methylnonacosane) is known as a male-enriched CHC, and peak 22 (3,7-, 3,9-, and 3,11-dimethylnonacosane) is a female-enriched CHC. 3,11-Dimethylnonacosane also serves as precursor to several components of the female contact sex pheromone.
- https://doi.org/10.7554/eLife.47490.028
-
Supplementary file 7
Primers used in RACE, RT-PCR, and qPCR in this study.
- https://doi.org/10.7554/eLife.47490.029
-
Transparent reporting form
- https://doi.org/10.7554/eLife.47490.030





