Evolution of (p)ppGpp-HPRT regulation through diversification of an allosteric oligomeric interaction
Figures
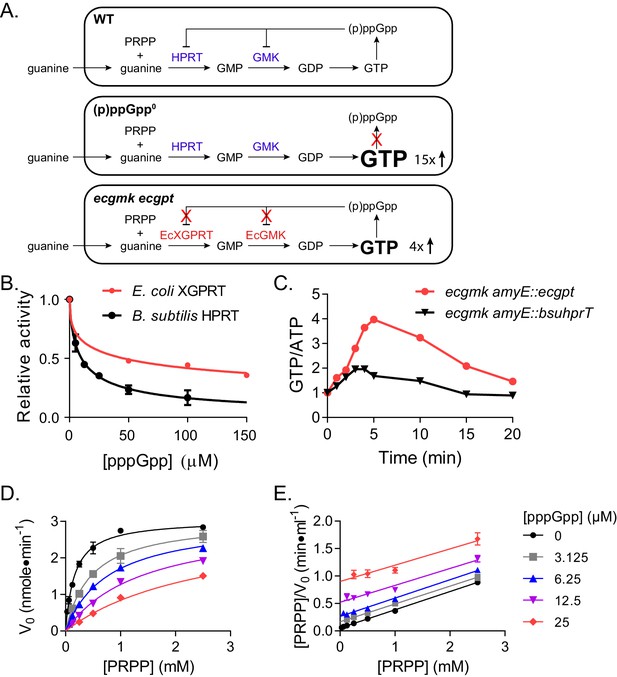
Regulation of HPRT by basal levels of (p)ppGpp is important for GTP homeostasis.
(A) Pathways showing the effect of extracellular guanine on B. subtilis GTP homeostasis. In WT, (p)ppGpp regulates HPRT and GMK. (p)ppGpp0 cannot produce (p)ppGpp (see B) and ecgmk ecgpt has enzymes resistant to (p)ppGpp regulation (see F). (B) E. coli XGPRT more weakly inhibited by pppGpp than B. subtilis HPRT. The IC50 for E. coli XGPRT is 45 µM compared to 10 µM for B. subtilis HPRT. Error bars represent SEM of triplicate. (C) Expression of E. coli XGPRT leads to imbalanced GTP/ATP homeostasis in B. subtilis. GTP/ATP ratio in B. subtilis treated with 1 mM guanosine as determined by thin layer chromatography of 32P-labeled cells. Time is minutes after guanosine treatment. In ecgmk, (p)ppGpp-insensitive E. coli gmk replaces B. subtilis gmk at its endogenous locus. ecgmk amyE::bsuhprT and ecgmk amyE::ecgpt express B. subtilis hprT and E. coli gpt, respectively, from an IPTG-dependent promoter at an exogenous locus. All strains grown with 1 mM IPTG. (D) Initial velocities of B. subtilis HPRT at varied PRPP and pppGpp concentrations. Data are fitted to a global competitive inhibition equation (r2 = 0.975), and Ki = 1.7 µM. (E) Data from (D) in a Hanes–Woolf transformation. Parallel lines indicate equal maximum velocities for each pppGpp concentration. Error bars represent SEM of at least three replicates.
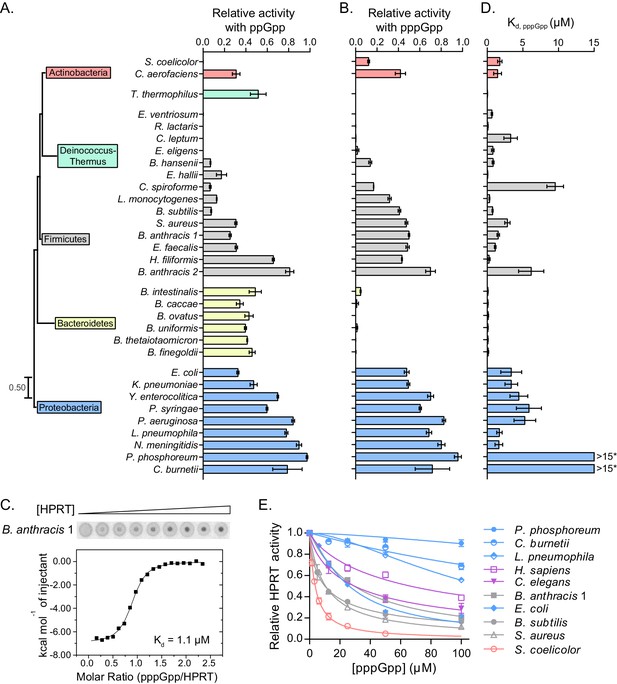
Conservation and variation in (p)ppGpp regulation of HPRTs across species.
(A–B) (p)ppGpp inhibits HPRTs across species. Representative phylogenetic tree constructed from 16S rRNA sequences of species shown with eukaryotic outgroup hidden. Branch length scale represents 0.5 substitutions per site. See Materials and methods for tree construction. (A) Relative activities of HPRTs with 25 μM ppGpp. (B) Relative activities of HPRTs with 25 μM pppGpp. Error bars represent SEM of triplicates. (C–D) pppGpp binds HPRTs across species. (C) Representative DRaCALA between B. anthracis Hpt-1 in cell lysate serially diluted 1:2 and 32P-labeled pppGpp. The central signal is proportional to pppGpp – HPRT interaction. Binding isotherm from isothermal titration calorimetry between B. anthracis Hpt-1 and pppGpp. See Figure 2—figure supplement 1 for energy isotherm and parameters. (D) The Kd between pppGpp and HPRTs obtained with DRaCALA from serially diluted cell lysates containing overexpressed HPRTs (see Materials and methods). Error bars represent SEM derived from one binding curve. P. phosphoreum and C. burnetii HPRTs with * have affinities too weak to calculate but are estimated to be >15 μM (see Figure 2—figure supplement 1). (E) Relative activities of HPRTs with increasing concentrations of pppGpp. See Table 1 for partial datasets for HPRTs from M. tuberculosis, C. crescentus, S. meliloti, R. torques, S. mutans, and C. gilvus.
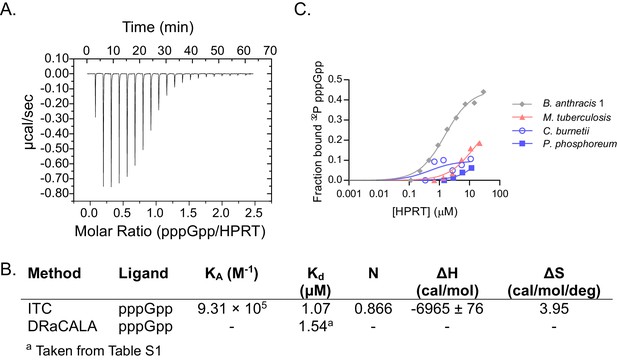
Characterizing pppGpp interaction with bacterial HPRTs.
(A) Energy isotherm of isothermal titration calorimetry between pppGpp and B. anthracis Hpt-1. (B) Parameters from isothermal titration calorimetry and DRaCALA with B. anthracis Hpt-1. DRaCALA Kd estimated from data in panel C. (C) DRaCALA binding curves between HPRT and 32P-labeled pppGpp performed with a single replicate. The interactions with M. tuberculosis, C. burnetii, and P. phosphoreum HPRTs were too weak to use for a Kd approximation. B. anthracis Hpt-1 is shown for comparison.
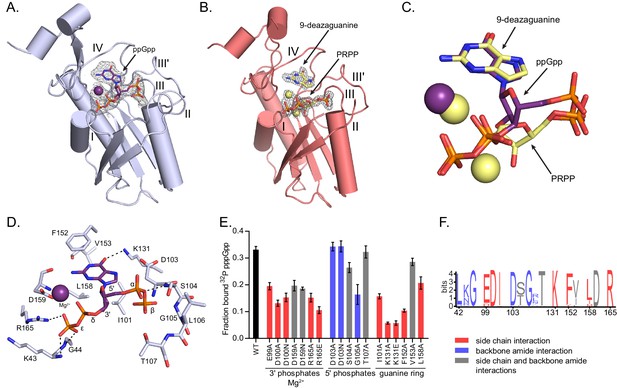
(p)ppGpp binds the HPRT active site and shares an almost identical binding pocket with substrates.
(A) B. anthracis Hpt-1 crystallized with ppGpp. Five loops (I, II, III, III′, IV) form the HPRT active site (Sinha and Smith, 2001). ppGpp and Mg2+ shown with omit electron density contoured at 2σ. Figure 3—figure supplement 1 shows that pppGpp competitively inhibits Hpt-1. Figure 3—figure supplement 4 shows omit electron density for the two ppGpp molecules crystallized in the asymmetric unit. (B) B. anthracis Hpt-1 crystallized with 9-deazaguanine, PRPP, and two Mg2+. Ligands shown with omit electron density contoured at 2.5σ. Residues Tyr70 – Ser76 in loop II were not resolved. See Figure 3—figure supplement 2 for asymmetric units of Hpt-1 crystallized with ppGpp, substrates, and sulfates. See Figure 3—figure supplement 3 for coordination of Mg2+ with ppGpp and PRPP. (C) Overlay of substrates (PRPP and 9-deazaguanine; yellow) and inhibitor (ppGpp; purple) bound to HPRT. Spheres represent Mg2+ and are colored according to their coordinating ligand. See Figure 3—figure supplement 6 for binding pocket comparison. (D) The ppGpp binding pocket on HPRT. Black dotted lines indicate select hydrogen bonds. The peptide backbone is shown for residues where the interactions are relevant. See Figure 3—figure supplement 5 for complete interaction maps. (E) DRaCALA of B. subtilis HPRT variants binding to 32P-labeled pppGpp, determined with cell lysates containing overexpressed HPRT variants. Disruption of side chain interactions weakened binding. For E and F, the residues are colored according to their interaction with ppGpp. Red = side chain interaction, blue = backbone amide interaction, and gray = both side chain and backbone interactions. Error bars represent SEM of three replicates. (F) Sequence frequency logo of the (p)ppGpp binding pocket from 99 bacterial HPRTs. Logo created using WebLogo from UC-Berkeley. See Figure 3—figure supplement 8 for binding residues from select bacterial and eukaryotic HPRTs.
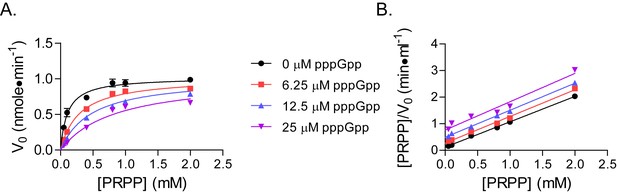
pppGpp competes with PRPP to inhibit B. anthracis Hpt-1.
(A) Initial velocities of B. anthracis Hpt-1 at varied PRPP and pppGpp concentrations. Data are fitted to a global competitive inhibition model (r2 = 0.97), and Ki = 3.6 µM. (B) Data from (A) in a Hanes–Woolf transformation. The slope of the transformed lines is a function of the maximum velocities with each pppGpp concentration. Parallel lines indicate equal maximum velocities. Error bars represent SEM of at least three replicates.

Asymmetric units of B.anthracis Hpt-1 structures.
(A) B. anthracis Hpt-1 crystallized as a tetramer in the asymmetric unit without ligands. Each color represents a separate monomer. Yellow spheres represent two sulfates bound to each monomer. (B) B. anthracis Hpt-1 crystallized with substrates (PRPP and 9-deazaguanine) as a dimer in the asymmetric unit. Red sticks represent the substrates and the green spheres represent Mg2+. (C) B. anthracis Hpt-1 crystallized with ppGpp as a dimer in the asymmetric unit. Red sticks represent pppGpp and the green sphere represents Mg2+. Two asymmetric units form a tetramer nearly identical to A.
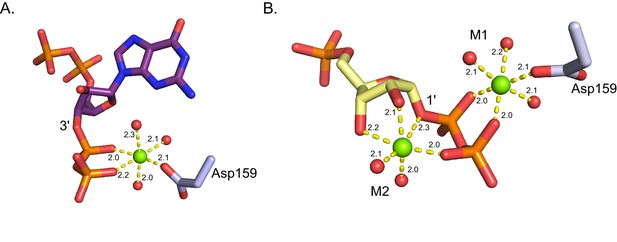
Coordination of Mg2+ in B. anthracis Hpt-1 crystallized with ppGpp and substrates.
(A) Octahedral coordination of Mg2+ (green sphere) crystallized with B. anthracis Hpt-1 and ppGpp (shown in purple sticks). The Mg2+ is coordinated by three water molecules (red spheres), the 3' phosphates of ppGpp, and the side chain of aspartate 159. Yellow dashes represent interaction and distances for each interaction are shown in Å. (B) Octahedral coordination of Mg2+ (green spheres) crystallized with B. anthracis Hpt-1 and substrates (9-deazaguanine not shown; PRPP shown in yellow sticks). M1 is coordinated by three water molecules (red spheres), the 1' phosphates of PRPP, and the side chain of aspartate 159. M2 is coordinated by two water molecules, the 1' phosphates of PRPP, and the 2' and 3' hydroxyls of PRPP’s ribose.Yellow dashes represent interaction and distances for each interaction are shown in Å.
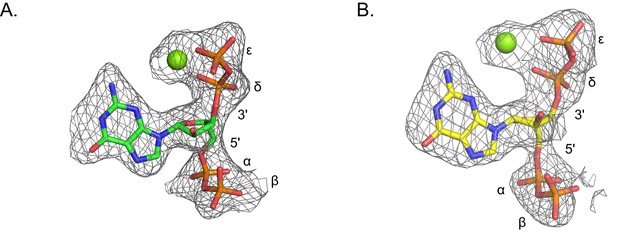
Omit electron densities of ppGpp crystallized with B. anthracis Hpt-1.
Omit electron densities contoured to 2σ of ppGpp crystallized with B. anthracis Hpt-1. ppGpp associated with molecule A of the asymmetric unit is shown in (A) and ppGpp associated with molecule B of the asymmetric unit is shown in (B). Green spheres represent Mg2+. Phosphates are labeled α through ϵ (α and β on 5′, δ and ϵ on 3′).
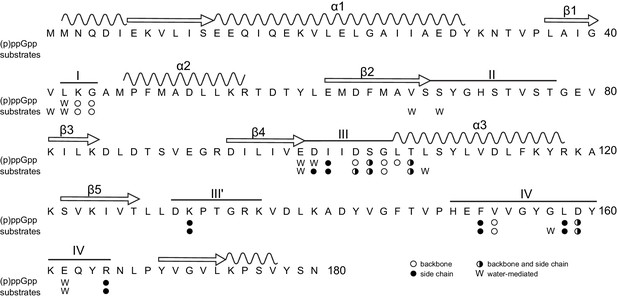
Primary structure of B. anthracis Hpt-1 showing ligand-interacting residues.
Primary structure of B. anthracis Hpt-1 with secondary structure elements annotated above (α1–3, β1–5, loops I – IV). Residues involved in (p)ppGpp and substrate binding are marked: empty circles denote residues with a backbone interaction with ligands; filled circles denote residues with a side chain interaction; half-filled circles represent residues with both backbone and side chain interactions; and ‘W’ represents water-mediated interactions.
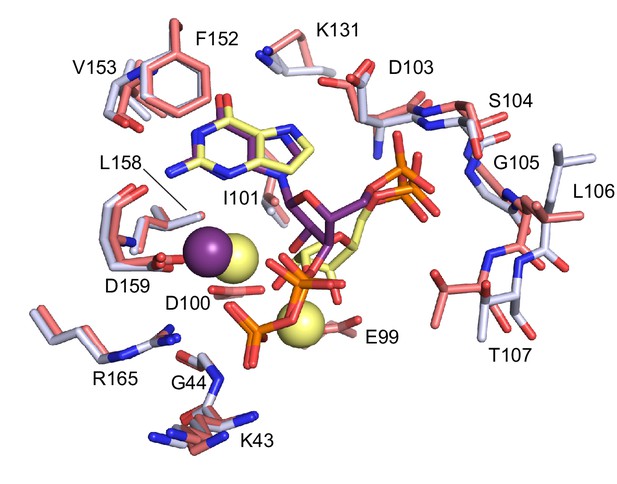
Comparison between ppGpp-bound and substrates-bound binding pocket.
Overlay of the active site of ppGpp-bound HPRT (silver) and substrates-bound HPRT (salmon). ppGpp is shown in purple and substrates are shown in yellow. Spheres represent Mg2+. E99 and D100, which interact with substrates, are not shown for ppGpp. Val67 and Ser69 on loop II, which interact with PRPP, are omitted.

LigPlots of ppGpp crystallized with B. anthracis Hpt-1.
Interaction diagrams generated by LigPlot for each ppGpp molecule in the asymmetric unit. Interactions for molecule A shown in (A) and interactions for molecule B shown in (B).

Conservation of the ppGpp-binding site across select bacteria and eukaryotes.
(p)ppGpp binding residues are conserved. Alignment of (p)ppGpp binding residues from representative bacterial and eukaryotic HPRTs. The bacteria are a subset of the 99 HPRTs used to make the frequency logo in Figure 3F. Numbering is according to B. anthracis Hpt-1.
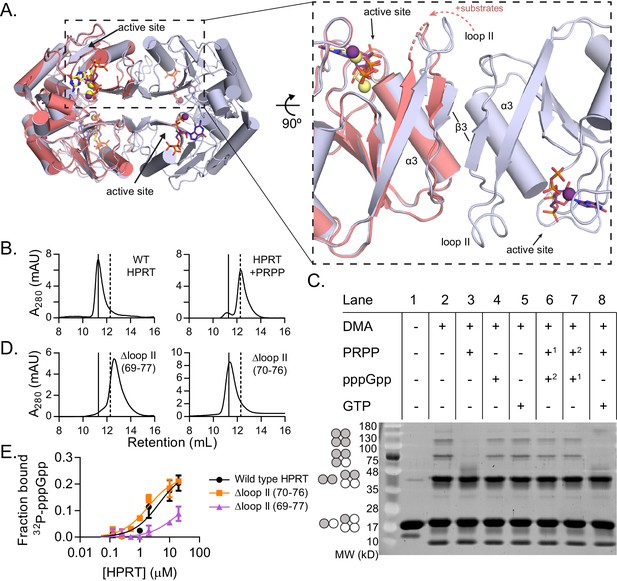
(p)ppGpp counteracts substrate-induced HPRT dimerization and HPRT tetramerization potentiates (p)ppGpp binding.
(A) Overlay of HPRT tetramer crystallized with ppGpp (silver) and HPRT dimer crystallized with substrates (salmon). ppGpp is purple and the substrates are yellow. Inset, View of the dimer–dimer interface within an HPRT tetramer. Secondary structure components at the interface are labeled β3, α3, and loop II. Figure 4—figure supplement 1 shows that changes in loop II are not induced by crystal contacts. (B) Size-exclusion chromatography of B. subtilis HPRT without ligand (left) and with PRPP (right). PRPP addition shifts the oligomeric state from tetramer to dimer. B. anthracis Hpt-1 is also a dimer with PRPP (see Figure 4—figure supplement 2). In B and D, solid line shows B. subtilis HPRT tetramer peak and dotted line shows dimer peak. PRPP must be in mobile phase to cause the shift (Figure 4—figure supplement 3), and the purine base does not affect the oligomeric state with PRPP (Figure 4—figure supplement 4). For molecular weight standards, see Figure 4—figure supplement 7. (C) B. subtilis HPRT crosslinked with dimethyl adipimidate (DMA). Crosslinked multimers were separated on SDS–PAGE. Predicted multimers are shown on the left. Shaded circles represent predicted to be crosslinked monomers, and white circles represent incomplete crosslinking. 1, 2 indicates order of incubation. GTP competition with PRPP did not recapitulate pppGpp blockage of PRPP-induced dimer–dimer dissociation which could be due to difference in affinity, whereby pppGpp binds strongly enough to outcompete PRPP but GTP does not. See Figure 4—figure supplement 5 for effect of crosslinked tetramer (Y117C) on HPRT activity and PRPP competition with pppGpp. See Figure 4—figure supplement 6 for ITC with PRPP. (D) Size-exclusion chromatography of B. subtilis HPRT with a partial loop II deletion (∆70–76) or a complete loop II deletion (∆ 69–77). The complete deletion results in dimerization of the protein. (E) Binding curves between 32P-labeled pppGpp and B. subtilis HPRT variants obtained with DRaCALA. The tetrameric Δloop II (70-76) variant (orange squares) binds as well as wild type HPRT. The dimeric Δloop II (69-77) variant (purple triangles) displays weaker binding. Error bars represent SEM of three replicates.
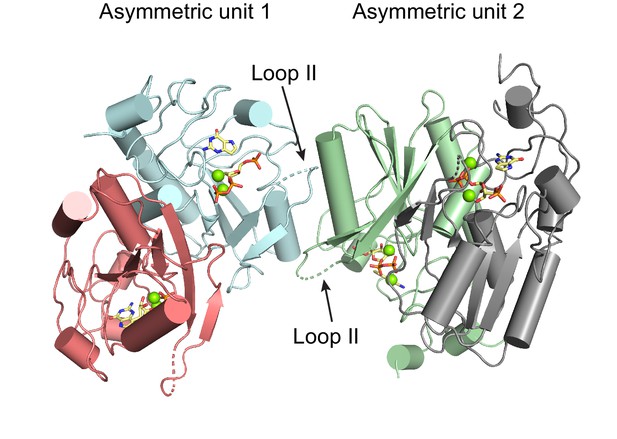
Crystal contacts do not induce changes in loop II in B. anthracis Hpt-1 crystallized with substrates.
Two asymmetric units of B. anthracis Hpt-1 crystallized with PRPP and 9-deazaguanine (substrates shown in yellow). Loop II from one asymmetric unit interacts with loop II and α3 from the neighboring asymmetric unit. Rather than inducing loop II movement toward the active site, these crystal contacts appear to prevent loop II from completely covering the active site, which has been observed in eukaryotic HPRTs crystallized with substrate analogs (Héroux et al., 2000; Shi et al., 1999). Loop II likely moves closer to the active site than captured in the crystal structure.
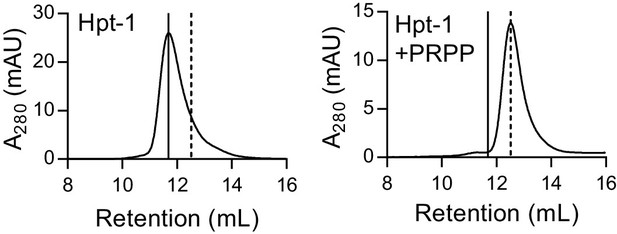
B. anthracis Hpt-1 is a tetramer without ligands and a dimer with PRPP.
Size-exclusion chromatography of B. anthracis Hpt-1 without ligands (left) and with 1 mM PRPP in the mobile phase (right). The solid vertical line represents a tetramer, and the dotted vertical line represents a dimer.
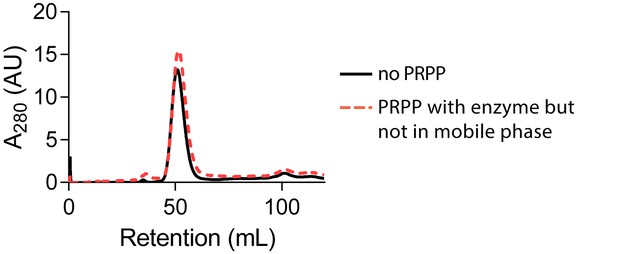
PRPP does not cause HPRT dimerization when not in the mobile phase.
Size-exclusion chromatography of B. subtilis HPRT without PRPP (black solid line) or with PRPP in the enzyme sample but not in the mobile phase (red dotted line). Chromatography was performed with a HiPrep Sephacryl S-100 HR column.
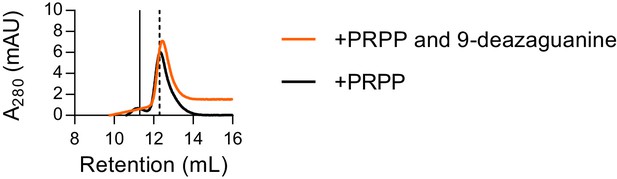
Size exclusion chromatography with both PRPP and 9-deazaguanine.
Size exclusion chromatography of B. subtilis HPRT with 500 µM PRPP (black) or 500 µM PRPP and 100 µM 9-deazaguanine (orange) shows that the purine base does not alter the enzyme’s oligomeric state in the presence of PRPP. Thus, PRPP alone is sufficient for dissociation of HPRT tetramers to dimers. We note that the small tetramer subpopulation present with PRPP alone is gone with both PRPP and the purine base analog present, suggesting that the addition of both substrates may further stabilize the dissociated dimeric state. Chromatogram for PRPP alone replicated from Figure 4B. Solid vertical line indicates apo HPRT and dashed vertical line indicates HPRT with PRPP.

Effect of crosslinked tetramer on activity and PRPP competition with pppGpp.
(B) subtilis HPRT variant Y117C is crosslinked across the dimer-dimer interface. (A) This crosslinked variant has reduced activity relative to WT, suggesting that PRPP is unable to dimerize the protein for activity. Reactions performed in duplicate with error bars representing the range. (B) DRaCALA of HPRT Y117C with 32P-labeled pppGpp in competition with PRPP. In WT HPRT, 1 mM PRPP outcompetes 32P-pppGpp, but 1 mM PRPP does not outcompete 32P-pppGpp for binding the crosslinked tetramer Y117C. Error bars represent SEM of triplicate.
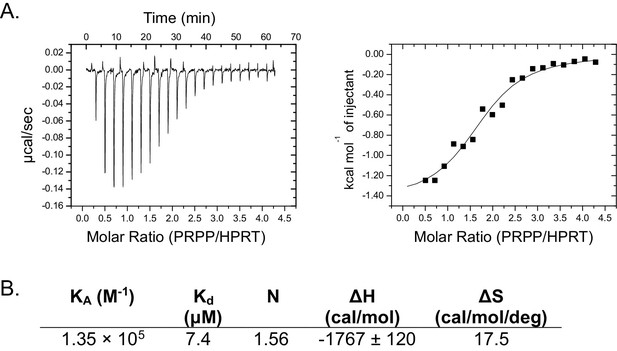
Isothermal titration calorimetry between PRPP and B. anthracis Hpt-1.
(A) Binding isotherm between PRPP and B. anthracis Hpt-1. Data are fitted to a single-site binding model. (B) Parameters from the binding isotherm in (A).
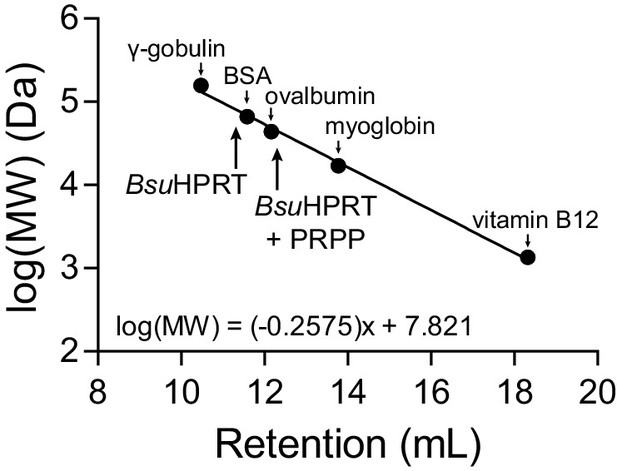
Molecular weight standards for size-exclusion chromatography.
Molecular weight standards for the Superose 12 10/300 GL size exclusion column. The standards are: γ-gobulin (158 kDa), bovine serum albumin (BSA) (66.5 kDa), ovalbumin (44 kDa), myoglobin (17 kDa), and vitamin B12 (1.35 kDa). The equation is a linear regression of the log molecular weight in Daltons versus the retention volume of each marker. Arrows point to the retention volumes of B. subtilis HPRT with and without PRPP.
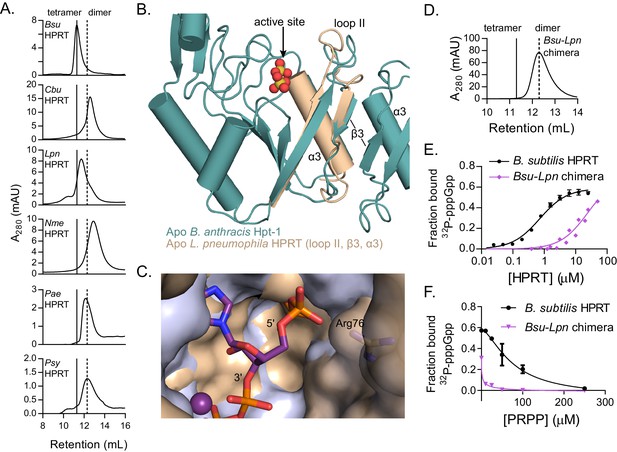
Dimer–dimer interaction holds loop II away from the (p)ppGpp binding pocket to potentiate (p)ppGpp binding.
(A) Size-exclusion chromatograms show that C. burnetii, L. pneumophila, N. meningitidis, P. aeruginosa, and P. syringae HPRTs are non-tetrameric without ligands. Vertical lines represent B. subtilis HPRT tetramer and dimer peaks. (B) Overlay of apo B. anthracis Hpt-1 (teal) and the L. pneumophila HPRT interface (wheat; PDB ID 5ESW). In L. pneumophila HPRT, loop II and β3 are further from the dimer-dimer interface and closer to the active site. See Figure 5—figure supplement 1 for overlay with substrates-bound HPRT. (C) Overlay of B. anthracis Hpt-1 – ppGpp (silver; ppGpp in purple) and L. pneumophila HPRT (wheat; PDB ID 5ESX) shows that the 5′ phosphate binding pocket is compressed by the conformation of loop II in the dimeric HPRT. Arg76 (side chain shown as sticks) in L. pneumophila HPRT forms part of the compressed pocket. Asp109 in L. pneumophila HPRT is hidden due to poor electron density. (D) Size-exclusion chromatogram of the dimeric Bsu-Lpn chimera. The chimera has 21 residues at the B. subtilis HPRT interface replaced with L. pneumophila HPRT residues (see Figure 5—figure supplement 2). (E) DRaCALA shows that the Bsu-Lpn chimera binds 32P-labeled pppGpp more weakly. Error bars represent SEM of three replicates. Bsu-Lpn chimera performed in duplicate. Figure 5—figure supplement 3 shows that stability of dimeric HPRTs is not compromised. (F) Competition between PRPP and 32P-labeled pppGpp shows that the dimeric Bsu-Lpn chimera is more sensitive to PRPP than the tetrameric wild type. Error bars are range of duplicate.
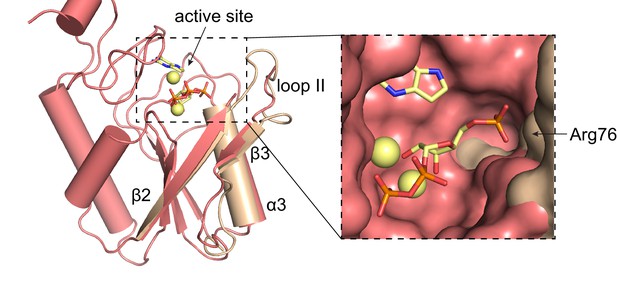
Overlay of L. pneumophila HPRT and substrates-bound B. anthracis Hpt-1.
Overlay of substrate-bound B. anthracis Hpt-1 (salmon) and loop II, β3, and α3 from apo L. pneumophila HPRT (wheat; PDB ID 5ESW). PRPP and 9-deazaguanine are shown as yellow sticks and Mg2+ as yellow spheres. Inset shows surface view of the binding pocket with the L. pneumophila HPRT interface components.
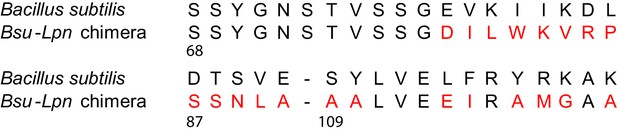
Residues replaces in B. subtilis HPRT for the Bsu-Lpn chimera.
Residues at the B. subtilis (Bsu) HPRT dimer–dimer interface replaced with corresponding residues from L. pneumophila (Lpn) HPRT (shown in red).
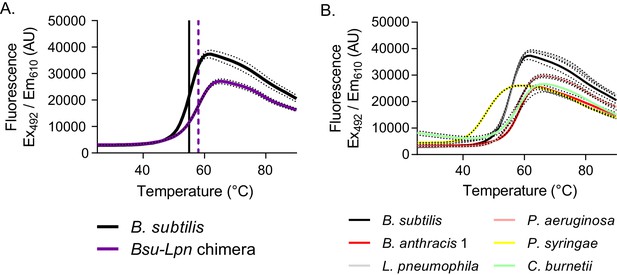
Stability of dimeric HPRTs is not compromised.
(A) Fluorescence intensity versus temperature of B. subtilis HPRT (black) and the Bsu-Lpn chimera (purple) in the presence of SYPRO orange. The vertical lines mark the melting temperature, determined from the first derivative of the melting curve (black solid line for B. subtilis HPRT and purple dashed line for the Bsu-Lpn chimera). Dashed lines represent SEM for reactions performed in triplicate. (B) Fluorescence intensity versus temperature of bacterial HPRTs in the presence of SYPRO orange. Increased melting temperature indicates increased stability, and protein stability is independent of oligomeric state among the HPRT homologs tested. Dashed lines represent SEM for reactions performed in triplicate. B. subtilis HPRT data are the same as in panel A.
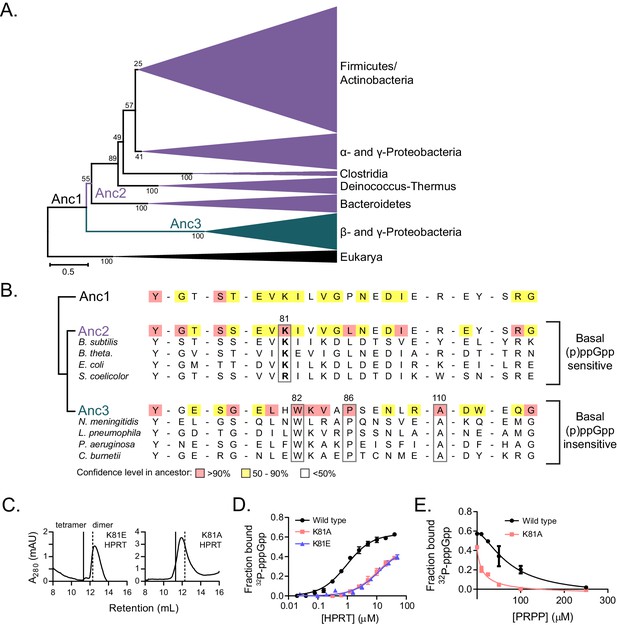
Coevolution of the HPRT dimer-dimer interface and regulation by basal levels of (p)ppGpp.
(A) A maximum likelihood phylogenetic tree of 430 HPRT amino acid sequences rooted on eukaryotic HPRTs. Bootstrap values from 100 replicates are shown. Anc1-3 refer to ancestral HPRTs. The tree reveals two main lineages: one with dimer-dimer interaction motifs (Anc2; purple) and one lacking the interaction motifs (Anc3; blue). Branch length scale indicates 0.5 substitutions per site. See Materials and methods for tree construction. See Source Data one for the full alignment of HPRTs. (B) Alignment of HPRT dimer-dimer interface from Anc1, Anc2, Anc3, and select extant HPRTs. Nearly all (p)ppGpp-regulated HPRTs share Lys81 found in Anc2. Dimeric HPRTs insensitive to basal (p)ppGpp lack Lys81, but share Trp82, Pro86, and Ala110 (B. anthracis Hpt-1 numbering). Figure 6—figure supplement 1 shows the role of these residues at the interface. For the ancestors, red indicates high (>90%), yellow moderate (50–90%), and no color low (<50%) confidence level in the residue identity. (C) B. subtilis K81E and K81A HPRTs have weakened tetramerization according to size-exclusion chromatography. Figure 6—figure supplement 2 shows Lys81 at the dimer–dimer interface. (D) Binding of 32P-labeled pppGpp to wild type (black circles), K81A (red squares), and K81E (blue triangles) HPRTs using DRaCALA. Error bars represent SEM of three replicates. (E) PRPP competition with 32P-labeled pppGpp for binding wild type (black circles) and K81A HPRTs (red squares). K81A is more sensitive to PRPP competition than the tetrameric wild type. Error bars are range of duplicate.
-
Figure 6—source data 1
Protein alignment of HPRTs.
- https://doi.org/10.7554/eLife.47534.036

Role of residues conserved in the dimer–dimer interface of dimeric HPRTs.
(A) The role of interface residues conserved in dimeric HPRTs. Overlay of B. anthracis Hpt-1 (teal) and L. pneumophila HPRT (wheat; PDB ID 5ESW). Ile82, Leu86, and Tyr110 are shown from B. anthracis Hpt-1, and the homologous residues Trp88, Pro92, and Ala116 are shown from L. pneumophila HPRT. Trp88 packs between loop II and α3. Ala116, which replaces Tyr110, may help accommodate Trp88. Pro86 introduces a kink below loop II that may promote the packing of Trp88 against α3. (B) Variants conserved in dimeric HPRTs weaken tetramerization. Size-exclusion chromatography of Y110A, I82W/Y110A, and I82W/L86P/Y110A B. subtilis HPRT variants. Solid vertical lines represent an expected HPRT tetramer and dotted vertical line represents an expected dimer.
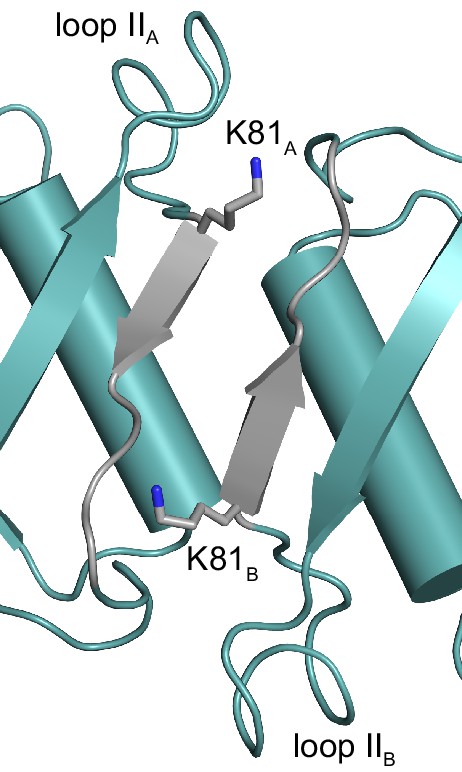
The conserved Lys81 bridges the dimer–dimer interface of tetrameric HPRTs.
The structure of the dimer-dimer interface of B. anthracis Hpt-1 with the side chain of Lys81 shown as sticks. Regions colored gray correspond with residues 81–87 (see Figure 6B). The subscripts A and B refer to the subunit contributing the residues to the interface interaction.
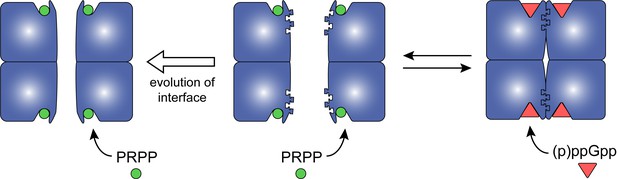
Model for how evolution of protein oligomerization affects ligand-mediated regulation.
For an enzyme such as HPRT, where the inhibitor (p)ppGpp binds to nearly identical sites as the substrates (PRPP), evolutionary plasticity of inhibition can be mediated through subunit oligomerization. Dimeric HPRTs present a smaller binding pocket that preferentially bind to the substrate than to (p)ppGpp. On the other hand, HPRTs with a dimer–dimer interface motif would allosterically open the pocket, thus favoring (p)ppGpp binding and inhibition than substrate-binding and activity.
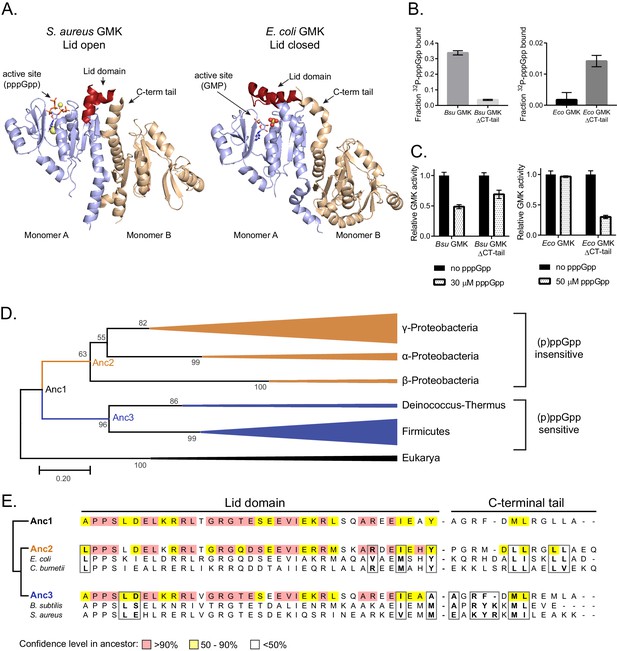
Protein–protein interface allosterically alters the binding of (p)ppGpp to guanylate kinase.
(A) In S. aureus GMK (left; PDB ID 4QRH), an open lid domain (red) allows (p)ppGpp binding at the active site, but in E. coli GMK (right; PDB ID 2ANB), the closed lid domain occludes (p)ppGpp binding at the active site (bound to GMP). A C-terminal tail in the second monomer of a GMK dimer interacts with the lid domain in both proteins. Nineteen residues in the lid domain and six residues in the C-terminal tail of S. aureus GMK are unresolved. (B) DRaCALA of full-length and ∆C-terminal tail (∆CT tail) B. subtilis and E. coli GMKs. Error bars are mean of duplicate. (C) Inhibition of GMKs from (D) relative to uninhibited activity. Error bars are SEM of triplicate. (D) Maximum likelihood phylogenetic tree of 41 GMKs rooted on eukaryotic GMKs as the outgroup. Bootstrap values from 1000 replicates are shown. Anc1-3 refer to ancestral HPRTs. See Materials and methods for tree construction. See Source Data one for the full alignment of GMKs. (E) Alignment of the lid domain and C-terminal tail of ancestral and select extant GMKs. Boxed residues are part of the lid domain - C-terminal tail interaction.
-
Figure 8—source data 1
Protein alignment of GMKs.
- https://doi.org/10.7554/eLife.47534.038
Tables
(p)ppGpp binding and inhibition of HPRT homologs.
https://doi.org/10.7554/eLife.47534.005| Organism | Relative activityd | Kd (pppGpp) (μM) | IC50 (pppGpp) (μM) | |
|---|---|---|---|---|
| 25 μM ppGpp | 25 μM pppGpp | |||
| Actinobacteria | ||||
| Streptomyces coelicolor | −0.01 ± 0.01 | 0.12 ± 0.01 | 1.77 ± 0.31 | 3.8 ± 0.2 |
| Collinsella aerofaciens | 0.31 ± 0.04 | 0.42 ± 0.05 | 1.46 ± 0.56 | |
| Cellulomonas gilvus | NAb | NAb | 0.6 ± 0.11 | |
| Mycobacterium tuberculosis | NAa | NAa | NAc | |
| Bacteroidetes | ||||
| Bacteroides intestinalis | 0.49 ± 0.06 | 0.05 ± 0.01 | 0.11 ± 0.03 | |
| Bacteroides caccae | 0.34 ± 0.03 | 0.01 ± 0.02 | 0.11 ± 0.02 | |
| Bacteroides ovatus | 0.43 ± 0.04 | −0.03 ± 0.02 | 0.15 ± 0.04 | |
| Bacteroides uniformis | 0.4 ± 0.01 | 0.01 ± 0.01 | 0.1 ± 0.04 | |
| Bacteroides thetaiotaomicron | 0.41 ± 0 | 0 ± 0.01 | 0.11 ± 0.04 | |
| Bacteroides finegoldii | 0.46 ± 0.03 | −0.01 ± 0.01 | 0.15 ± 0.06 | |
| Firmicutes | ||||
| Eubacterium ventriosum | −0.03 ± 0.03 | −0.07 ± 0.03 | 0.66 ± 0.1 | |
| Ruminococcus lactaris | −0.01 ± 0 | 0 ± 0 | 0.11 ± 0.03 | |
| Ruminococcus torques | NAb | NAb | 0.22 ± 0.07 | |
| Clostridium leptum | −0.01 ± 0.01 | 0 ± 0 | 3.29 ± 0.94 | |
| Eubacterium eligens | −0.03 ± 0.03 | 0.02 ± 0.01 | 0.79 ± 0.18 | |
| Blautia hansenii | 0.07 ± 0 | 0.14 ± 0.01 | 0.83 ± 0.14 | |
| Eubacterium hallii | 0.17 ± 0.05 | −0.06 ± 0.05 | 0.07 ± 0.02 | |
| Clostridium spiroforme | 0.07 ± 0.01 | 0.17 ± 0 | 9.54 ± 1.17 | |
| Listeria monocytogenes | 0.13 ± 0 | 0.32 ± 0.02 | 0.32 ± 0.06 | |
| Bacillus subtilis | 0.08 ± 0.01 | 0.41 ± 0.01 | 0.75 ± 0.07 | 10.1 ± 1.3 |
| Staphylococcus aureus | 0.31 ± 0.01 | 0.48 ± 0.01 | 2.83 ± 0.38 | 10.6 ± 0.3 |
| Streptococcus mutans | NAb | NAb | 5.12 ± 1.45 | |
| Bacillus anthracis 1 | 0.25 ± 0.01 | 0.5 ± 0.01 | 1.54 ± 0.18 | 29.5 ± 1.2 |
| Enterococcus faecalis | 0.31 ± 0.01 | 0.49 ± 0.02 | 1.12 ± 0.11 | |
| Holdemania filiformis | 0.66 ± 0.01 | 0.43 ± 0 | 0.17 ± 0.11 | |
| Bacillus anthracis 2 | 0.81 ± 0.04 | 0.7 ± 0.05 | 6.18 ± 1.77 | |
| Deinococcus-Thermus | ||||
| Thermus thermophilus | 0.52 ± 0.07 | −0.01 ± 0.01 | 0.04 ± 0.01 | |
| Proteobacteria | ||||
| Caulobacter crescentus | 0.33 ± 0.05 | 0.47 ± 0.07 | 0.24 ± 0.09 | |
| Escherichia coli | 0.33 ± 0.01 | 0.48 ± 0.02 | 3.38 ± 1.47 | 26.2 ± 2.0 |
| Klebsiella pneumoniae | 0.47 ± 0.03 | 0.49 ± 0.01 | 3.38 ± 0.89 | |
| Sinorhizobium meliloti | NAb | NAb | 3.08 ± 0.92 | |
| Yersinia enterocolitica | 0.7 ± 0.01 | 0.7 ± 0.03 | 4.44 ± 1.25 | |
| Photobacterium phosphoreum | 0.97 ± 0.01 | 0.96 ± 0.03 | NAc | >100 |
| Pseudomonas syringae | 0.6 ± 0.01 | 0.6 ± 0.01 | 5.85 ± 1.76 | |
| Pseudomonas aeruginosa | 0.84 ± 0.01 | 0.83 ± 0.02 | 5.28 ± 1.52 | |
| Legionella pneumophila | 0.78 ± 0.01 | 0.69 ± 0.03 | 1.72 ± 0.38 | >100 |
| Coxiella burnetii | 0.79 ± 0.14 | 0.72 ± 0.16 | NAc | >100 |
| Neisseria meningitidis | 0.9 ± 0.02 | 0.8 ± 0.03 | 1.64 ± 0.55 | |
| Eukarya | ||||
| Saccharomyces cerevisiae | 0.64 ± 0.02 | 0.96 ± 0.01 | 8.08 ± 8.58 | |
| Caenorhabditis elegans | NAb | NAb | 12.77 ± 7.68 | 29.6 ± 2.8 |
| Homo sapiens | NA | NA | NA | 66.6 ± 5.4 |
-
± standard error of the mean.
NA = no data obtained.
-
aProtein not purified.
bProtein did not have activity.
-
cInteraction with pppGpp too weak to estimate Kd.
dRelative to activity without ppGpp or pppGpp.
X-ray data collection and structure determination statistics.
https://doi.org/10.7554/eLife.47534.015| Data collection | |||
|---|---|---|---|
| Structure | HPRT with sulfates (PDB 6D9Q) | HPRT with substrates (PDB 6D9R) | HPRT with ppGpp (PDB 6D9S) |
| Wavelength | 0.9786 | 0.97872 | 0.9786 |
| Resolution range (highest resolution bin) (Å) | 41.3–2.06 (2.13–2.06) | 50–1.64 (1.67–1.64) | 40.2–2.11 (2.18–2.11) |
| Space group | P 31 2 1 | P 32 2 1 | P 31 2 1 |
| Unit cell | |||
| a, b, c (Å) | 82.597, 82.597, 242.416 | 113.758, 113.758, 56.731 | 82.61, 82.61, 174.92 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 |
| Completeness (%) | 99.16 (95.52) | 99.8 (99.0) | 98.92 (94.51) |
| Unique reflections | 60264 | 51750 | 40413 |
| Redundancy | 10.8 (6.4) | 21.5 (14.4) | 8.1 (6.2) |
| I/σI | 24.8 (1.74) | 31.09 (2.93) | 16.41 (1.22) |
| Rmeas | 0.124 | 0.137 (1.376) | 0.164 |
| Rpim | 0.037 | 0.029 (0.35) | 0.06 |
| CC 1/2 | (0.791) | (0.81) | (0.724) |
| Refinement | |||
| Resolution range (highest resolution bin) (Å) | 41.3–2.06 (2.11–2.06) | 37.19–1.64 (1.70–1.64) | 40.2–2.11 (2.16–2.11) |
| Rwork/Rfree a (%) | 18.2/21.7 | 16.5/19.6 | 20.2/24.4 |
| r.m.s.b deviations | |||
| Bonds (Å) | 0.004 | 0.01 | 0.003 |
| Angles (Å) | 1.03 | 1.41 | 0.812 |
| Ramachandran statistics (%) | |||
| Favored | 97.90 | 98.52 | 96.63 |
| Allowed | 2.10 | 1.48 | 3.37 |
| Disallowed | 0.00 | 0.00 | 0.00 |
| Rotamer outliers (%) | 0.78 | 0.30 | 0.63 |
| No. of atoms | |||
| Macromolecules | 5742 | 2881 | 2837 |
| Ligands | 46 | 161 | 116 |
| Solvent | 440 | 317 | 118 |
| B factor (Å2) | |||
| Macromolecules | 50.63 | 24.06 | 76.03 |
| Ligands | 68.83 | 37.76 | 161.17 |
| Solvent | 52.15 | 35.53 | 70.22 |
-
aRwork/Rfree = Σ||Fobs|-|Fcalc||/|Fobs|, where the working the free R factors are calculated by using the working and free reflection sets, respectively. The free R reflections were held aside throughout refinement. bRoot mean square. See Table 2—source datas 1–3 for PDB validation reports.
-
Table 2—source data 1
Validation report for PDB ID 6D9Q.
- https://doi.org/10.7554/eLife.47534.016
-
Table 2—source data 2
Validation report for PDB ID 6D9R.
- https://doi.org/10.7554/eLife.47534.017
-
Table 2—source data 3
Validation report for PDB ID 6D9S.
- https://doi.org/10.7554/eLife.47534.018
Dynamic light scattering of B. anthracis Hpt-1 with pppGpp and PRPP.
https://doi.org/10.7554/eLife.47534.027| Sample | RHa (nm) | Polydb | % Polyd | Est. MW (kDa) |
|---|---|---|---|---|
| HPRT | 4.1 | 0.9 | 23.1 | 90 |
| HPRT + PRPP | 3.1 | 0.7 | 22.5 | 47 |
| HPRT + pppGpp | 4.1 | 1.0 | 24.9 | 91 |
| HPRT + both | 4.2 | 1.2 | 27.8 | 97 |
-
PRPP = phosphoribosyl pyrophosphate.
aRH = hydrodynamic radius.
-
bPolyd = polydispersity.
HPRT kinetic parameters.
https://doi.org/10.7554/eLife.47534.032| HPRT | Km (guanine) (μM) | Km (PRPP) (μM) | kcat (s−1) |
|---|---|---|---|
| B. anthracis 1 | 5.2 ± 1.8 | 111.6 ± 16.8 | 22 |
| B. subtilis | -- | 165.6 ± 11.9 | 25.4 |
| C. burnetii | 17.6 ± 2.0 | 44 ± 3.3 | 12.9 |
| P. phosphoreum | -- | 81 ± 12.5a | 30.4a |
| N. meningitidis | 68 ± 7.0 | 103 ± 12 | 9.6 |
| E. colib | -- | 192 ± 7.0a | 59a |
| L. pneumophilac | 10.5 ± 1.1 | 60 ± 9 | -- |
| M. tuberculosisd | 10 ± 1 | 650 ± 70 | 0.193 |
-
PRPP = phosphoribosyl pyrophosphate.
aobtained with hypoxanthine as the purine base.
-
± standard error.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Bacillus subtilis) | gmk::ecgmk | (Liu et al., 2015b) | N/A | |
| Strain, strain background (B. subtilis) | gmk::ecgmk amyE::Phyperspac-ecgpt | This work | N/A | |
| Strain, strain background (B. subtilis) | gmk::ecgmk amyE::Phyperspac-bshprT | This work | N/A | |
| Recombinant DNA reagent | pLIC-trPC-HA (plasmid) | (Stols et al., 2002) | N/A | |
| Recombinant DNA reagent | pHM1381 (plasmid) | (Mechold et al., 2002) | N/A | |
| Recombinant DNA reagent | pDR90 (plasmid) | (González-Pastor et al., 2003) | N/A | |
| Peptide, recombinant protein | human HPRT1 | Novoprotein | Novoprotein #C294 | |
| Chemical compound, drug | 5-phospho-D -ribose 1-diphosphate pentasodium salt (PRPP) | MilliporeSigma | MilliporeSigma # P8296 | |
| Chemical compound, drug | dimethyl adipimidate (DMA) | ThermoFisher | ThermoFisher # 5001417627 | |
| Chemical compound, drug | 9-deazaguanine | Santa Cruz Biotechnology | Santa Cruz #sc-217528 | |
| Chemical compound, drug | SYPRO Ruby | Bio-Rad | Bio-Rad #1703125 | |
| Chemical compound, drug | SYPRO Orange | MilliporeSigma | MilliporeSigma #S5692 | |
| Chemical compound, drug | guanine | MilliporeSigma | MilliporeSigma #G6779 | |
| Software, algorithm | GraphPad Prism v. 5.02 (software) | https://www.graphpad.com/ | RRID:SCR_002798 | |
| Software, algorithm | ImageQuant (software) | https://www.gelifesciences.com | RRID:SCR_014246 | |
| Software, algorithm | MicroCal Origin 5.0 (software) | https://www.malvernpanalytical.com | RRID:SCR_002815 | |
| Software, algorithm | MEGA X (software) | https://www.megasoftware.net/ | RRID:SCR_000667 | |
| Software, algorithm | Phenix (software) | https://www.phenix-online.org/ | RRID:SCR_014224 | |
| Software, algorithm | Coot (software) | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ | RRID:SCR_014222 | |
| Software, algorithm | Dynamics v. 5.25.44 (software) | https://www.wyatt.com/products/software/dynamics.html | N/A |
Additional files
-
Supplementary file 1
Primers, plasmids, and strains.
- https://doi.org/10.7554/eLife.47534.040
-
Transparent reporting form
- https://doi.org/10.7554/eLife.47534.041





