Protein engineering expands the effector recognition profile of a rice NLR immune receptor
Figures
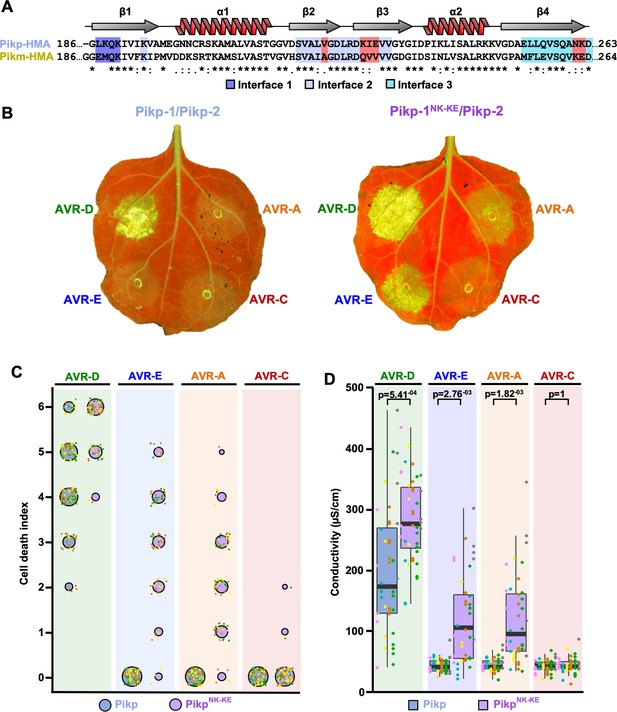
Structure-informed engineering expands Pikp-mediated effector recognition in N. benthamiana.
(A) Sequence alignment of the Pikp-1 and Pikm-1 HMA domains. Secondary structure features of the HMA fold are shown above, and the residues that are located at binding interfaces are as colored. Key residues from interface 2 and interface 3 involved in this study are highlighted in red. (B) Representative leaf images showing Pikp- (left) or Pikp-1NK-KE (right)-mediated cell death in response to AVR-Pik variants as autofluorescence under UV light. (C) Autofluorescence intensity is scored as previously described (Maqbool et al., 2015; De la Concepcion et al., 2018). Cell death assay scores are represented as dot plots for Pikp and PikpNK-KE (blue and purple, respectively). For each sample, all of the data points are represented as dots with a distinct color for each of the three biological replicates; these dots are plotted around the cell death score for visualization purposes. The size of the centre dot at each cell death value is directly proportional to the number of replicates in the sample with that score. The total number of repeats was 80. Data for Pikp have been previously shown (De la Concepcion et al., 2018), but was acquired at the same time as those for PikpNK-KE. The estimation methods used to visualize differences in the data sets are shown in Figure 1—figure supplement 3. (D) Conductivity measurements showing ion leakage as a quantitative measure of cell death. The centre line represents the median, the box limits are the upper and lower quartiles, the whiskers extend to the largest value within Q1 – 1.5x the interquartile range (IQR) and the smallest value within Q3 + 1.5x IQR. All the data points are shown as dots with distinct colors for each biological replicate. For each experiment, six biological replicates with 5 or 10 internal repeats were performed (total data points = 40). ‘p’ is the p-value obtained from statistical analysis and Tukey’s HSD (honestly significant difference) test.
-
Figure 1—source data 1
Cell death scoring data used in the preparation of Figure 1C.
- https://doi.org/10.7554/eLife.47713.009
-
Figure 1—source data 2
Conductivity measurements used in the preparation of Figure 1D and Figure 1—figure supplement 2B.
- https://doi.org/10.7554/eLife.47713.010
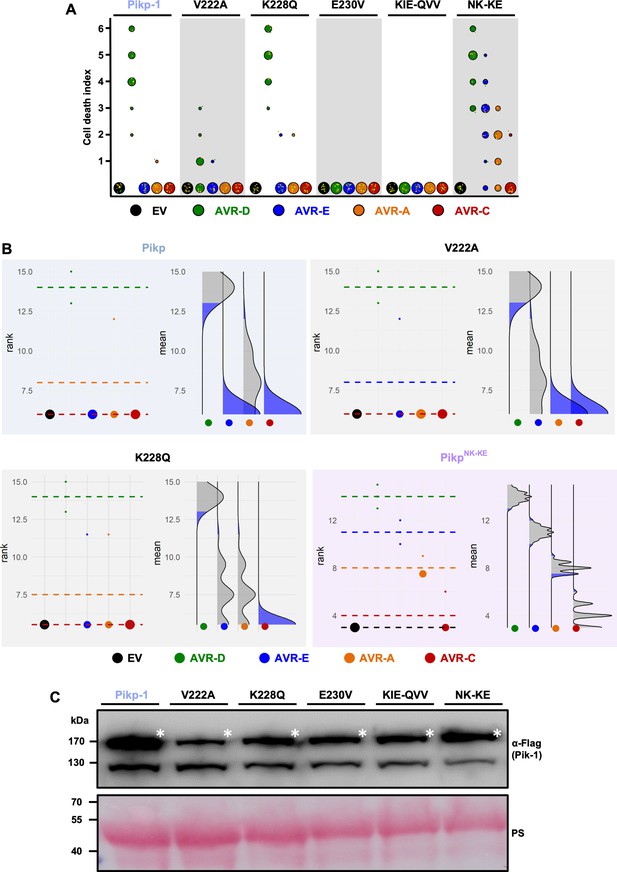
Mutations at interface 2 of the Pikp-1 HMA domain compromise the response to AVR-Pik effectors.
(A) Cell-death assay scoring represented as dot plots for Pikp-1 mutants with mutations on HMA interface 2 and 3. For each sample, all of the data points are represented as dots with a distinct color for each of the three biological replicates; these dots are plotted around the cell death score for visualization purposes. The size of the central dot at each cell death value is proportional to the number of replicates of the sample with that score. The number of repeats was 18 for each mutant. (B) Statistical analysis by estimation methods of the cell death assay for Pikp-1 mutants. The left panel represents the ranked data (dots) for each effector, and their corresponding mean (dotted line). The size of the dots is proportional to the number of observations with that value. The right panel shows the distribution of 1000 bootstrap sample rank means for each effector. The blue areas represent the 0.025 and 0.975 percentiles of the distribution. A sample (effector) score is considered significantly different from the control (EV) when the control rank mean (dotted line on the left) falls beyond the blue regions of the effector mean distribution. When the rank means for different effectors have the same value, only one dotted line is visible (EV, AVR-E and AVR-C for Pikp and K228Q, and EV, AVR-A and AVR-C for V222A). (C) Western blot analysis confirming similar levels of Pik-1 protein accumulation in N. benthamiana. The asterisks mark the Pik-1 band, PS = Ponceau Stain.
-
Figure 1—figure supplement 1—source data 1
Cell death scoring data used in the preparation of Figure 1—figure supplement 1A.
- https://doi.org/10.7554/eLife.47713.004
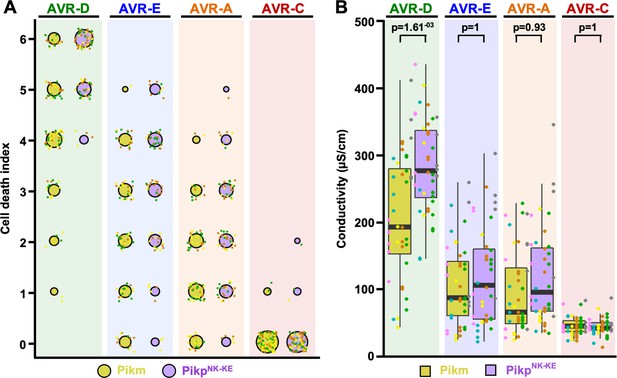
Response of PikpNK-KE to AVR-Pik effectors compared to that of Pikm.
(A) Cell-death autofluorescence scoring represented as dot plots for Pikm and PikpNK-KE (yellow and purple, respectively). The numbers of repeats were 80 and 90 for PikpNK-KE and Pikm, respectively. For each sample, all of the data points are represented as dots with a distinct color for each of the three biological replicates; these dots are plotted around the cell-death score for visualization purposes. The size of the central dot at each cell death value is proportional to the number of replicates of the sample with that score. Data for Pikm have been previously shown (De la Concepcion et al., 2018), but were acquired at the same time as those for PikpNK-KE. The estimation methods used to visualize differences in the data sets are shown in Figure 1—figure supplement 4. (B) Conductivity measurements showing ion leakage as a quantitative measure of cell death. The centre line represents the median, the box limits are the upper and lower quartiles, the whiskers extend to the largest value within Q1 – 1.5x the interquartile range (IQR) and the smallest value within Q3 + 1.5x IQR. All of the data points are shown as dots with distinct colors for each biological replicate. For each experiment, six biological replicates with 5 or 10 internal repeats were performed (total data points = 40). ‘p’ is the p-value obtained from statistical analysis and Tukey’s HSD.
-
Figure 1—figure supplement 2—source data 1
Cell-death scoring data used in the preparation of Figure 1—figure supplement 2A.
- https://doi.org/10.7554/eLife.47713.006
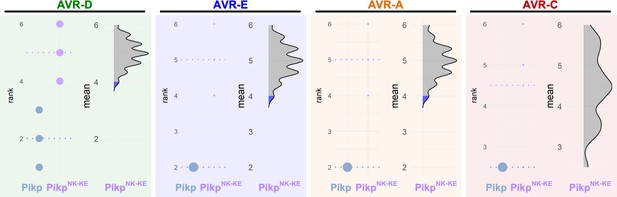
Estimation graphics for cell death, Pikp vs PikpNK-KE.
Statistical analysis by estimation methods of the cell-death assay for Pikp and PikpNK-KE. For each effector, the panel on the left represents the ranked data (dots) for each NLR, and their corresponding mean (dotted line). The size of the dots is proportional to the number of observations with that specific value. The panel on the right shows the distribution of 1000 bootstrap sample rank means for PikpNK-KE. The blue areas represent the 0.025 and 0.975 percentiles of the distribution. The response of Pikp and PikpNK-KE are considered significantly different if the Pikp rank mean (dotted line, left panel) falls beyond the blue regions of the PikpNK-KE mean distribution.
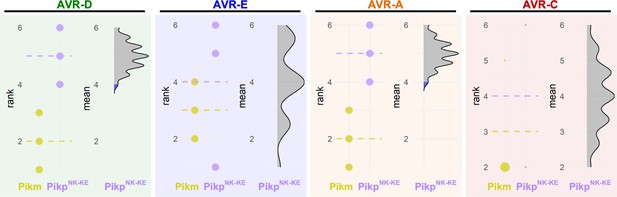
Estimation graphics for cell death, Pikm vs PikpNK-KE.
Statistical analysis by estimation methods of the cell-death assay for Pikm and PikpNK-KE. For each effector, the panel on the left represents the ranked data (dots) for each NLR, and their corresponding mean (dotted line). The size of the dots is proportional to the number of observations with that specific value. The panel on the right shows the distribution of 1000 bootstrap sample rank means for PikpNK-KE. The blue areas represent the 0.025 and 0.975 percentiles of the distribution. The responses of Pikm and PikpNK-KE are considered significantly different if the Pikp rank mean (dotted line, left panel) falls beyond the blue regions of the PikpNK-KE mean distribution.
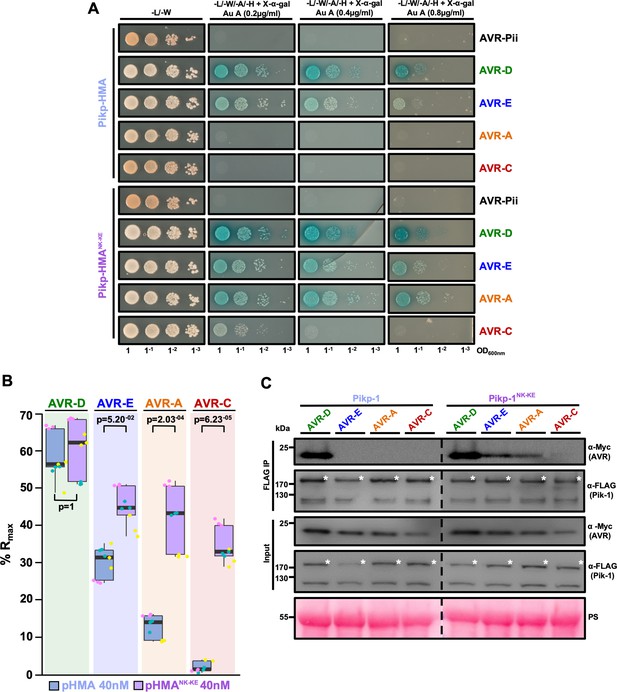
PikpNK-KE shows increased binding to effector variants in vivo and in vitro when compared to wild type Pikp.
(A) Yeast-two-hybrid assay of Pikp-HMA and Pikp-HMANK-KE with AVR-Pik alleles. Control plates for yeast growth are on the left, with quadruple dropout media supplemented with X-α-gal and increasing concentrations of Aureobasidin A on the right for each combination of HMA/AVR-Pik. The unrelated M. oryzae effector AVR-Pii was used as a negative control. Growth and the development of blue coloration in the selection plate are both indicative of protein–protein interaction. HMA domains were fused to the GAL4 DNA binding domain, and AVR-Pik alleles to the GAL4 activator domain. Each experiment was repeated a minimum of three times, with similar results. (B) Box plots showing %Rmax, as measured by surface plasmon resonance, for Pikp-HMA and Pikp-HMANK-KE with the AVR-Pik effectors alleles at an HMA concentration of 40 nM. Pikp-HMA and Pikp-HMANK-KE are represented by blue and purple boxes, respectively. The centre line within each box represents the median, the box limits are the upper and lower quartiles, the whiskers extend to the largest value within Q1 – 1.5 × the interquartile range (IQR) and the smallest value within Q3 + 1.5 × IQR. All of the data points are represented as dots with distinct colors for each biological replicate. For each experiment, three biological replicates with three internal repeats were performed. ‘p’ is the p-value obtained from statistical analysis and Tukey’s HSD. For results of experiments with 4 nM and 100 nM HMA protein concentration, see Figure 2—figure supplement 2. (C) Co-immunoprecipitation of full length Pikp-1 and Pikp-1NK-KE with AVR-Pik variants. N-terminally 4xMyc tagged AVR-Pik effectors were transiently co-expressed with Pikp-1:6xHis3xFLAG (left) or with Pikp-1NK-KE:6xHis3xFLAG (right) in N. benthamiana. Immunoprecipitates (IPs) obtained with anti-FLAG antiserum, and total protein extracts, were probed with appropriate antisera. The dashed line indicates a crop site on the same blot used to compose the figure. Each experiment was repeated at least three times, with similar results. The asterisks mark the Pik-1 band. PS = Ponceau Stain.
-
Figure 2—source data 1
Surface plasmon resonance measurements used in the preparation of Figure 2B.
- https://doi.org/10.7554/eLife.47713.018
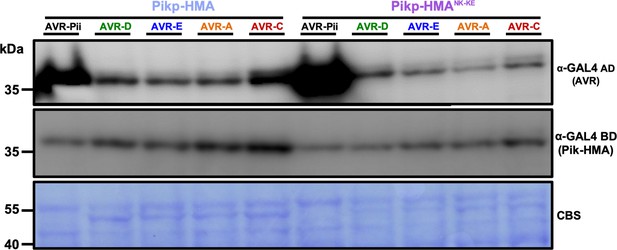
Western blot confirming the accumulation of proteins in yeast.
Yeast lysate was probed for the expression of HMA domains with anti-GAL4 DNA-binding domain (BD) or AVR-Pik/AVR-Pii effectors anti-GAL4 activation domain (AD). Total extract was colored with Ponceau Stain (PS). The experiment was repeated a minimum of 3 times, with similar results. PS = Ponceau Stain.
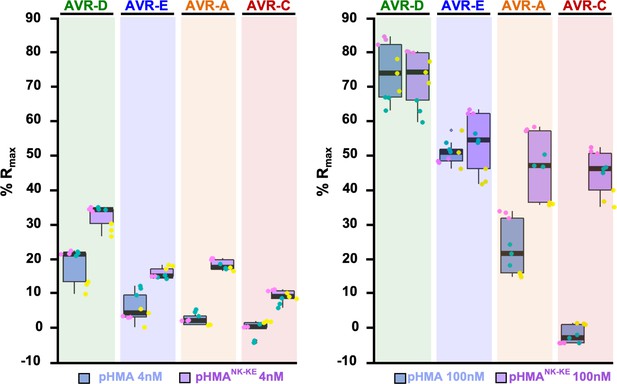
Binding of the Pikp-HMANK-KE domain to the AVR-Pik effectors is consistently greater than that of Pikp-HMA.
%Rmax of Pikp-HMA and Pikp-HMANK-KE with each AVR-Pik effectors allele with HMA concentrations of 4 nM (left) and 100 nM (right). The centre line within each box represents the median, the box limits are the upper and lower quartiles, the whiskers extend to the largest value within Q1 – 1.5 × the interquartile range (IQR) and the smallest value within Q3 + 1.5 × IQR. All of the data points are represented as dots with distinct colors for each biological replicate. For each experiment, three biological replicates with three internal replicates were performed.
-
Figure 2—figure supplement 2—source data 1
Surface plasmon resonance measurements used in the preparation of Figure 2—figure supplement 2, left panel.
- https://doi.org/10.7554/eLife.47713.014
-
Figure 2—figure supplement 2—source data 2
Surface plasmon resonance measurements used in the preparation of Figure 2—figure supplement 2, right panel.
- https://doi.org/10.7554/eLife.47713.015
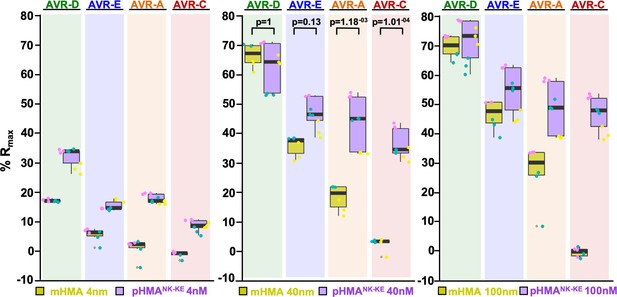
Binding of the Pikp-HMANK-KE domain to the AVR-Pik effectors is consistently greater than that of the Pikm-HMA domain.
Surface plasmon resonance %Rmax values for Pikm-HMA and Pikp-HMANK-KE with the AVR-Pik effector alleles. The Pikm-HMA and Pikp-HMANK-KE results are represented by yellow and purple boxes, respectively. The centre line within each box represents the median, the box limits are the upper and lower quartiles, the whiskers extend to the largest value within Q1 - 1.5 × the interquartile range (IQR) and the smallest value within Q3 + 1.5 × IQR. All the data points are represented as dots with distinct colors for each biological replicate. For each experiment, we performed at least two biological replicates with three internal repeats. Results obtained using HMA protein concentrations of 4, 40 and 100 nM are plotted in the left, middle and right panels, respectively. (‘p’ is the p-value obtained from statistical analysis and Tukey’s HSD.)
-
Figure 2—figure supplement 3—source data 1
Surface plasmon resonance measurements used in the preparation of Figure 2—figure supplement 3.
- https://doi.org/10.7554/eLife.47713.017
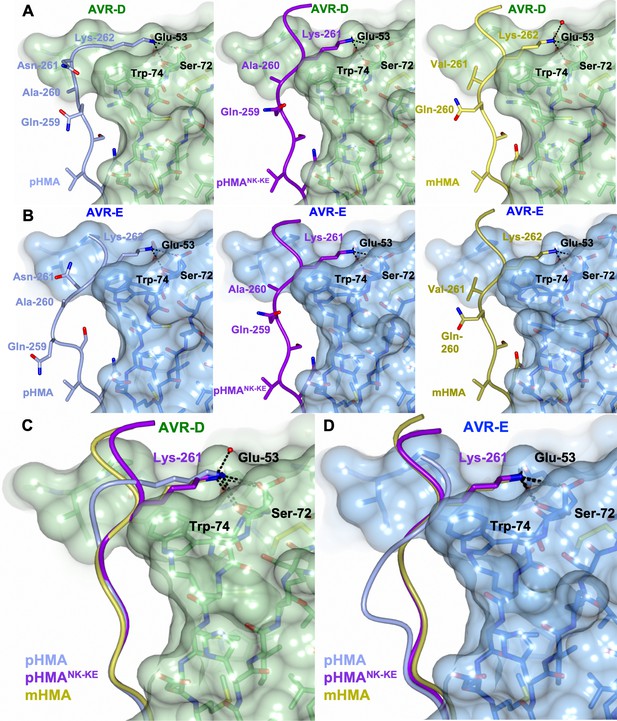
The PikpNK-KE-HMA mutant adopts a Pikm-like conformation at the effector-binding interface.
Schematic view of the different conformations adopted by Pikp-HMA, Pikp-HMANK-KE and Pikm-HMA at interface 3 when in complex with AVR-PikD or AVR-PikE. In each panel, the effector is shown as sticks with the molecular surface also shown and colored as labeled. Pik-HMA residues are colored as labeled and shown in the Cα-worm with side-chain representation. (A) Schematic of Pikp-HMA (left), Pikp-HMANK-KE (middle) and Pikm-HMA (right) bound to AVR-PikD. Important residues in the HMA–effector interaction are labeled as shown. (B) Schematic of HMA residues as for panel (A), but bound to AVR-PikE. (C) Superposition showing Pikp-HMA, Pikp-HMANK-KE and Pikm-HMA chains (colored in blue, purple and yellow, respectively) bound to AVR-PikD. For clarity, only the Lys-261/262 side chain is shown. (D) Superposition as described before, but bound to AVR-PikE.
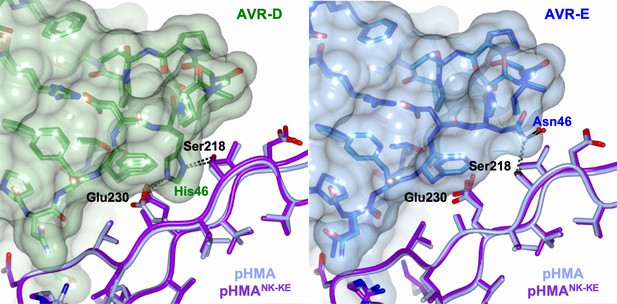
Interface 2 is essentially identical in the complexes comprising Pikp-HMA and Pikp-HMANK-KE bound to AVR-PikD or AVR-PikE.
Schematic view of the conformations adopted by Pikp-HMA and Pikp-HMANK-KE at interface 2 in complex with AVR-PikD or AVR-PikE. In each panel, the effector is shown in sticks, with the molecular surface also shown and colored as labeled. Pik-HMA residues are colored as labeled and shown in the Cα-worm with side-chain representation. The structures were overlaid on the effectors. (A) Pikp-HMA and Pikp-HMANK-KE (colored in blue and purple, respectively) bound to AVR-PikD (light and dark green). (B) Pikp-HMA and Pikp-HMANK-KE bound to AVR-PikE (light and dark blue).
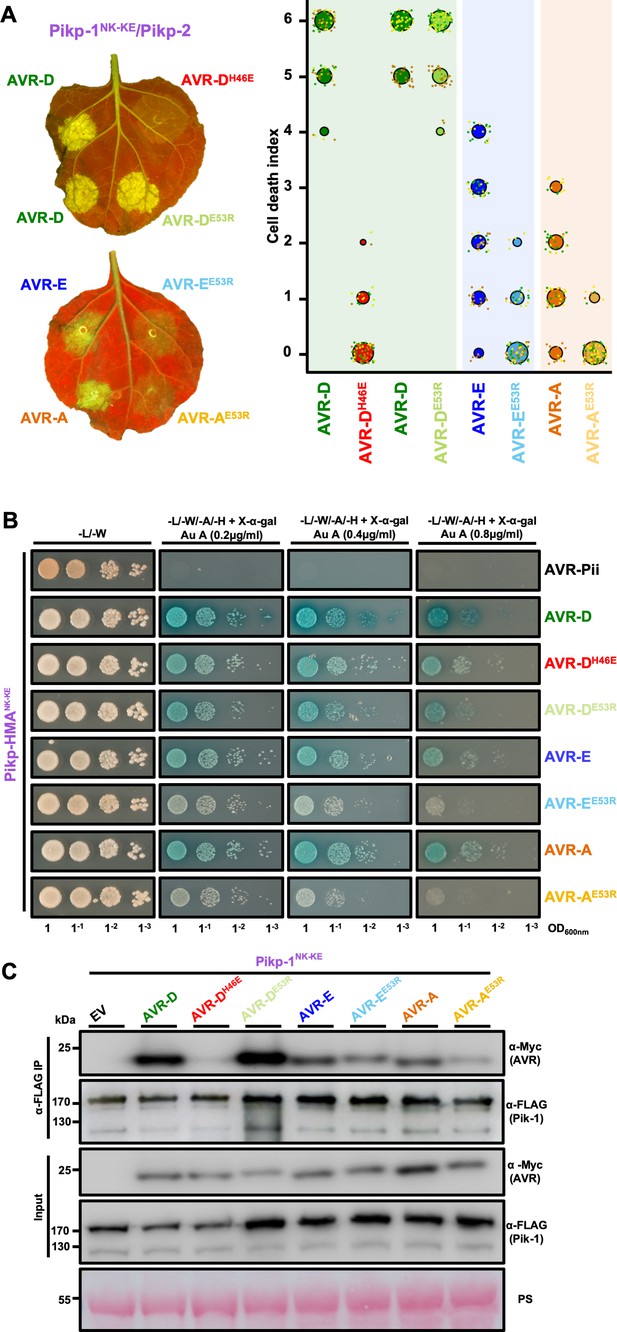
Mutation of AVR-Pik effectors at the engineered binding interface compromises binding and response.
(A) (Left) A representative leaf image showing Pikp-1NK-KE-mediated cell death induced by AVR-Pik variants and mutants as autofluorescence under UV light (the AVR-PikDH46E mutant is located at interface 2, whereas the AVR-PikDE53R, AVR-PikEE53R, and AVR-PikAE53R mutants are located at interface 3). Autofluorescence intensity is scored as in Figure 1. (Right) PikpNK-KE cell-death assay quantification in the form of dot plots. For each sample, the data points are represented as dots with a distinct color for each of the three biological replicates; these dots are plotted around the cell death score for visualization purposes. The size of the central dot at each cell death value is proportional to the number of replicates of the sample with that score. The number of repeats was 90. (B) Yeast-two-hybrid assay of Pikp-HMANK-KE with AVR-Pik variants and mutants. Control plate for yeast growth is on the left with quadruple dropout media supplemented with X-α-gal and increasing concentrations of Aureobasidin A on the right for each combination of HMA/AVR-Pik. The unrelated M. oryzae effector AVR-Pii was used as a negative control. Growth and the development of blue coloration in the selection plate are both indicative of protein–protein interaction. HMA domains were fused to the GAL4 DNA binding domain, and AVR-Pik alleles to the GAL4 activator domain. Each experiment was repeated a minimum of three times, with similar results. (C) Co-immunoprecipitation of full-length Pikp-1NK-KE with AVR-Pik variants and mutants. N-terminally 4xMyc tagged AVR-Pik effectors were transiently co-expressed with Pikp-1NK-KE:6xHis3xFLAG in N. benthamiana leaves. Immunoprecipitates (IPs) obtained with anti-FLAG antiserum, and total protein extracts, were probed with appropriate antisera. Each experiment was repeated at least three times, with similar results. The asterisks mark the Pik-1 band. PS = Ponceau Stain.
-
Figure 4—source data 1
Cell-death scoring data used in the preparation of Figure 4A.
- https://doi.org/10.7554/eLife.47713.025
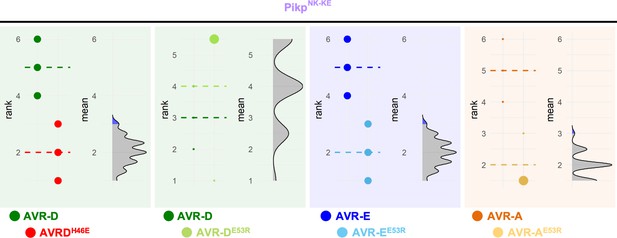
Estimation graphics for cell death induced by PikpNK-KE with AVR-Pik variants and mutants.
Statistical analysis by estimation methods of the cell-death assay for PikpNK-KE with AVR-Pik variants and mutants. The response to each mutant is compared with the response to the corresponding wild-type variant. For each couple, the panel on the left represents the ranked data (dots) for each effector, and their corresponding mean (dotted line). The size of the dots is proportional to the number of observations with that specific value. The panel on the right shows the distribution of 1000 bootstrap sample rank means for the mutant. The blue areas represent the 0.025 and 0.975 percentiles of the distribution. The responses to PikpNK-KE for the wild-type effectors and their corresponding effector mutants are considered to be significantly different if the wild-type rank mean (dotted line, left panel) falls beyond the blue regions of the mutant mean distribution.
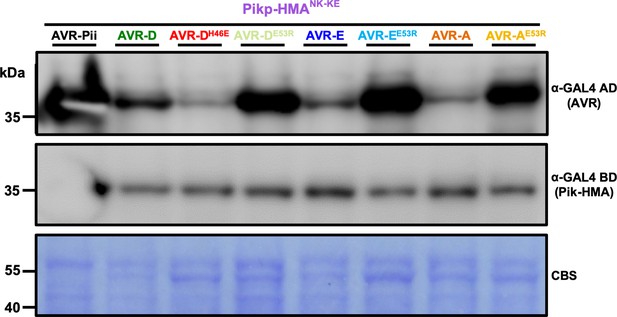
Western blot analysis confirming the accumulation of proteins in yeast.
Yeast lysate was probed for the expression of the HMA domain using the anti-GAL4 DNA binding domain (BD) or the AVR-Pik/AVR-Pii effectors anti-GAL4 activation domain (AD). The experiment was repeated a minimum of three times, with similar results. CBS = Coomassie Blue Stain. In each case, the effector mutants consistently accumulate to higher levels than the wild-type proteins, but result in weaker readouts of protein–protein interaction in the Y2H assay.
Tables
Data collection and refinement statistics
https://doi.org/10.7554/eLife.47713.019| PikpNK-KE–AVR-PikD | PikpNK-KE–AVR-PikE | |
|---|---|---|
| Data collection statistics | ||
| Wavelength (Å) | 0.9763 | 0.9763 |
| Space group | P 21 21 21 | P 21 21 21 |
| Cell dimensions | ||
| a, b, c (Å) | 29.79, 65.33, 75.86 | 66.46, 80.70, 105.58 |
| Resolution (Å)* | 32.80–1.60 (1.63–1.60) | 29.50–1.85 (1.89–1.85) |
| Rmerge (%)# | 8.1 (97.1) | 5.2 (75.1) |
| I/σI# | 16.1 (2.6) | 31.0 (4.1) |
| Completeness (%)# | 100 (100) | 99.8 (97.8) |
| Unique reflections# | 20,294 (978) | 49337 (2963) |
| Redundancy# | 12.8 (13.3) | 18.3 (17.8) |
| CC(1/2) (%)# | 99.9 (86.6) | 100 (95.2) |
| Refinement and model statistics | ||
| Resolution (Å) | 32.82–1.60 (1.64–1.60) | 29.52–1.85 (1.90–1.85) |
| Rwork/Rfree (%)^ | 19.7/23.2 (25.5/27.3) | 18.6/23.0 (29.1/35.0) |
| No. atoms (Protein) | 1277 | 3604 |
| B-factors (Protein) | 25.6 | 39.7 |
| R.m.s. deviations^ | ||
| Bond lengths (Å) | 0.009 | 0.012 |
| Bond angles (°) | 1.5 | 1.4 |
| Ramachandran plot (%)** | ||
| Favored | 98.1 | 97.3 |
| Outliers | 0 | 0.2 |
| MolProbity Score | 1.41 (93th percentile) | 1.59 (91st percentile) |
-
*The highest resolution shell is shown in parenthesis.
#As calculated by Aimless, ^As calculated by Refmac5, **As calculated by MolProbity.
Additional files
-
Supplementary file 1
Table of p-values for all pairwise comparisons of the ion-leakage data in N. benthamiana.
Underlined values are those presented in the respective figures.
- https://doi.org/10.7554/eLife.47713.026
-
Supplementary file 2
Table of p-values for all pairwise comparisons of the SPR data including Pikp and PikpNK-KE.
Underlined values are those presented in the respective figures.
- https://doi.org/10.7554/eLife.47713.027
-
Transparent reporting form
- https://doi.org/10.7554/eLife.47713.028





