Cytoplasmic protein misfolding titrates Hsp70 to activate nuclear Hsf1
Figures
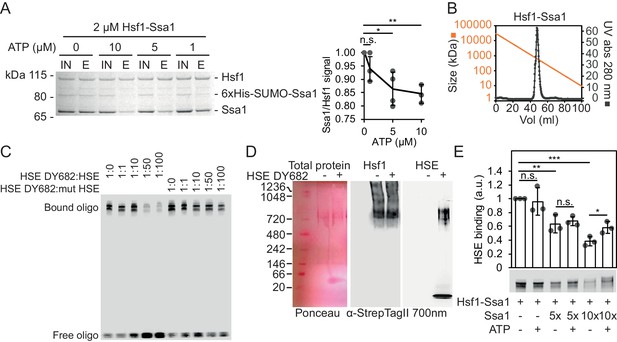
Reconstitution of large ATP-sensitive Hsf1-Hsp70 complexes with chaperone regulated HSE-binding activity.
(A) Recombinant Hsf1-Ssa1 complexes (Input; IN) were immobilized onto Strep-Tactin Sepharose (StrepTag II in Hsf1) in the presence of 0, 1, 5 or 10 µM ATP and after washing, bound protein was eluted (E) with desthiobiotin. The Ssa1/Hsf1 densitometric signal ratios from silver staining were quantified. Error bars indicate standard deviation from at least three experiments. (B) Hsf1-Ssa1 complexes subjected to size exclusion chromatography. Molecular weight as a function of elution volume plotted in orange. (C) EMSA of fluorescently labeled HSE (HSE DY682) mixed with recombinant Hsf1-Ssa1complexes. The complexes were competed with unlabeled (HSE) or unlabeled and inactive mutant HSE (mut HSE). (D) Hsf1-Ssa1 complexes were incubated with and without HSE DY682 and were separated by native gel electrophoresis. HSE binding was assessed using 700 nm in-gel fluorescence and the Hsf1-Ssa1 complexes were visualized following membrane transfer (Ponceau S staining and α-StrepTag II). (E) Formation of Hsf1-Ssa1-HSE complexes (0.25 µM Hsf1-Ssa1) was assessed by EMSA in the presence of increasing concentrations of extra added Ssa1 (5x, +1.25 µM and 10x +2.5 µM) with or without 1 mM ATP. Error bars show standard deviation of at least three independent experiments.
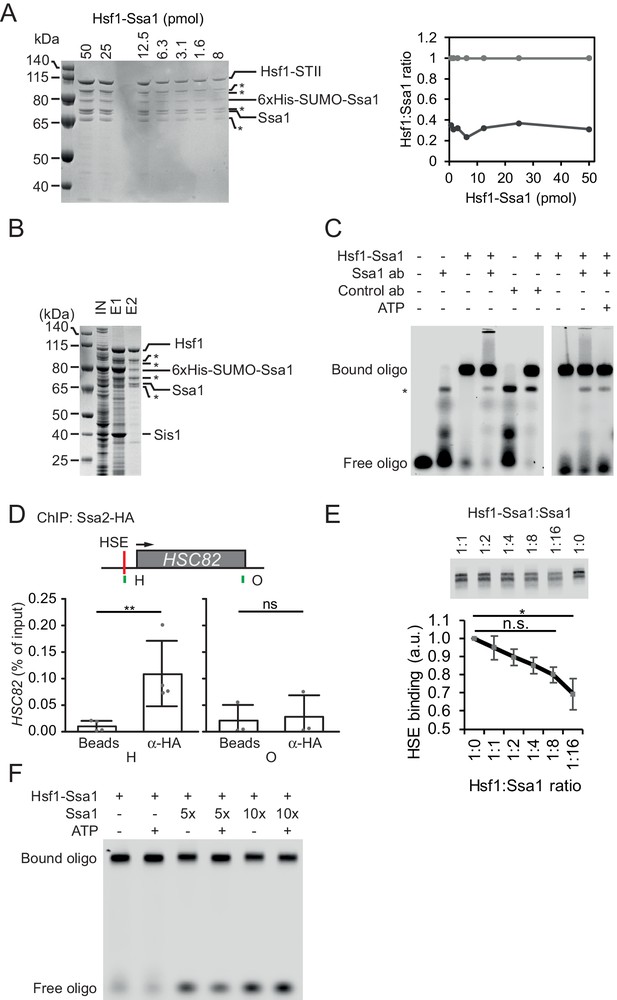
The DNA-binding activity of recombinant Hsf1-Ssa1 complexes is inhibited by Hsp70.
(A) Serial dilution of purified Hsf1-Ssa1 analyzed by SDS-PAGE for stoichiometric analysis. Densitometric quantification of the Coomassie stained gel is presented normalized to the level of total Hsf1 signal. Hsf1 species lacking parts of its flexible N-terminal due to proteolysis during expression and purification is marked with *. (B) SDS-PAGE analysis (silver staining) of input lysate (IN) of E. coli expressing Hsf1-StrepTag II, 6xHis-SUMO-Ssa1 and Sis1 loaded onto Protino Ni-IDA. The eluted protein (E1) from Ni-IDA resin was digested with Ulp1 SUMO protease, loaded onto a Strep-Tactin Sepharose column and was after washing eluted with desthiobiotin-containing buffer (E2). (C) EMSA analysis of 0.25 µM Hsf1-Ssa1 binding to HSE oligos. Hsf1-Ssa1 complexes were preincubated with rabbit serum reactive (Ssa1 Ab) or not reactive (Control Ab) to Ssa1, or ATP. The preincubation included a condition with 0.5 mM ATP added. (D) ChIP analysis of Ssa1-HA binding to the HSE region of the HSC82 promoter (P)-HSC82). Graphical representation of the HSC82 locus with the position of the HSE marked. Positions of the ChIP amplicons overlapping with the HSE (H) and the end of the ORF (O) are marked. The qPCR signals ChIP experiments with control magnetic beads or beads coated with anti-HA antibody are presented as % of input. Error bars show standard deviation of three independent experiments. (E) Formation of Hsf1-Ssa1-HSE complexes (0.25 µM Hsf1-Ssa1) was assessed by EMSA in the presence of increasing concentrations of extra added Ssa1 (molar ratio Hsf1-Ssa1:Ssa1 1:0, + 0 µM Ssa1; 1:1 + 0.25 µM; 1:2 + 0.50 µM; 1:4 + 1.0 µM; 1:8 + 2.0 µM; 1:16 + 4.0 µM). Error bars show standard deviation of three independent experiments. (F) EMSA assay of Hsf1-Ssa1-HSE complexes as above performed under modified conditions (see Materials and methods) that better visualizes the unbound HSE oligonucleotides (Free oligo).
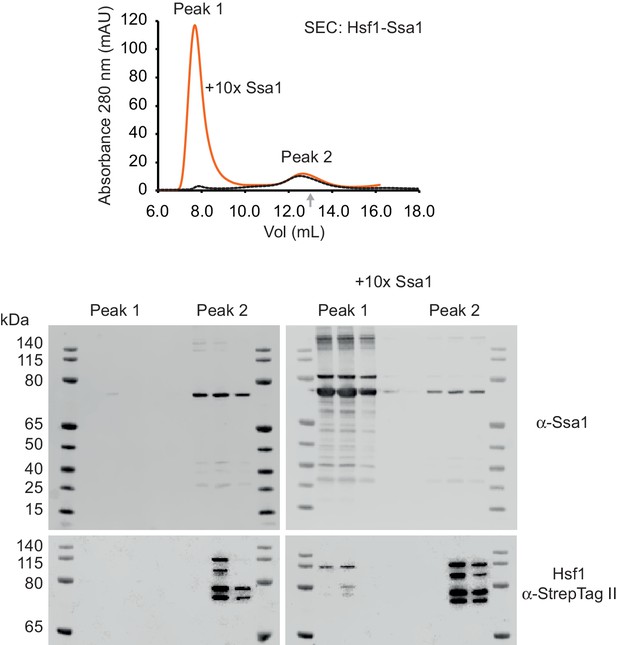
Hsf1 forms supercomplexes upon supplementation with excess Hsp70.
0.34 nmol Hsf1-Ssa1 complexes were mixed with buffer (dotted line) or preincubated with 10-fold molar excess of Ssa1-ATP (orange line) and were loaded onto a Superose 6 10/300 GL column. Two distinct subpopulations (Peak one and Peak2) defined by 280 nm UV absorbance are indicated. The elution volume of a 669 kDa size standard is indicated (gray arrow). Western analysis (α-Ssa1 and α-StrepTag II) was performed on three collected peak fractions from Peak one and Peak 2.
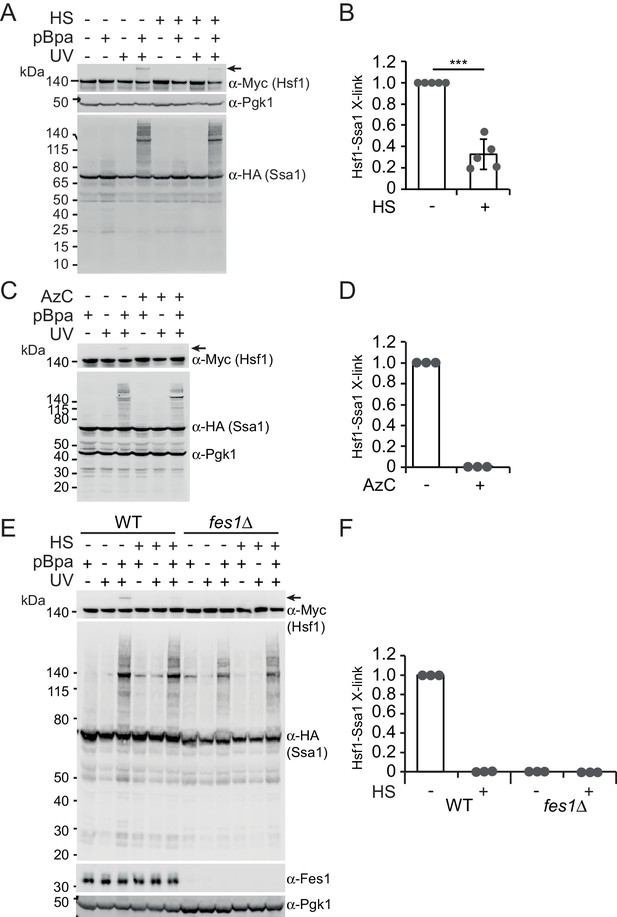
Heat shock negatively regulates Hsp70 binding of Hsf1 via its SBD.
(A) Photocrosslinking (UV) of yeast cells grown at 30°C or subjected to a 3 min heat shock (HS) at 43°C that express Ssa1-HA with photoreactive Bpa (pBPa) incorporated at amino acid position 423 of the Hsp70 SBDβ in cells. Proteins and crosslinking-products were visualized by western analysis using α-HA and α-Myc antibodies with α-Pgk1 as a loading control. Representative data from five independent experiments are shown and the relative levels of Hsf1-Ssa1 crosslinking products marked with an arrow were quantified in (B) with error bars showing standard deviation. (C) and (D) The experiment in A was performed with a 2 hr AzC treatment at 25°C to induce the misfolding of newly synthesized proteins. (E) and (F) The experiment in A was performed using fes1Δ cells.
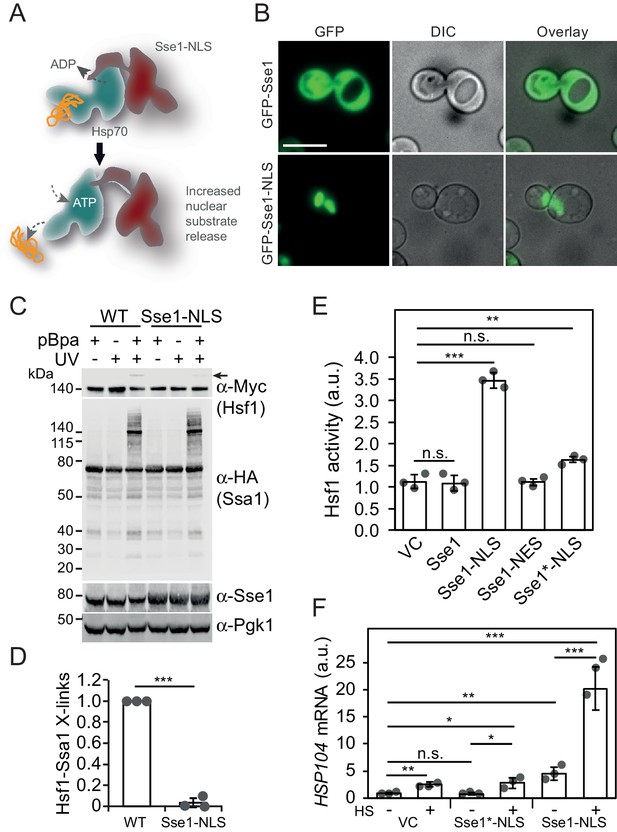
Accelerating Hsp70 substrate release in the nucleus activates Hsf1 and potentiates the heat-shock response.
(A) Schematic representation of Sse1-NLS binding to nuclear Hsp70. Binding accelerates Hsp70 nucleotide exchange and triggers substrate release. (B) Micrographs of the subcellular localization of control GFP-Sse1 and GFP-Sse1-NLS. The scale bar corresponds to 5 μm. (C) Photocrosslinking (UV) of yeast cells grown at 30°C or subjected to a 3 min heat shock (HS) at 43°C that express Ssa1-HA with photoreactive Bpa (pBPa) incorporated at amino acid position 423 of the Hsp70 SBDβ in cells. Proteins and crosslinking-products were visualized by western analysis using α-HA and α-Myc antibodies with α-Pgk1 as a loading control. (D) The relative levels of Hsf1-Ssa1 crosslinking products (marked with an arrow in C) were quantified. (E) Hsf1 activity at 30°C in cells carrying vector control (VC) or plasmid derivatives that express Sse1, Sse1-NLS, Sse1-NES and Sse1*-NLS. Hsf1-activity was determined using a bioluminescent reporter construct. (F) Analysis of HSP104 mRNA levels normalized to TAF10 mRNA levels at 25°C and after a 30 min heat shock (HS) at 37°C in cells carrying the same plasmids as in E. All experiments were repeated three times with error bars showing standard deviation.
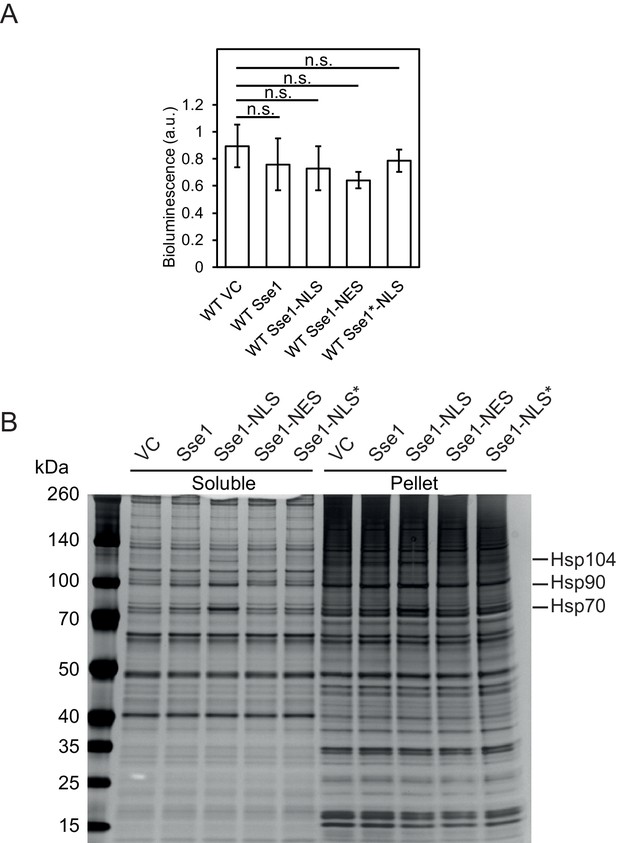
Expression of Sse1-NLS does not affect Msn2/4 activity but strongly induces the expression of Hsp70, Hsp90 and Hsp104.
(A) Msn2/4 activity determined by a bioluminescent reporter in cells carrying vector control (VC) or expressing Sse1, Sse1-NLS, Sse1-NES, Sse1*-NLS at 30°C. Data from triplicate experiments with error bars showing standard deviation. (B) Protein aggregation analysis of extracts from cells carrying plasmids as in A. Cells we grown at 30°C and the proteins were visualized by silver staining. The proteins bands corresponding to Hsp70, Hsp90 and Hsp104 are marked.
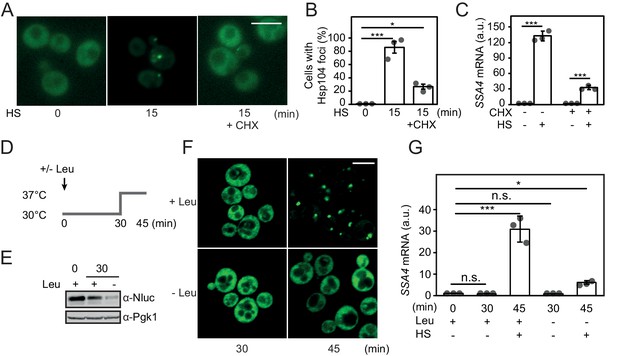
Heat shock activates Hsf1 by the misfolding of newly synthesized proteins.
(A) Micrographs of Hsp104-GFP in cells grown at 25°C and heat shocked (HS) for 15 min at 37°C either with or without the addition of CHX. The white scale bar is 5 μm. (B) Quantification of the fraction of cells with Hsp104-GFP foci in A. At least 260 cells were counted in three biological replicates for each condition. Error bars denote standard error. (C) Quantification of SSA4 mRNA levels in cells treated as in A. (D) Outline of the experimental setup for acute leucine starvation of leu2Δ0 cells in E-G. (E) Steady state levels of the rapidly turned-over protein yNlucPEST expressed from the strong TDH3 promoter in leu2Δ0 cells grown in complete synthetic media or acutely starved for leucine for 30 min. (F) Micrographs of Hsp104-GFP in leu2Δ0 cells grown under leucine-rich (+Leu) and acute leucine starvation (-Leu) conditions before (30 min) and after (45 min) a 15 min heat shock. The white scale bar is 5 μm. (G) Quantification of SSA4 mRNA levels in cells grown at 30°C and heat-shocked (HS) for 15 min at 37°C with or without 30 min of leucine starvation. All qPCRs were normalized to TAF10 mRNA levels and all bioluminescence measurements to OD600. Error bars show standard deviation unless otherwise stated.
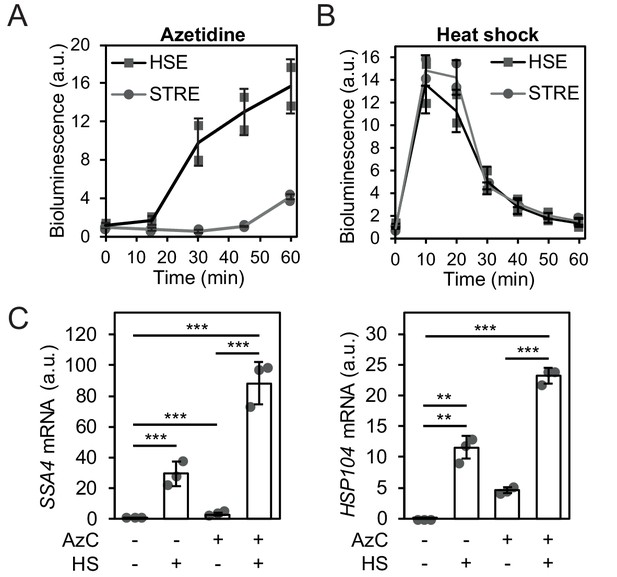
Heat shock activates Hsf1 by the misfolding of newly synthesized proteins.
(A) Hsf1 and Msn2/4 activity was followed over time in cells exposed to 10 mM AzC using bioluminescent transcriptional reporters promoted by HSE (Hsf1) or STRE (Msn2/4). (B) Hsf1 and Msn2/4 activity was monitored as in A but in cells exposed to a heat shock (HS) at 37°C. (C) SSA4 and HSP104 mRNA levels in cells grown at 25°C and heat-shocked (HS) 15 min at 37°C, exposed to 10 mM AzC for 60 min or exposed to 10 mM AzC for 60 min where the last 15 min included a heat shock at 37°C.
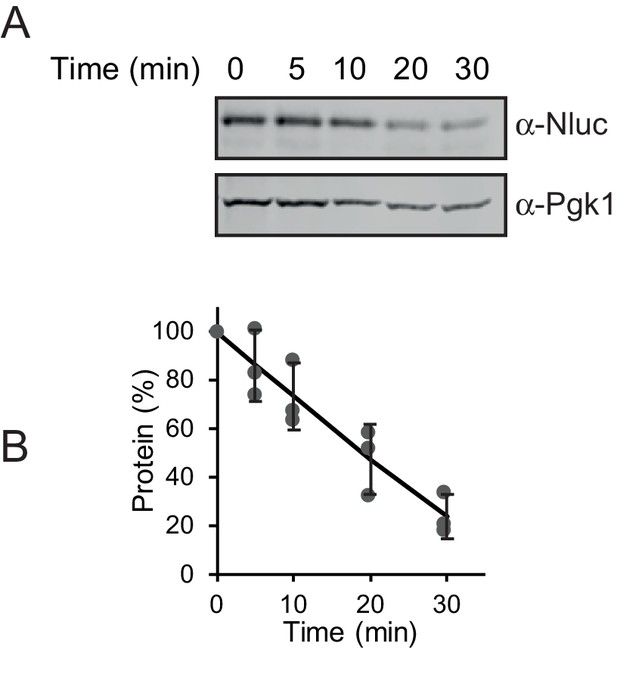
Half-life of NlucPEST.
(A) Western analysis of NlucPEST levels following the addition of CHX to arrest translation. (B) Quantification of NlucPEST levels in A. Protein levels were normalized to the loading control Pgk1. All experiments were performed in triplicates with error bars showing standard deviation.
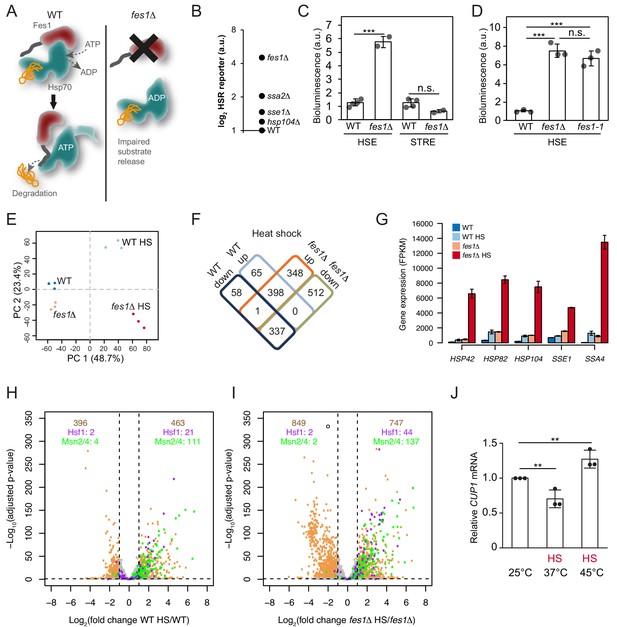
Limiting the available pool of Hsp70 by impairing cytosolic substrate release unleashes a Hsf1 hyper-stress program.
(A) Schematic representation of Fes1-accelerated release of persistent substrates from Hsp70. In fes1Δ cells persistent substrates remain associated with Hsp70. (B) Hsf1 activity was determined in cells with chaperone mutations (hsp104Δ, sse1Δ, ssa2Δ and fes1Δ) grown at 25°C a using HSE bioluminescent reporter. (C) Hsf1 and Msn2/4 activities in WT and fes1Δ cells grown at 25°C were measured using HSE and STRE bioluminescent reporters. (D) Hsf1 activity was determined in WT, fes1Δ and fes1-1 cells as in B. (E) Principal component analysis of transcriptome RNA-seq data of WT and fes1Δ cells grown at 25°C or subjected to heat shock (HS) for 30 min at 37°C. (F) Venn diagram of differentially expressed (DE) genes following heat shock in WT and fes1Δ cells. (G) Transcript levels (FPKM) of the Hsf1 target genes HSP42, HSP82, HSP104, SSE1 and SSA4. (H) Volcano plot showing gene expression changes following heat shock of WT cells. The Hsf1 and Msn2/4 targets are colored in purple and green, respectively. Genes with adjusted p value < 0.05 and absolute log2 fold change >1 are considered significant. These thresholds are indicated by the dotted black lines. The number of genes that are up- and down-regulated are written in tan color at the top right and top left of the plot, respectively. Among these DE genes, the number of Hsf1 and Msn2/4 targets are showed below in purple and green color. (I) Volcano plot as in H but for fes1Δ cells. (J) Relative CUP1 mRNA levels in WT cells grown at 25°C or subjected to heat shock (HS) at 37°C or 45°C for 30 min. Experiments were performed in triplicates with error bars showing standard deviation.
-
Figure 5—source data 1
FKPM values from RNA seq analysis of heat-shocked WT and fes1Δ cells.
Data from three independent cultures of each strain (I, II, III) are provided.
- https://doi.org/10.7554/eLife.47791.016
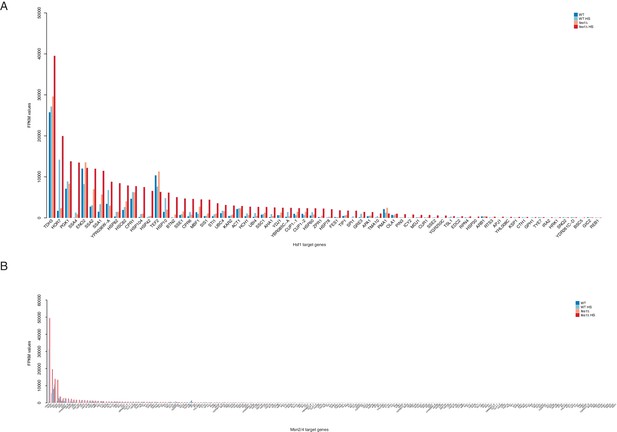
Induction of Hsf1 and Msn2/4 target genes under heat shock and hyper-stress conditions.
(A) Bar plot showing mean FPKM values of Hsf1 target genes (Pincus et al., 2018) before and after heat shock in WT and fes1Δ cells. (B) Bar plot showing mean FPKM values of Msn2/4 target genes (Solís et al., 2016) before and after heat shock in WT and fes1Δ cell.
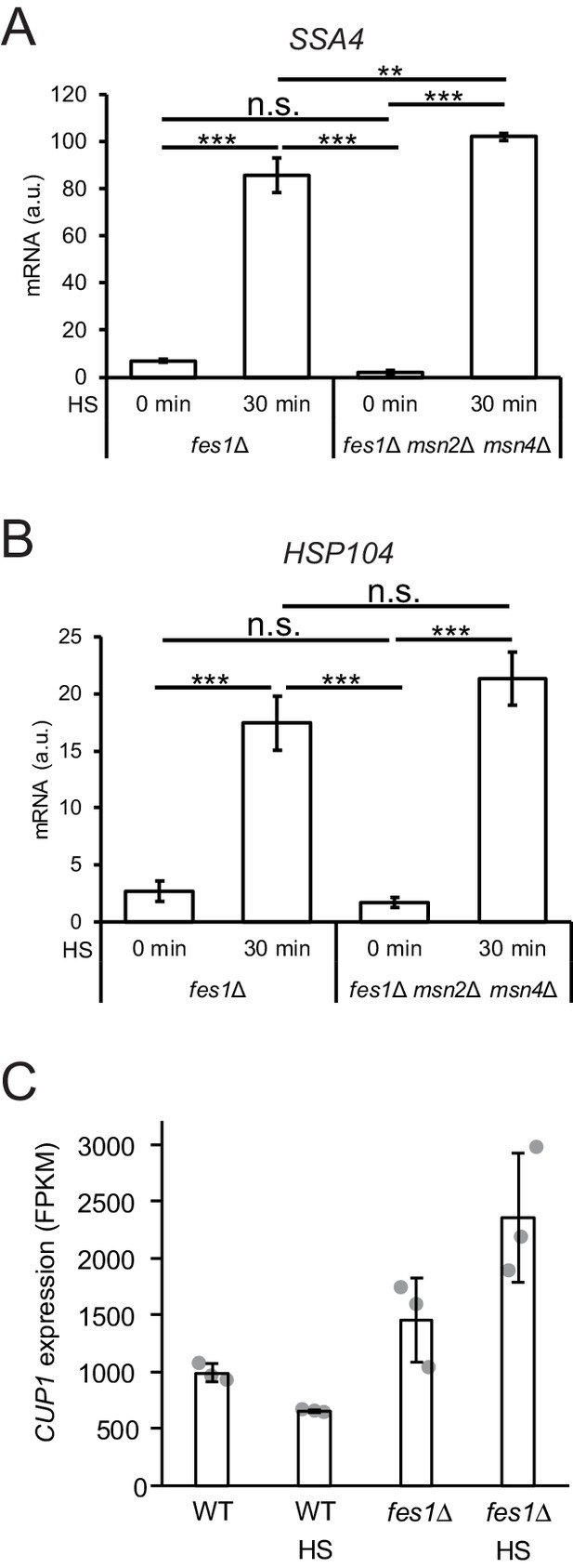
Msn2/4 is dispensable for hyperinduction of the HSR and expression of the CUP1 gene.
(A) Induction of SSA4 and (B) HSP104 in fes1Δ and fes1Δ msn2/4Δ cells before and after a 30 min heat shock (HS). levels of mRNA were determined by qPCR and normalized to TAF10. (C) FKPM values (RNA seq) for the expression of CUP1. All experiments were performed in triplicates with error bars showing standard deviation.
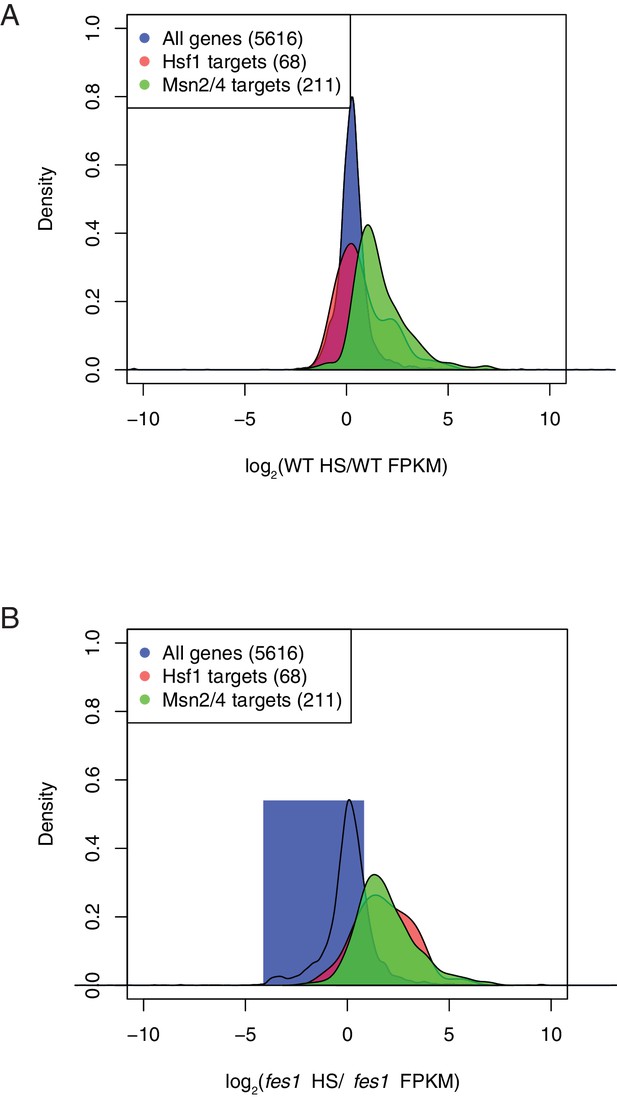
Density plot obtained from DESeq2 analysis showing gene expression changes following heat shock in (A) WT and (B) fes1Δ cells.
Hsf1 and Msn2/4 target genes has been described earlier (Pincus et al., 2018; Solís et al., 2016).
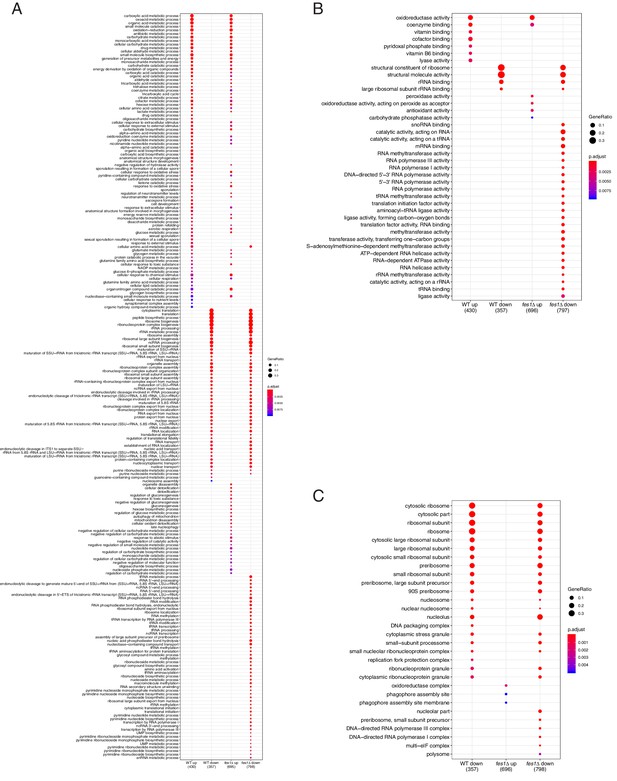
GO analysis of differentially expressed genes in heat shocked cells.
(A) Significantly enriched GO terms related to biological process. (B) Significantly enriched GO terms related to molecular function. (C) Significantly enriched GO terms related to cellular component.
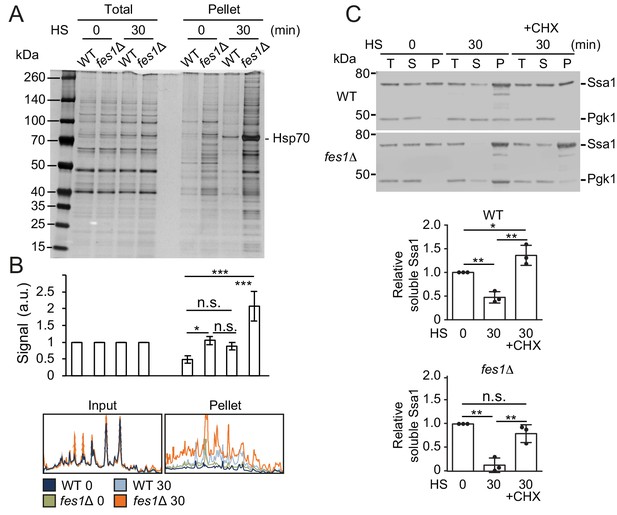
The soluble pool of Hsp70 is titrated to the aggregate fraction by heat shock.
(A) Total lysates and the aggregate pellet fractions of protein lysates from WT and fes1Δ cells grown at 25°C and heat shocked for 30 min at 37°C (HS). Proteins were visualized by silver staining. (B) Densitometric measurements of triplicate experiments presented in A. The signals in the aggregate pellets are normalized to the respective signals in the total lysate. (C) Western analysis of lysates from cells as in A. Cycloheximide (CHX) was added right before the heat shock. Total lysates (T), the aggregate pellet (P) and the remaining soluble (S) fraction were analyzed with anti-Ssa1 and anti-Pgk1 antibodies. Relative Ssa1 signals in the soluble fractions are presented. All experiments were performed in triplicates with error bars showing standard deviation.
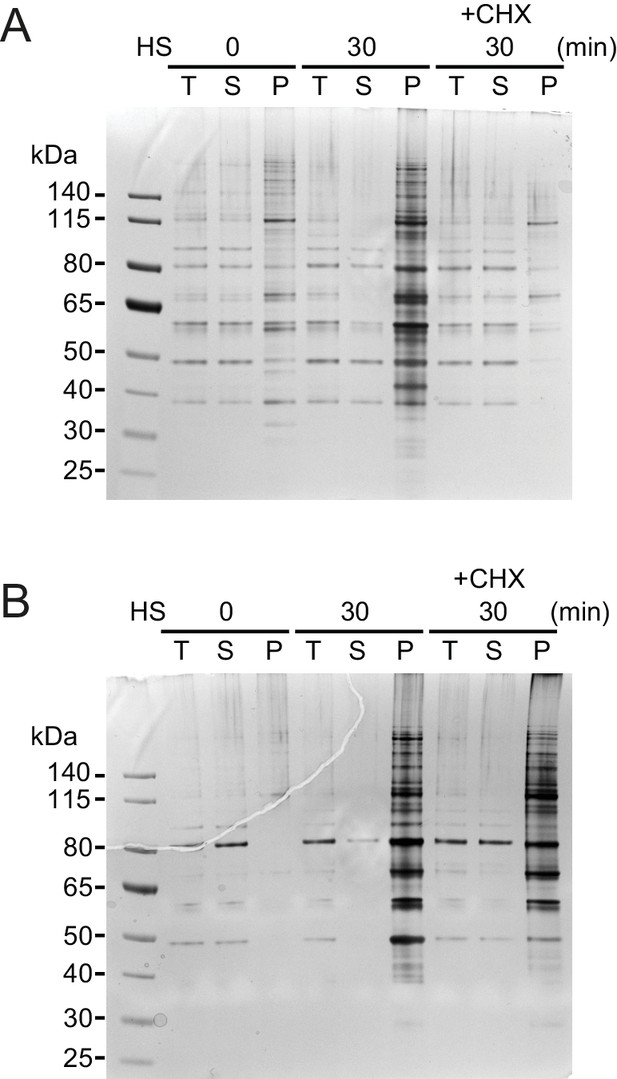
Heat-shock-induced protein aggregation depends on translation in WT but not fes1Δ cells.
(A) Total lysates (T), the aggregate pellet (P) and the remaining soluble (S) fractions following centrifugation of protein lysates from WT and (B) fes1Δ cells grown at 25°C and heat shocked for 30 min at 37°C (HS). Cycloheximide (CHX) was added right before the heat shock. Proteins were visualized by silver staining.
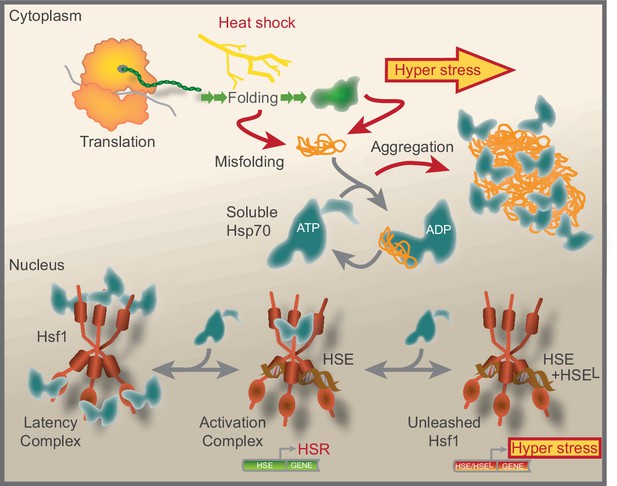
Model that show how Hsf1 is negatively regulated by the nuclear pool of Hsp70.
Summative graphical model of our current understanding of Hsf1 regulation. See Discussion for details.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (S. cerevisiae) | AMY31 | This paper | For details see Table 1. Dr. C Andréasson, Stockholm University, Sweden | |
| Strain, strain background (S. cerevisiae) | AMY41 | This paper | For details see Table 1. Dr. C Andréasson, Stockholm University, Sweden | |
| Strain, strain background (S. cerevisiae) | AMY46 | This paper | For details see Table 1. Dr. C Andréasson, Stockholm University, Sweden | |
| Strain, strain background (S. cerevisiae) | AMY62 | This paper | For details see Table 1. Dr. C Andréasson, Stockholm University, Sweden | |
| Strain, strain background (S. cerevisiae) | BY4721 | EUROSCARF; PMID: 9483801 | Y00000 | Distributed by EUROSCARF |
| Strain, strain background (S. cerevisiae) | CAY1005 | EUROSCARF | Y02146 | Distributed by EUROSCARF |
| Strain, strain background (S. cerevisiae) | CAY1015 | PMID: 23530227 | For details see Table 1. Dr. C Andréasson, Stockholm University, Sweden | |
| Strain, strain background (S. cerevisiae) | CAY1038 | EUROSCARF | Y01514 | Distributed by EUROSCARF |
| Strain, strain background (S. cerevisiae) | CAY1057 | EUROSCARF | Y1512 | Distributed by EUROSCARF |
| Strain, strain background (S. cerevisiae) | CAY1140 | This paper | For details see Table 1. Dr. C Andréasson, Stockholm University, Sweden | |
| Strain, strain background (S. cerevisiae) | CAY1221 | PMID: 26912797 | For details see Table 1. Dr. C Andréasson, Stockholm University, Sweden | |
| Strain, strain background (S. cerevisiae) | CAY1255 | PMID: 14562095 | HSP104-GFP | Dr. T Nyström, University of Gothenburg, Sweden |
| Strain, strain background (S. cerevisiae) | CAY1257 | PMID: 23530227 | For details see Table 1. Dr. C Andréasson, Stockholm University, Sweden | |
| Strain, strain background (S. cerevisiae) | NY137 | This paper | For details see Table 1. Dr. C Andréasson, Stockholm University, Sweden | |
| Recombinant DNA reagent | pAM14 | This paper | For details see Table 2. Dr. C Andréasson, Stockholm University, Sweden | |
| Recombinant DNA reagent | pAM17 | This paper | For details see Table 2. Dr. C Andréasson, Stockholm University, Sweden | |
| Recombinant DNA reagent | pAM23 | This paper | For details see Table 2. Dr. C Andréasson, Stockholm University, Sweden | |
| Recombinant DNA reagent | pCA502 | PMID: 18948593 | For details see Table 2. Dr. C Andréasson, Stockholm University, Sweden | |
| Recombinant DNA reagent | pCA503 | PMID: 18948593 | For details see Table 2. Dr. C Andréasson, Stockholm University, Sweden | |
| Recombinant DNA reagent | pCA901 | This paper | For details see Table 2. Dr. C Andréasson, Stockholm University, Sweden | |
| Recombinant DNA reagent | pCA926 | This paper | For details see Table 2. Dr. C Andréasson, Stockholm University, Sweden | |
| Recombinant DNA reagent | pCA955 | PMID:26860732 | For details see Table 2. Dr. C Andréasson, Stockholm University, Sweden | |
| Recombinant DNA reagent | pCA970 | PMID: 28289075 | For details see Table 2. Dr. C Andréasson, Stockholm University, Sweden | |
| Recombinant DNA reagent | pCA1026 | This paper | For details see Table 2. Dr. C Andréasson, Stockholm University, Sweden | |
| Recombinant DNA reagent | pJK001 | PMID:28289075 | For details see Table 2. Dr. C Andréasson, Stockholm University, Sweden | |
| Recombinant DNA reagent | pJK010 | PMID: 28289075 | For details see Table 2. Dr. C Andréasson, Stockholm University, Sweden | |
| Recombinant DNA reagent | pJK011 | This work | For details see Table 2. Dr. C Andréasson, Stockholm University, Sweden | |
| Recombinant DNA reagent | pJK070 | PMID: 29323280 | For details see Table 2. Dr. C Andréasson, Stockholm University, Sweden | |
| Recombinant DNA reagent | ECYRS-BpA (plasmid) | PMID: 17560600 | Dr. PG Schultz, The Scripps Research Institute, CA |
Yeast strains.
https://doi.org/10.7554/eLife.47791.020| Strain | Genotype | Reference/source |
|---|---|---|
| AMY31 | MATa his3Δ1 leu2Δ0 ura3Δ0 HSF1-EGFP-hphMX SSA2-HA-kanMX | This work |
| AMY41 | MATa his3Δ1 leu2Δ0 ura3Δ0 msn2::hphMX4 msn4::natMX4 fes1Δ::LEU2 | This work |
| AMY46 | MATa his3Δ1 leu2Δ0 ura3Δ0 msn2::hphMX4 msn4::natMX4 | This work |
| AMY62 | MATa his3Δ1 leu2Δ0 ura3Δ0 trp1Δ::natMX Hsf1-13*Myc-kanMX | This work |
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | (Brachmann et al., 1998) |
| CAY1005 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 sse1Δ::kanMX4 | EUROSCARF |
| CAY1015 | MATa his3Δ1 leu2Δ0 ura3Δ0 | (Gowda et al., 2013) |
| CAY1038 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hsp104Δ::kanMX4 | EUROSCARF |
| CAY1057 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ssa2Δ::kanMX4 | EUROSCARF |
| CAY1140 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 fes1Δ::LEU2 | This work |
| CAY1221 | MATa his3Δ1 leu2Δ0 ura3Δ0 fes1Δ::ura3 | (Gowda et al., 2016) |
| CAY1255 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 HSP104::-GFP-his3M × 6 | This work |
| CAY1267 | MATa his3Δ1 leu2Δ0 ura3Δ0 fes1-1 (A79R, R195A) | (Gowda et al., 2013) |
| NY137 | MATa leu2Δ0 ura3Δ0 trp1Δ::natMX Hsf1-13*Myc-kanMX his3Δ1::[SSE1-NLS HIS3] | This work |
Plasmids.
https://doi.org/10.7554/eLife.47791.021| Plasmid | Description | Type | Reference/source |
|---|---|---|---|
| pAM14 | URA3 PCYC1-4xSTRE-yNlucPEST | CEN/ARS | This work |
| pAM17 | URA3 PCYC1-3xHSE-yNlucPEST | CEN/ARS | This work |
| pAM23 | URA3 PTHD3-yNlucPEST | CEN/ARS | This work |
| pCA502 | HIS3 VC | CEN/ARS | (Andréasson et al., 2008) |
| pCA503 | HIS3 SSE1 | CEN/ARS | (Andréasson et al., 2008) |
| pCA901 | HIS3 SSE1*-NLS | CEN/ARS | This work |
| pCA926 | HIS3 SSE1-NLS | YIP | This work |
| pCA955 | URA3 PCYC1-HSE-yNlucPEST | CEN/ARS | (Masser et al., 2016) |
| pCA970 | HIS3 SSE1-NES | CEN/ARS | (Kaimal et al., 2017) |
| pCA1026 | PT7-lacO-6xHis-SUMO-Ssa1-S/D-Hsf1-StrepTag II PT7-Sis1 lacI | E. coli | This work |
| pJK001 | HIS3 ysfGFP-SSE1 | CEN/ARS | (Kaimal et al., 2017) |
| pJK010 | HIS3 SSE1-NLS | CEN/ARS | (Kaimal et al., 2017) |
| pJK011 | HIS3 ysfGFP-Sse1-NLS | CEN/ARS | This work |
| pJK070 | URA3 SSA1E423TAG-HA | 2 μ | (Gowda et al., 2018) |
| ECYRS-BpA | TRP1 BPa system | 2 μ | (Chen et al., 2007) |
qPCR primers.
https://doi.org/10.7554/eLife.47791.022| Gene | Sequence 5’−3’ | Reference/source |
|---|---|---|
| CUP1 | GTGCCAATGCCAATGTGGTAG CATTTCCCAGAGCAGCATGAC | This work |
| HSC82 H | CTCGTTTTCTCGAACTTC CAAATCTCCTCCCTCATTAC | This work |
| HSC82 O | GAGAGTTGATGAGGGTGGTG GTTAGTCAAATCTTTGACGGTC | This work |
| TAF10 | ATATTCCAGGATCAGGTCTTCCGTAGC GTAGTCTTCTCATTCTGTTGATGTTGTTGTTG | (Teste et al., 2009) |
| SSA4 | CCAAGAGGCGTACCACAAAT GCTTCTTGTTCATCTTCGGC | This work |
| HSP104 | GTCGCTGAACCAAGTGTGAG CTCTTGCGACGGCGACACCA | This work |
EMSA oligos.
https://doi.org/10.7554/eLife.47791.023| Name | Sequence 5’−3’ | Reference/source |
|---|---|---|
| HSE-DY682 | TCGATTTTCCAGAACGTTCCATCGGC GCCGATGGAACGTTCTGGAAAATCGA | This work |
| HSE | TCGATTTTCCAGAACGTTCCATCGGC GCCGATGGAACGTTCTGGAAAATCGA | This work |
| HSE* | TCGATGTGCCAGTACGTAGCATCGGC GCCGATGCTACGTACTGGCACATCGA | This work |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.47791.024





