Cardiac neural crest contributes to cardiomyocytes in amniotes and heart regeneration in zebrafish
Figures
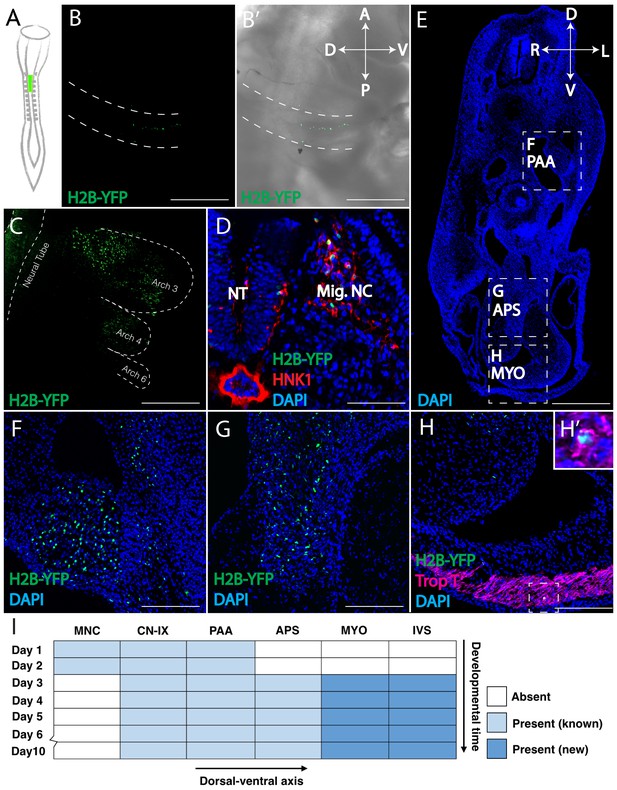
Retrovirally mediated fate mapping of cardiac neural crest reveals novel derivatives.
(A) Schematic diagram of the approach: Replication Incompetent Avian (RIA) retrovirus encoding nuclear H2B-YFP was injected into the lumen of the hindbrain from which cardiac neural crest arises. (B) One day post-infection (HH14), whole mount image (lateral view) showing virally labeled progeny (green) in the cardiac migration stream en route to pharyngeal arch 3. (B’) Brightfield image to show anatomical information. A, anterior; P, posterior; D, dorsal; V, ventral. (C) Two days post-infection (HH18), virally labeled cardiac crest has populated pharyngeal arches 3, 4 and 6, highlighted with dashed line. (D) Transverse section showing that labeled cardiac crest expresses neural crest marker HNK-1 (red). D, dorsal; V, ventral; L, left; R, right. (E) Low magnification transverse section of an E6 embryo (DAPI, blue). Dashed boxes show relative positions of cardiac crest-derived populations. (F–H) High magnification image of selected regions in E: pharyngeal arch arteries (F); aorticopulmonary septum (G); Neural crest derivatives located in the outflow tract express Troponin T (magenta), a myocardium marker (H, H’). (I) Temporal map of the establishment of distinct cardiac neural crest derivatives. Labeled cells initially are in the migration stream, cranial nerve IX (CN-IX) and mesenchyme around pharyngeal arch arteries (PAA). Subsequently, they populate the aorticopulmonary septum (APS), myocardium (MYO) and interventricular septum (IVS). Separate channels are displayed in Figure 1—figure supplement 1. Light blue indicates known neural crest derivatives. Dark blue reflects newly discovered neural crest derivatives. Scale bars: B, C, E 400 μm; D, F, G, H 100 μm.
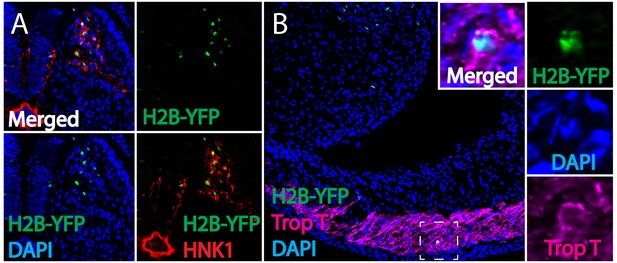
Separate channels for retroviral lineage analysis and immunohistochemistry.
(A) Separate channels of Figure 1D. All H2B-YFP labeled cells that migrate out of the neural tube are HNK1-positive cardiac neural crest cells (H2B-YFP, green; HNK1, red; DAPI, blue). (B) Separate channels of Figure 1H. H2B-YFP labeled cardiac neural crest is present in myocardium of the outflow tract (H2B-YFP, green; Troponin T, magenta; DAPI, blue).
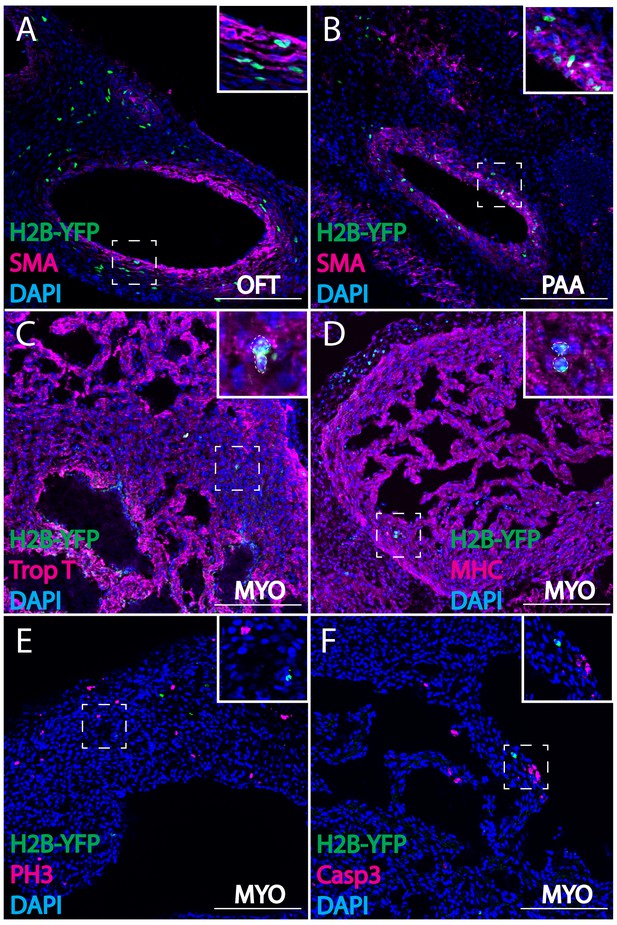
Cardiac crest-derived cells differentiate into smooth muscle and cardiomyocytes in avian embryos.
(A, B) Retrovirally labeled cardiac crest cells (H2B-YFP, green) that migrate into the outflow tract (A, OFT) and pharyngeal arch arteries (B) express smooth muscle actin (SMA, magenta) marker. (C, D) Labeled cardiac crest cells that enter the ventricle express myocardial marker Troponin T (magenta) (C), and myocardial terminal differentiation marker Myosin Heavy Chain (MHC, magenta) (D) enclosed in dashed line. (E, F) Neural crest-derived cardiomyocytes are not actively dividing or undergoing apoptosis, as demonstrated by phosphohistone H3 staining (PH3, magenta) (E) and Caspase 3 staining (magenta) (F). Transverse view of E6 embryos. Separate channels are displayed in supplement 1. Scale bars: 100 μm.
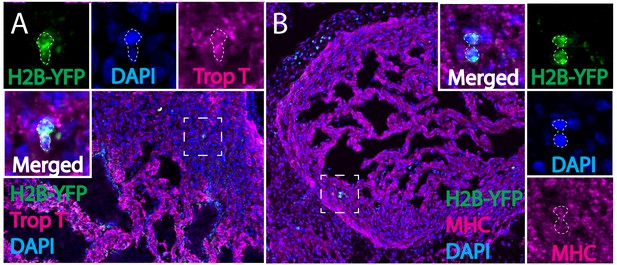
Separate channels for retroviral lineage analysis and immunohistochemistry.
(A) Separate channels of Figure 2C. H2B-YFP virus labeled cardiac neural crest is present in myocardium of the ventricle and expresses cardiomyocyte marker Troponin T (H2B-YFP, green; Troponin T, magenta; DAPI, blue; enclosed in dashed line). (B) Separate channels of Figure 2D. H2B-YFP virus labeled cardiac neural crest is present in myocardium of the ventricle and expresses cardiomyocyte terminal differentiation marker myosin heavy chain (MHC) (H2B-YFP, green; MHC, magenta; DAPI, blue; enclosed in dashed line).
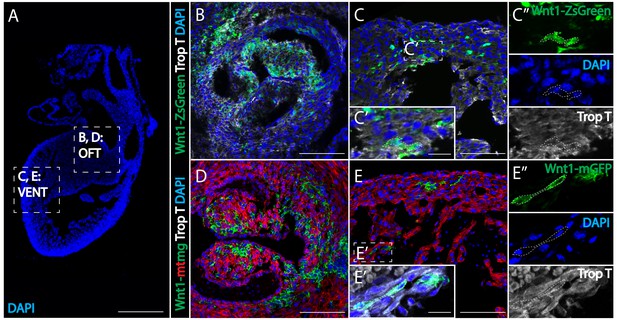
Wnt1-Cre fate mapping in mice confirms the presence of cardiac crest-derived myocardium.
(A) Low magnification image to show the relative anatomical positions of a mouse heart at E15.5 (sagittal view, DAPI-blue). (B, C) In Wnt1-Cre; ZsGreenfl/fl mice, neural crest-derived cells (green, Wnt1-Cre driven ZsGreen expression is abbreviated as Wnt1-ZsGreen, enclosed in dashed line) were observed in myocardium (Troponin T, gray) of the outflow tract (B), and ventricle (VENT) (C, C’’: separate channels of inset C’). (D, E) Similar results were obtained from Wnt1-Cre2+; R26mTmG mice (Wnt1-Cre2+ driven replacement of membrane localized tdTomato (mT) by EGFP (mG) (abbreviated as Wnt1-mtmg), where cardiac crest-derived cells (green, enclosed in dashed line) were present in myocardium of the outflow tract (D) and ventricle (Troponin T, gray) (E, E’’: separate channels of inset E’). Transverse view. Spatial-temporal information and antibody staining are displayed in supplement 1. Scale bars: A 400 μm; B-E 100 μm; C’, E’ 10 μm.
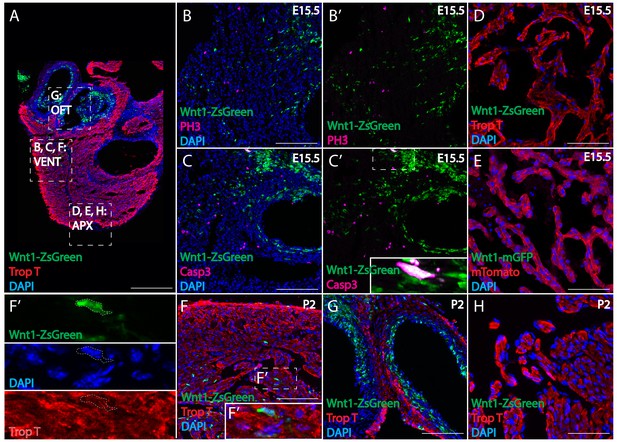
Spatial-temporal distribution of neural crest-derived cardiomyocytes in Wnt1-Cre mouse.
(A) In E15.5 Wnt1-Cre; Zsgreenfl/fl mouse heart, cardiac neural crest progenies were distributed in the outflow tract, valves and myocardium of both ventricles. Density of Zsgreen+ cells decreased along the proximal-to-distal axis, thus no cell contributed to the apex (Wnt1-Zsgreen, green; Troponin T, red; DAPI, blue). (B) Proliferation marker PH3 did not selectively colocalize with Zsgreen+ cells (PH3, magenta; B’, separate channels for colocalization). (C) Apoptosis marker Caspase3 was not selectively expressed in Zsgreen+ cells (Caspase 3, magenta; C’, separate channels for colocalization). (D) Cardiac neural crest did not contribute to apical myocardium in Wnt1-Cre; Zsgreenfl/fl mouse (Wnt1-Zsgreen, green; Troponin T, red; DAPI, blue), or (E) Wnt1-Cre; R26mtmg mouse (Wnt1-membrane GFP, green; membrane Tomato, red; DAPI, blue). (F–H) On postnatal day 2, cardiac neural crest progenies in Wnt1-ZsGreen mouse persisted in the myocardium of the ventricle (F). (separate channels in F’), outflow tract (G), but not the apex (H) (Wnt1-Zsgreen, green; Troponin T, red; DAPI, blue), similar to what was observed in E15.5 hearts. Scale bars: A 400 μm; B, C, F, G 100 μm; D, E, H 160 μm.
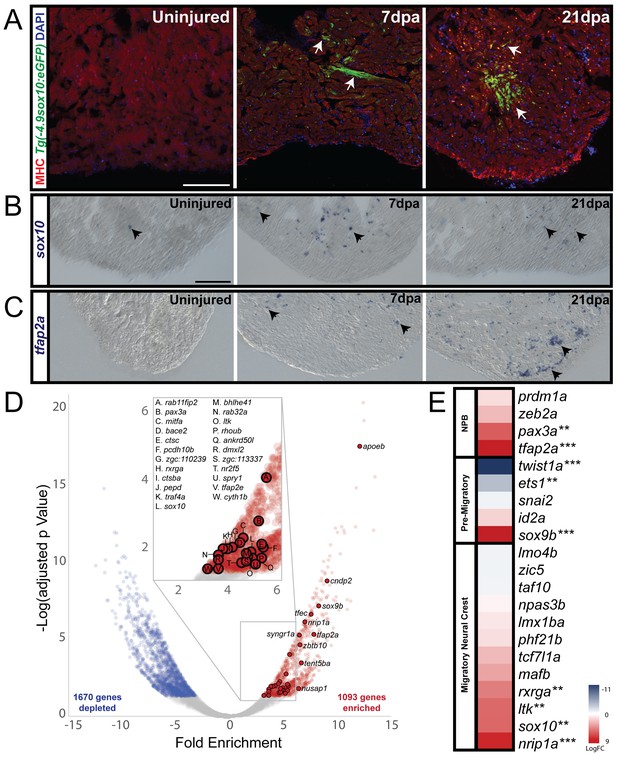
Cardiac neural crest contributes to heart regeneration in zebrafish.
(A) In sham-operated adult zebrafish hearts from a transgenic line expressing GFP under the control of a sox10 promoter, very few cells expressed Tg(−4.9sox10:eGFP) (green) (n = 3). 7 days-post amputation (dpa), the sox10 promoter was reactivated as shown by GFP+ cells in the trabeculated myocardium near the site of injury (Tg(−4.9sox10:eGFP), green) (n = 6). 21dpa, when the resected apex regenerated, more GFP+ cells were observed in sagittal sections within and surrounding the site of injury (n = 6). Sections in A are counterstained with DAPI in blue and Myosin Heavy Chain in red. (B) Endogenous sox10 mRNA expression was observed by paraffin section in situ hybridization in uninjured, 7dpa, and 21dpa hearts. Arrows denote cells with sox10 expression. From these results, we conclude that sox10 is reactivated after injury. (C) Along with sox10, expression of neural crest marker, tfap2a, was also enriched after injury. Arrows label areas of expression in the myocardium. (D) Differential gene expression analysis of FACS-sorted Tg(sox10:mRFP)+ and FACS-sorted Tg(sox10:mRFP)- transcriptomes show n = 1093 genes are enriched at 21dpa in the sox10+ cells compared to the rest of the ventricular tissue (n = 12 ventricles per replicate). Zebrafish neural crest genes as determined by GO analysis are highlighted on the volcano plot. (E) Upregulation of neural crest gene regulatory network genes was also observed from our differential expression analysis (**p<0.05, ***p<0.001). Co-localization of sox10 mRNA expression with Tg(sox10:GAL4-UAS-Cre;ubi:Switch)+ neural crest-derived cardiomyocytes is presented in supplement 1; schematic diagram of experimental design for obtaining the regenerating neural crest transcriptome and further analysis of gene enrichments is presented in supplement 2. Scale bars: 100 μm.
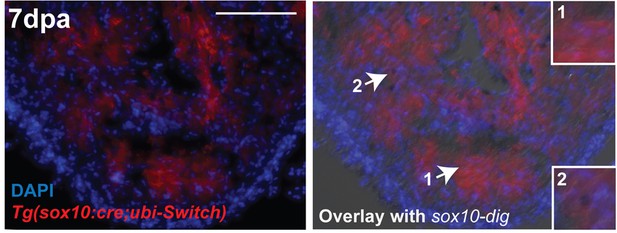
Co-localization of sox10 mRNA expression with Tg(sox10:GAL4-UAS-Cre;ubi:Switch)+ neural crest-derived cardiomyocytes.
sox10 transcript in adult zebrafish regenerating hearts (7dpa are reactivated in cells labeled by neural crest lineage transgenic line, Tg(sox10:GAL4-UAS-Cre;ubi:Switch) that permanently marks all neural crest-derived cells with mCherry (red). mCherry+ neural crest-derived cardiomyocytes were located in both the compacted and trabeculated layers of the ventricle. While not all neural crest-derived cells expressed sox10 (1), sox10 transcripts were only present in mCherry positive cells (2). Scale bar: 100 µm.
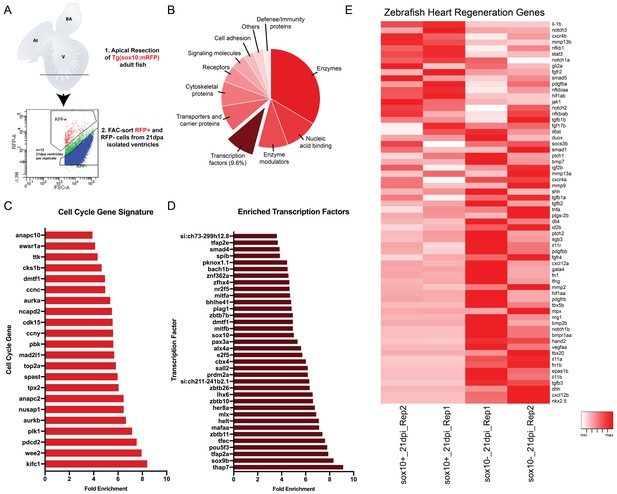
Analysis of regenerating neural crest transcriptome.
(A) Experimental design for the transcriptome analysis of regenerating neural crest cells. 1.Hearts were resected and ventricles were collected and dissociated at 21dpa, 2. mRFP+ and mRFP- cells were collected by FAC-sorting (FACS plot shows gating of populations). (B) Panther classification of proteins encoded by upregulated genes in the sox10+ cells. (C) Bar plot showing enriched mitotic cell cycle genes. (D) Bar plot showing enriched transcription factors. (E) Heatmap of normalized counts for known genes implicated in heart regeneration.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Mus musculus) | Wnt1-Cre; ZsGreenfl/fl | PMID:10725243 | Jackson Laboratories, Stock# 003829 | Drs. Xia Han and Yang Chai at University of Southern California, Center for Craniofacial Molecular Biology |
| Genetic reagent (Mus musculus) | Wnt1-Cre2+((129S4-Tg(Wnt1-cre)1Sor/J));R26mTmG | PMID: 23648512 | Jackson Laboratory, Stock# 22137 | Dr. Jeffrey Bush at University of California, San Francisco |
| Genetic reagent (Danio rerio) | Tg(−4.9sox10:eGFP) | PMID: 17065232 | ZFIN ID: ZDB-TGCONSTRCT-070117–69 | |
| Genetic reagent (Danio rerio) | Tg(sox10:GAL4-UAS-Cre;ubi-Switch) | PMID: 26086691 | Drs. Ann M. Cavanaugh and Jau-Nian Chen at Department of Molecular, Cell and Developmental biology, University of California, Los Angeles | |
| Genetic reagent (Danio rerio) | Tg(sox10:mRFP) | PMID: 18176560 | ZFIN ID: ZDB-TGCONSTRCT-080321–2 | |
| Cell line (Galllus gallus DF1) | UMNSAH/DF-1 fibroblast spontaneously transformed | ATCC | #CRL-12203, Lot number 62712171; RRID:CVCL_0570 | |
| Recombinant DNA reagent | RES-H2B-YFP-DD | Addgene | RRID:Addgene_96893 | |
| Antibody | Mouse monoclonal anti-bovine Troponin T, IgG2a (CT3) | DSHB | RRID:AB_528495 | Dilution (1:10) |
| Antibody | Mouse monoclonal anti-chicken Myosin Heavy Chain, IgG1 kappa light chain (ALD58) | DSHB | RRID:AB_528361 | Dilution (1:10) |
| Antibody | Mouse monoclonal anti- chicken Myosin Heavy Chain, IgG1 kappa light chain (F59) | DSHB | RRID:AB_528373 | Dilution (1:10) |
| Antibody | Mouse monoclonal anti-NH2 terminal synthetic decapeptide of alphasmooth muscle actin, IgG2a | Sigma | # A5228 | Dilution (1:500) |
| Antibody | Mouse monoclonal anti-human phospho-histone H3, IgG1 | Abcam | #ab14955 | Dilution (1:500) |
| Antibody | Rabbit polyclonal anti-human Caspase 3, IgG | R and D systems | #AF835 | Dilution (1:500) |
| Antibody | Goat polyclonal anti -GFP, IgG | Abcam | #ab6673 | Dilution (1:500) |
| Antibody | Goat polyclonal anti-mouse IgG2a Alexa-568 | Molecular Probes | RRID:AB_2535773 | Dilution (1:1000) |
| Antibody | Goat polyclonal anti-mouse IgG1 Alexa-568 | Molecular Probes | RRID:AB_2535766 | Dilution (1:1000) |
| Antibody | Goat polyclonal anti-rabbit IgG Alexa-568 | Molecular Probes | RRID:AB_2534121 | Dilution (1:1000) |
| Antibody | Donkey polyclonal anti-goat IgG Alexa-488 | Molecular Probes | RRID:AB_2534102 | Dilution (1:1000) |
| Software, algorithm | Image processing software FIJI | https://imagej.net/Fiji | ||
| Software, algorithm | R v3.6.1 | https://www.r-project.org/ | ||
| Software, algorithm | DESeq2 | PMID: 25516281 | RRID:SCR_015687 | |
| Software, algorithm | Bowtie2 | PMID: 22388286 | RRID:SCR_005476 | |
| Software, algorithm | featureCounts (Subread) | PMID: 24227677 | RRID:SCR_009803 | |
| Other | Accumax | Innovative Cell Technologies, Inc | #AM105 | |
| Commercial assay or kit | SMART-seq Ultra Low Input RNA Kit V4 | Takara Clontech | #634891 |
Additional files
-
Supplementary file 1
Quantification of cardiac neural crest contribution to heart development in amniotes and sox10:eGFP+ cells in zebrafish heart regeneration.
(a, b) Quantification of cardiac neural crest contribution to the heart in chick and mouse.Table presents virally labeled cardiac neural crest derivatives at MNC (migratory neural crest), CN-IX (cranial nerve nine), PAA (pharyngeal arch arteries), APS (aorticopulmonary septum), MYO (myocardium of ventricle) and IVS (interventricular septum) at day 1–6 and day 10 post injection in chick. The bottom part presents number of Wnt1+ cells in E15.5 Wnt1-Cre mouse. Percentage in parentheses represents the proportion of the population among all NC-derived cells in cardiovascular structure (including MYO, APS, and IVS). % Neural crest contribution to ventricle, the proportion of Wnt1+ cells (including MYO and IVS) among all cells in the ventricle is about 16.8%. Supplementary file 1b shows the raw data of each embryo from which data in Supplementary file 1a was generated. (c) Quantification of sox10:eGFP+ cells in the apex during zebrafish heart regeneration. Average number of Sox10-eGFP+ cells per 2 × 105 µm2 in one section through the middle of the apex of 7dpa (n = 3) and 21 dpa (n = 3) hearts after resection. Standard deviation is presented in parentheses next to the cell number. GFP expression was negligible in sham operated hearts (n = 3) at the same time points.
- https://doi.org/10.7554/eLife.47929.012
-
Transparent reporting form
- https://doi.org/10.7554/eLife.47929.013





