One-step efficient generation of dual-function conditional knockout and geno-tagging alleles in zebrafish
Figures
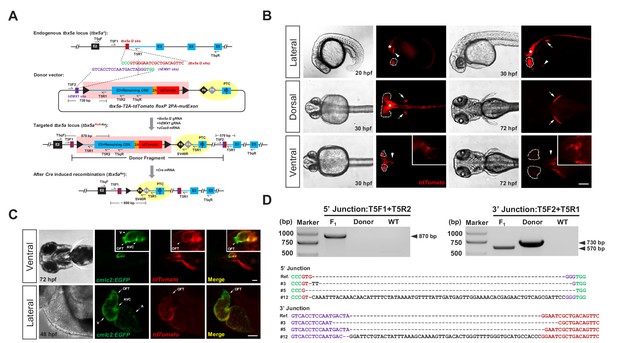
Generation of CKO coupled with gene labeling dual-function alleles through targeted insertion in zebrafish at the tbx5a locus.
(A) Schematic diagram of the KI strategy based on the dual-cassette PoNe donor tbx5a-T2A-tdTomato floxP 2PA-mutExon (the tbx5a PoR-Ne donor), consisting of a Po-cassette and a Ne-cassette (highlighted by pink and yellow shadows, respectively). The target sequences of hEMX1 and tbx5a are shown in purple and brown, respectively, and the PAMs are in shown green. Black triangles represent loxP. Black and gray diamonds indicate polyadenylation (PA) signals. The black bar in the third exon (E3) indicates the in-frame premature termination codon (PTC). Primers T5qF and T5qR are used for qRT-PCR in Figure 2—figure supplement 1E and F. (B) Images of F1 larvae from an outcross of a tbx5a PoR-Ne donor knockin founder (#12), showing tdTomato expression in the pectoral fins (white arrows), heart (white arrowheads), eyes (white dotted circle) and nervous system (white asterisk). Scale bar, 200 μm. (C) Images of F1 progeny from Tg(cmlc2:EGFP) transgenic zebrafish crossed with the tbx5a KI founder (#12), showing an antero-posterior gradient of tbx5a expression in the ventricle. Upper panel: Ventral view of a 72 hpf embryo. Lower panel: Z-stack confocal images of the heart region from a 48 hpf embryo. A: atrium. AVC: atrioventricular canal. OFT: outflow track. V: ventricle. Scale bar, 50 μm. (D) Junction PCR and direct sequencing results of individual positive F1 progeny from outcrosses of each of the three positive founders (#3, #5 and #12). Due to an extra copy of the T5R1 primer sequence in the PoR-Ne donor, PCR with the primer pair T5F2 and T5R1 targeting the donor plasmid results in a larger fragment than that of the F1 progeny. F1: an F1 embryo from F0 #12. Donor: tbx5a PoR-Ne donor plasmid. WT: pooled genomic DNA of five wild-type embryos.
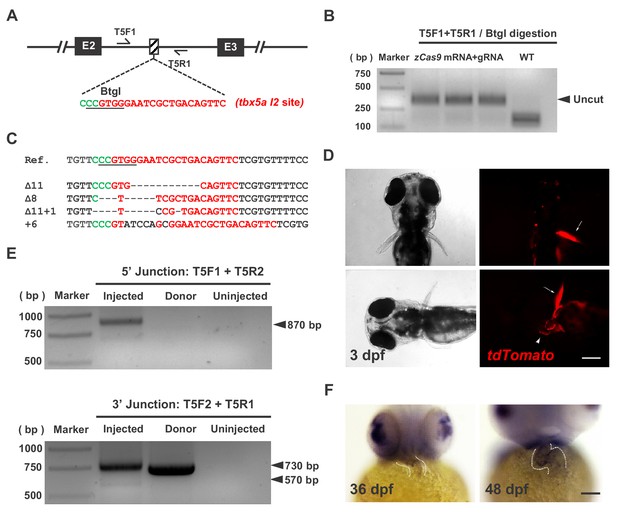
Evaluation of the expression of tbx5a and targeted insertion of the CKO + gene labeling PoNe donor at the tbx5a I2 target site in founder embryos.
(A) The position and sequence of the tbx5a intron 2 (I2) target site designed for the Cas9/gRNA system. The protospacer sequence is shown in red, and the PAM is shown in green. (B) Indel efficiency evaluated by PCR and BtgI restriction endonuclease digestion. (C) Sequencing results of the uncut PCR products (corresponding to indel mutations) from B after cloning. (D) Mosaic expression of tdTomato in the heart (white arrowhead) and fin (white arrows) in a founder embryo after the injection of gRNAs purified by LiCl precipitation together with zCas9 mRNA and the tbx5a PoR-Ne donor from Figure 1A. Scale bar, 200 μm. (E) Junction PCR to detect NHEJ-mediated knockin events in founder embryos. The expected products (870 bp and 570 bp) were obtained by amplification with the corresponding primers shown in Figure 1A. Injected: Donor+Cas9/gRNA-injected embryos. Donor: tbx5a PoR-Ne donor plasmid. Uninjected: Uninjected embryos. (F) The expression of tbx5a in the zebrafish heart shown by whole-mount in situ hybridization (ventral view). The dotted lines denote the outline of the heart. Scale bar, 100 μm.
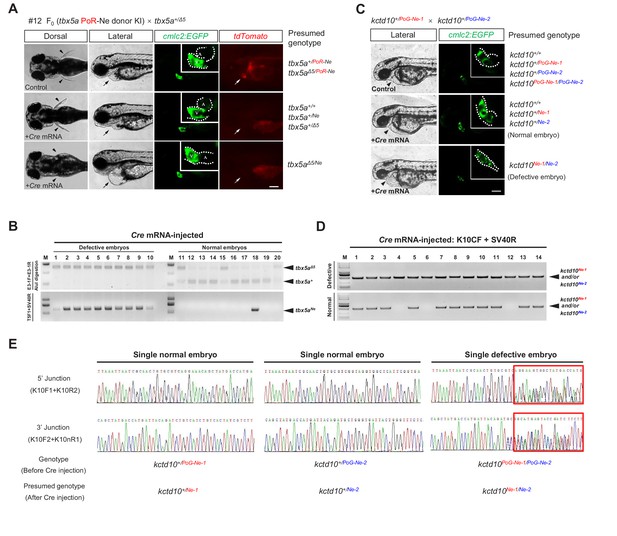
The tbx5a and kctd10 conditional alleles are responsive to Cre recombinase.
(A) Images of the 72 hpf progeny (F1) from a tbx5a+/Δ5 heterozygote crossed with the F0 #12 (mosaic for the tbx5aPoR-Ne allele) against the Tg(cmlc2:EGFP) transgenic background with or without Cre mRNA injection. The fluorescent images were obtained in the lateral view. Black arrowheads indicate pectoral fins, and black or white arrows indicate the heart. A: atrium. V: ventricle. Scale bar, 200 μm. (B) Genotyping results of the individual Cre mRNA-injected embryos obtained from the cross in A. (C) Images of the 48 hpf progeny (F2) derived from the cross of two kctd10 KI heterozygotes (F1), each carrying a different KI allele (kctd10PoG-Ne-1 from #32 and kctd10PoG-Ne-2 from #5) with the Tg(cmlc2:EGFP) transgenic background, to reveal the morphology of the heart. The white dotted line indicates the outline of the heart. The hearts in the upper and middle panels developed normally, showing obvious heart looping. In contrast, the heart in the lower panel shows defective development, exhibiting AVC malformations and heart looping failure. Black arrowheads indicate the heart. Scale bar, 200 μm. (D) Genotyping by PCR amplification of the region flanking the loxP recombination site of the Cre mRNA-injected individual embryos obtained from the cross in C. (E) Representative junction PCR and direct sequencing results of the Cre mRNA-injected individual embryos showing normal or defective heart development obtained from the cross in C. As expected, the results indicate that the embryo showing the heart phenotype (labeled as ‘Single defective embryo’ in the figure) was a kctd10PoG-Ne-1/PoG-Ne-2 compound heterozygote (F2) before Cre mRNA injection since it showed overlapping peaks (red boxed region) in the sequencing results of the PCR products at both the 5’ and 3’ junctions (right panel), as the two alleles have different indel sequences at the junction sites. In contrast, the normal embryos (labeled as ‘Single normal embryos’ in the figure) were either kctd10+/PoG-Ne-1 or kctd10+/PoG-Ne-2 heterozygotes before Cre mRNA injection and therefore displayed uniform sequencing results corresponding to either the kctd10PoG-Ne-1 (or kctd10Ne-1) or kctd10PoG-Ne-2 (or kctd10Ne-2) allele, respectively. The expected corresponding sequences can be found in Figure 2—figure supplement 2J and K.
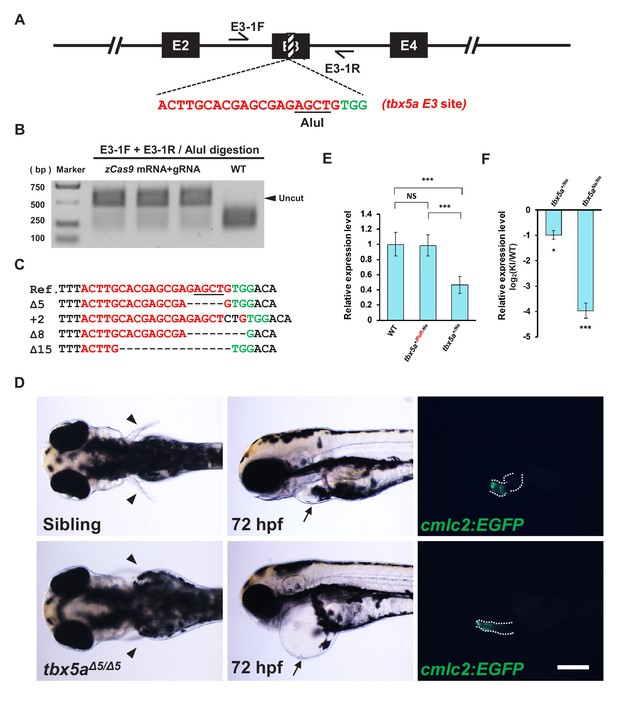
Evaluation of the indel efficiency of the tbx5a E3 target site and phenotype analysis of the tbx5a indel mutation.
(A) The position and sequence of the tbx5a exon 3 (E3) target site designed for the Cas9/gRNA system. The protospacer sequence is shown in red, and the PAM is shown in green. (B) Targeting efficiency evaluated by PCR and AluI restriction endonuclease digestion. The result indicates that the indel efficiency is nearly 90%. (C) Sequencing results of the uncut PCR products (corresponding to indel mutations) from B after cloning. (D) Approximately 25% of embryos from the incross of tbx5a+/Δ5 heterozygotes showed defects in heart (black arrows) and pectoral fins (black arrowheads). Genotyping results revealed that all the defective embryos were tbx5aΔ5/Δ5 homozygotes (lower panel), while the siblings showed a normal morphology. The Tg(cmlc2:EGFP) transgenic background was introduced to reveal the heart morphology, and all the defective embryos also showed failure of cardiac looping. The dotted lines denote the outline of the heart. Scale bar, 200 μm. (E) qRT-PCR results showing the transcription level of the tbx5a locus in wild-type (WT) and tbx5a PoR-Ne donor KI zebrafish embryos at 72 hpf, using T5qF and T5qR primers. The tbx5a+/Ne and tbx5a+/PoR-Ne embryos were obtained from the crossing of the tbx5a PoR-Ne/PoR-Ne homozygotes with wild-type zebrafish with or without injection of Cre mRNA, respectively. The average expression level of wild-type embryos was set as 1. (F) qRT-PCR results using T5qF and T5qR primers, showing the transcription level of the tbx5a locus in the tbx5a+/Ne and tbx5aNe/Ne embryos derived from the Cre mRNA-injected tbx5a+/PoR-Ne and tbx5aPoR-Ne/PoR-Ne embryos, respectively. The original embryos were obtained from the crossing of tbx5aPoR-Ne/PoR-Ne homozygotes with tbx5a+/PoR-Ne heterozygote zebrafish. The expression levels in the KI embryos were normalized to the WT ones. Data are presented as the mean ±s.d., and a two-tailed Student’s t-test was applied to calculate p values in all the experiments. *: p<0.05. ***: p<0.001. NS: Not significant.
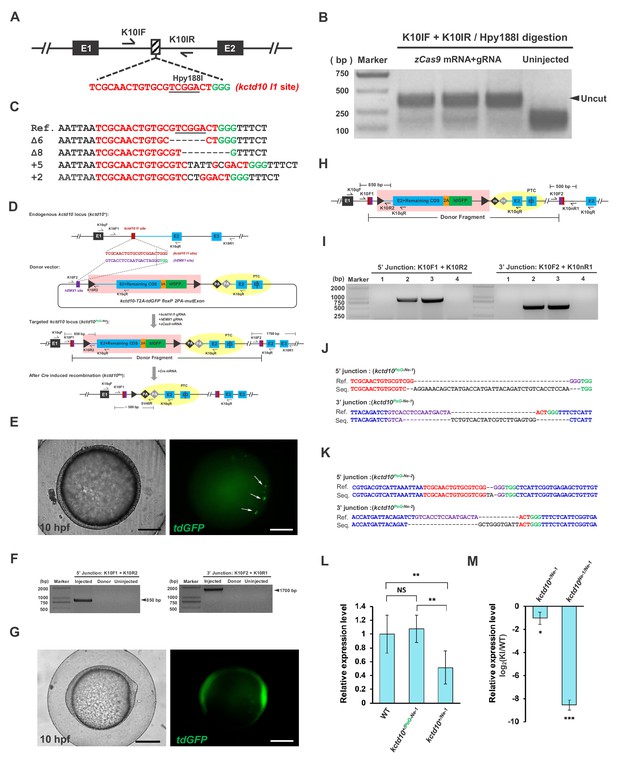
Strategy and evaluation of the targeted insertion of the PoG-Ne donor at the kctd10 locus.
(A) The position and sequence of the kctd10 intron 1 (I1) target site designed for the Cas9/gRNA system. The protospacer sequence is shown in red, and the PAM is shown in green. (B) Targeting efficiency evaluated by PCR and Hpy188I restriction endonuclease digestion. (C) Sequencing results of the uncut PCR products (corresponding to indel mutations) from B after cloning. (D) Schematic diagram of the kctd10-2A-td GFP floxP 2PA-mutExon PoNe donor (abbreviated as kctd10 PoG-Ne donor) and the strategy of targeted insertion and conditional knockout using the CRISPR/Cas system. Primers K10qF and K10qR are used for qRT-PCR in L and M. (E) Images of a 10 hpf F0 zebrafish embryo after the injection of the kctd10 PoG-Ne donor together with zCas9 mRNA and corresponding gRNAs. White arrows indicate tdGFP signals. Scale bar, 200 μm. (F) Junction PCR to detect NHEJ-mediated knockin events in the injected founder embryos. Injected: Donor+Cas9/gRNA-injected embryos. Donor: kctd10 PoG-Ne donor plasmid. Uninjected: Uninjected embryos. (G) Images of a 10 hpf F1 zebrafish embryo from an outcross of the kctd10 PoG-Ne donor KI-positive F0 female (#32) shown in Supplementary file 4, bearing the kctd10PoG-Ne-1 allele. Strong maternal expression of tdGFP can be clearly observed in this F1 embryo. Scale bar, 200 μm. (H) Schematic diagram of the kctd10 KI allele, showing the position of the primers used for junction PCR in I-K and qRT-PCR in L. A new primer pair was used to amplify the 3’ junction of the F1 embryos. (I) Junction PCR to detect the knockin allele in individual F1 embryos (1-4) from the cross in G. Note that not all of the embryos inherited the knockin allele from the F0 female, indicating germline mosaicism of this adult fish. (J) Sequencing results of the PCR products from the two positive embryos (2 and 3) in I, which showed the same junction sequence of the kctd10PoG-Ne-1 allele. (K) Sequencing results of the PCR products (using the same primer pair as in I and J) from an EGFP-positive F1 zebrafish embryo obtained from an outcross of the positive F0 male (#5), representing the junction sequence of the kctd10PoG-Ne-2 allele. (L) qRT-PCR results showing the transcription level of the kctd10 locus in wild-type (WT) and kctd10 PoG-Ne donor KI zebrafish embryos at 72 hpf, using K10qF and K10qR primers. The kctd10+/Ne-1 and kctd10+/PoG-Ne-1 embryos were obtained from the cross of kctd10 PoG-Ne-1/PoG-Ne-1 homozygotes with wild-type zebrafish with or without the injection of Cre mRNA, respectively. The average expression level of wild-type embryos was set as 1. (M) qRT-PCR results using K10qF and K10qR primers, showing the transcription level of the kctd10 locus in the kctd10+/Ne-1 and kctd10Ne-1/Ne-1 embryos derived from the Cre mRNA-injected kctd10+/PoG-Ne-1 and kctd10PoG-Ne-1/PoG-Ne-1 embryos, respectively. The original embryos were obtained from the crossing of kctd10PoG-Ne-1/PoG-Ne-1 homozygotes with kctd10+/PoG-Ne-1 heterozygote zebrafish. The expression levels in the KI embryos were normalized to the WT ones. Data are presented as the mean ±s.d., and a two-tailed Student’s t-test was applied to calculate p values in all the experiments. *: p<0.05. **: p<0.01. ***: p<0.001. NS: Not significant.
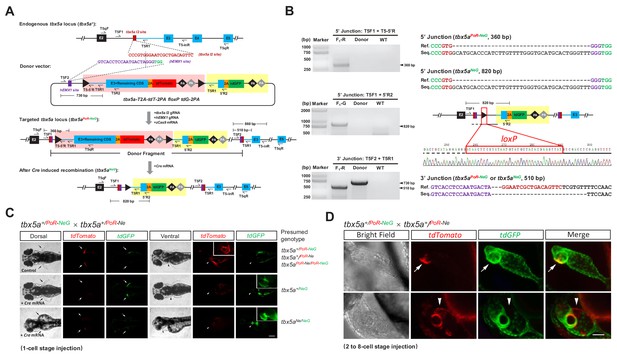
Generation of geno-tagging alleles by improving the dual-cassette donor strategy at the zebrafish tbx5a locus.
(A) Schematic diagram of the KI strategy for geno-tagging based on the tbx5a-T2A-tdT-2PA floxP tdG-2PA donor (or abbreviated as tbx5a PoR-NeG donor). Primers T5qF and T5qR are used for qRT-PCR in Figure 3—figure supplement 1E and F. (B) Results of junction PCR and direct sequencing to detect the tbx5a geno-tagging donor knockin and Cre-induced recombination events in 48 hpf embryos obtained from the cross in Figure 3—figure supplement 1B, that is the F1 embryos from the #42 positive F0 outcrossed with a wild-type zebrafish before or after Cre mRNA injection. Note that the sequences of both the T5F2 and T5R1 primers are also present in the donor vector, flanking the upstream loxP site (as shown in panel A); therefore, a 730 bp product could be amplified in the lane with the donor as the template. F1-R: an F1 embryo showing a red fluorescent signal (before Cre mRNA injection). F1-G: an F1 embryo showing a green fluorescent signal (after Cre mRNA injection). Donor: tbx5a PoR-NeG geno-tagging donor plasmid. WT: pooled genomic DNA of five wild-type embryos. (C) Phenotype analysis of the 72 hpf embryos from tbx5a+/PoR-NeG heterozygotes (derived from F0 #42 in Supplementary file 5) crossed with a tbx5a+/PoR-Ne heterozygote (derived from F0 #12 in Supplementary file 3) after the injection of Cre mRNA at the 1 cell stage. The upper panel represents an uninjected control embryo showing only a red fluorescent signal, whose genotype should be either tbx5a+/PoR-Ne, tbx5a+/PoR-NeG or tbx5a PoR-Ne/PoR-NeG. The middle panel represents a Cre mRNA-injected embryo showing normal development, whose genotype is expected to be tbx5a+/NeG. The lower panel represents a Cre mRNA-injected embryo showing a typical tbx5a mutant phenotype, including heart region defects and a lack of pectoral fins, whose genotype is expected to be tbx5aNe/NeG. Arrows indicate the pectoral fins, and arrowheads indicate the heart region. The boxed insets show a higher magnification of the corresponding heart region, for better comparison of heart morphology. Scale bar, 200 μm. (D) Z-stack confocal images of two representative 48 hpf embryos after the injection of Cre mRNA at the 2- to 8 cell stage from the same cross as in C. The white arrows indicate the colocalization of the tdGFP and tdTomato signals, and the white arrowheads indicate the mutually exclusive expression of the tdGFP and tdTomato signals. Scale bar, 50 μm.
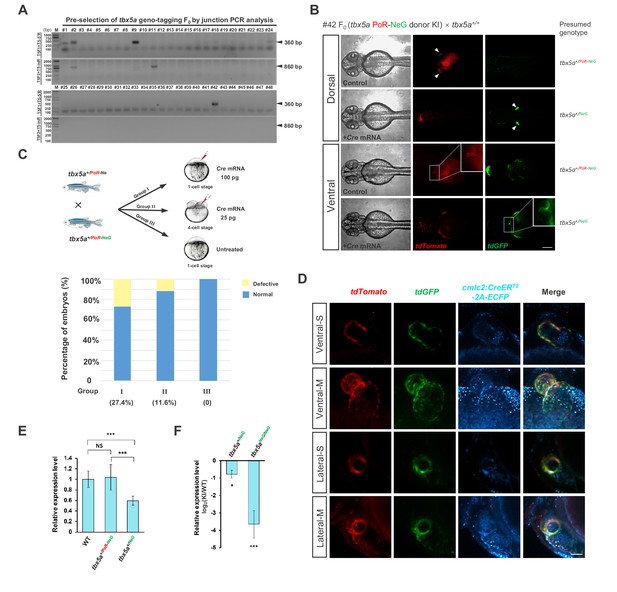
Evaluation of the tbx5a geno-tagging effect.
(A) Preselection of tbx5a geno-tagging F0 individual by junction PCR analysis. 5’ or 3’ junctions were amplified by PCR using genomic DNA extracted from fin clips of the #1, #2, #9, #11, #24 and #42 F0 adult fish. The corresponding primer pairs are shown on the left side of the gel images, and the positions of these primers can be found in Figure 3A. (B) Switching of fluorescent signals achieved from the tbx5a geno-tagging allele after Cre mRNA injection into the F1 progeny from #42 positive F0 outcrossed with a wild-type zebrafish. The arrowheads indicate pectoral fins. The outlined boxed areas indicate the heart region, showing the change in the fluorescent signals in the heart before and after Cre mRNA injection. Scale bar, 200 μm. (C) The experimental design for the functionality test of the tbx5a geno-tagging allele. The progeny from the cross of a tbx5a+/PoR-NeG heterozygote with a tbx5a+/PoR-Ne heterozygote were divided into three groups: Group I was injected with 100 pg Cre mRNA at the one-cell stage, Group II was injected with 25 pg Cre mRNA in a single cell at the 4 cell stage, and Group III remained untreated as a control. The histogram shows the ratio of defective embryos after Cre mRNA injection in different groups. (D) Confocal images of the heart regions of two embryos from the cross of Tg(cmlc2:zCreERT2-2A-ECFP) transgenic fish with tbx5a PoR-NeG/PoR-NeG after 4-HT treatment, showing a red to green change in the fluorescent signals upon Cre induction. -S: Single-plane view, -M: Maximum intensity projection view of z-stack images. Scale bar, 50 μm. (E) qRT-PCR results showing the transcription level of the tbx5a locus in wild-type (WT) and tbx5a PoR-NeG geno-tagging donor KI zebrafish embryos at 72 hpf, using T5qF and T5qR primers. The tbx5a+/NeG and tbx5a+/PoR-NeG embryos were obtained from crosses of tbx5aPoR-NeG/PoR-NeG homozygotes with wild-type zebrafish with or without the injection of Cre mRNA, respectively. The average expression level of wild-type embryos was set as 1. (F) qRT-PCR results using T5qF and T5qR primers, showing the transcription level of the tbx5a locus in the tbx5a+/NeG and tbx5aNeG/NeG embryos derived from the Cre mRNA-injected tbx5a+/PoR-NeG and tbx5aPoR-NeG/PoR-NeG embryos, respectively. The original embryos were obtained from the crossing of tbx5aPoR-NeG/PoR-NeG homozygotes with tbx5a+/PoR-NeG heterozygote zebrafish. The expression levels in the KI embryos were normalized to the WT ones. Data are presented as the mean ±s.d., and a two-tailed Student’s t-test was applied to calculate p values in all the experiments. *: p<0.05. ***: p<0.001. NS: Not significant.
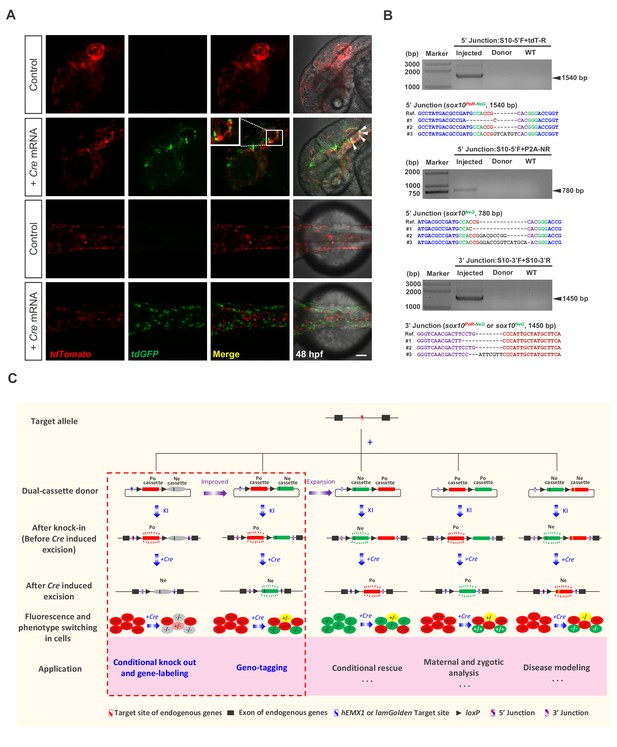
Generation of geno-tagging at the sox10 locus and summary of our dual-cassette donor KI strategy.
(A) Z-stack confocal images of 48 hpf zebrafish embryos after the injection of the sox10 geno-tagging donor knockin system at the one-cell stage (Control) followed by the further injection of 25 pg Cre mRNA into a single cell at the 4 cell stage (+Cre mRNA). White arrowheads indicate the colocalization of the tdGFP and tdTomato signals. Scale bar, 100 μm. (B) Junction PCR and clonal sequencing to detect the knockin and Cre-induced recombination events in the injected embryos showing mosaic double-fluorescence signals. Injected: Embryos injected with the donor, Cas9/gRNA system and Cre mRNA. Donor: sox10 geno-tagging donor plasmid. Uninjected: Uninjected embryos. (C) Summary of the applications and potential expansion of our dual-cassette donor KI strategy.
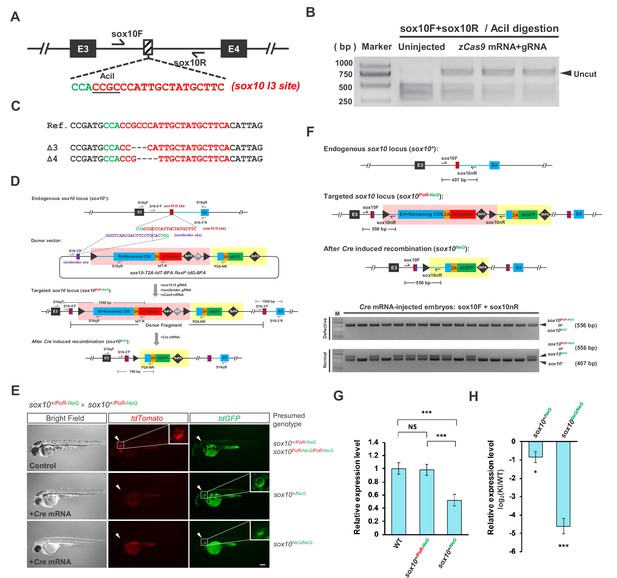
Generation and evaluation of the sox10 geno-tagging allele.
(A) The position and sequence of the sox10 intron 3 (I3) target site designed for the Cas9/gRNA system. The protospacer sequence is shown in red, and the PAM is shown in green. (B) Targeting efficiency evaluated by PCR and AciI restriction endonuclease digestion. The result indicates that the indel efficiency is nearly 85%. (C) Sequencing results of the uncut PCR products (corresponding to indel mutations) from B after cloning. (D) The donor design and geno-tagging KI strategy at the sox10 locus. Primers S10qF and S10qR are used for qRT-PCR in G and H. (E) Phenotype analysis of the 48 hpf F2 embryos from the incrossing of sox10+/PoR-NeG heterozygotes (derived from #6 F0) after the injection of Cre mRNA at the one-cell stage. The upper panel shows an uninjected control embryo bearing red fluorescent signals with normal pigmentation, whose genotype should be either sox10+/PoR-NeG or sox10PoR-NeG/PoR-NeG. The middle panel represents one Cre-injected embryo showing slightly less pigmentation but with only green fluorescent signals, indicating an efficient switch to the expression of tdGFP from that of tdTomoto after Cre injection; therefore, the genotype should be sox10+/NeG. The lower panel shows a Cre-injected embryo devoid of body pigmentation that faithfully recapitulates the expected phenotype of the sox10 loss-of-function mutation. Similar to the previous embryo, this embryo shows only green fluorescent signals due to the Cre-induced efficient switch of the expression of the fluorescent reporter gene; therefore, the genotype is most likely tbx5aNeG/NeG. The white arrowheads indicate otic vesicles, whose detailed structure can be seen under higher magnification of the boxed areas. Scale bar, 200 μm. (F) Genotyping results of the injected F2 embryos in E determined via 5’ junction PCR analysis. Since all the defective embryos showed only green (tdGFP) and no red (tdTomoto) fluorescent signal, the PCR products are most likely derived from the amplification of the sox10NeG allele. (G) qRT-PCR results showing the transcription level of the sox10 locus in wild-type (WT) and sox10 PoR-NeG geno-tagging donor KI zebrafish embryos at 72 hpf, using S10qF and S10qR primers. The sox10+/NeG and sox10+/PoR-NeG embryos were obtained from the crossing of sox10PoR-NeG/PoR-NeG homozygotes with wild-type zebrafish with or without the injection of Cre mRNA, respectively. The average expression level of wild-type embryos was set as 1. (H) qRT-PCR results using S10qF and S10qR primers, showing the transcription level of the tbx5a locus in the sox10+/NeG and sox10NeG/NeG embryos derived from the Cre mRNA-injected sox10+/PoR-NeG and sox10PoR-NeG/PoR-NeG embryos, respectively. The original embryos were obtained from the crossing of sox10PoR-NeG/PoR-NeG homozygotes with sox10+/PoR-NeG heterozygote zebrafish. The expression levels in the KI embryos were normalized to the WT ones. Data are presented as the mean ±s.d., and a two-tailed Student’s t-test was applied to calculate p values in all the experiments. *: p<0.05. ***: p<0.001. NS: Not significant.
Tables
KI efficiency using different gRNA purification methods
https://doi.org/10.7554/eLife.48081.004| hEMX1 gRNA purification | tbx5a I2 gRNA purification | Ratio of fluorescence- positive F0 embryos |
|---|---|---|
| LiCl | LiCl | 18/119 (15.1%) |
| LiCl | Ethanol | 8/103 (7.8%) |
| Ethanol | LiCl | 0/94 (0.0%) |
| Ethanol | Ethanol | 0/43 (0.0%) |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Danio rerio) | tbx5a | Gene: 30071 | ENSDARG00000024894 | |
| Gene (Danio rerio) | kctd10 | Gene: 406787 | ENSDARG00000017115 | |
| Gene (Danio rerio) | sox10 | Gene: 140616 | ENSDARG00000077467 | |
| Strain, strain background (Danio rerio) | Tübingen (TU) | Our lab | A commonly used wild-type zebrafish strain | |
| Strain, strain background (Danio rerio) | Tg(cmlc2:EGFP) | Tong et al., 2014 | PMID: 24430697 | |
| Strain, strain background (Danio rerio) | Tg(cmlc2:zCreERT2-T2A-ECFP) | This paper | A transgenic zebrafish expressing zebrafish codon-optimized CreERT2 and ECFP driven by the heart-specific cmlc2 promoter | |
| Recombinant DNA reagent | pMD18-T vector (plasmid) | TAKARA | Cat#: 6011 | |
| Recombinant DNA reagent | pMD19-T simple vector (plasmid) | TAKARA | Cat#: 3271 | |
| Recombinant DNA reagent | pminiTol2 (plasmid) | Balciunas et al., 2006 | PMID: 17096595 | |
| Recombinant DNA reagent | pGH-T7-zCas9 (plasmid) | Our lab | PMID: 24480746 | Liu et al., 2014 |
| Recombinant DNA reagent | pMD18T-tdGFP (plasmid) | Dr. Yulong Li | ||
| Recombinant DNA reagent | pUC19-scaffold (plasmid) | Dr. Jingwei Xiong | PMID: 23528705 | Chang et al., 2013 |
| Recombinant DNA reagent | bait otx2 CreERT2 (plasmid) | Dr. Michael Brand | PMID: 29435650 | Kesavan et al., 2018 |
| Recombinant DNA reagent | pX-T7-Cre (plasmid) | Dr. Yao Zu | ||
| Commercial assay or kit | Gibson assembly | NEB | E5510S | |
| Commercial assay or kit | mMessage mMachine T7 kit | Ambion | AM1344 | |
| Chemical compound, drug | 4-HT (Hydroxyltamoxifen) | Sigma | H6278-10MG | |
| Chemical compound, drug | Tricaine (ethyl 3-aminobenzoate methanesulfonate salt) | Sigma | E10521 | |
| Software, algorithm | AxioVision Rel.4.8 | Zeiss | RRID: SCR_002677 | |
| Software, algorithm | ZEN 2009 | Zeiss | RRID: SCR_013672 | |
| Other | TRIzol reagent | Invitrogen | Cat#: 10296028 | |
| Other | 5x All-In-One RT MasterMix | abm | G485 | |
| Other | EvaGreen 2x qRT-PCR Mastermix | abm | Mastermix-S |
Additional files
-
Supplementary file 1
The Cas9/gRNA target sequences used in this study.
- https://doi.org/10.7554/eLife.48081.012
-
Supplementary file 2
Indel and knockin efficiencies in founder embryos.
- https://doi.org/10.7554/eLife.48081.013
-
Supplementary file 3
Germline mosaicism of the tbx5a PoR-Ne donor KI in each positive F0.
- https://doi.org/10.7554/eLife.48081.014
-
Supplementary file 4
Germline mosaicism of the kctd10 PoG-Ne donor KI in each positive F0.
- https://doi.org/10.7554/eLife.48081.015
-
Supplementary file 5
Germline mosaicism of the tbx5a geno-tagging PoR-NeG donor KI in F0.
- https://doi.org/10.7554/eLife.48081.016
-
Supplementary file 6
Germline mosaicism of the sox10 geno-tagging PoR-NeG donor KI in F0.
- https://doi.org/10.7554/eLife.48081.017
-
Supplementary file 7
The sequences of the primers used for PCR and qRT-PCR analyses.
- https://doi.org/10.7554/eLife.48081.018
-
Transparent reporting form
- https://doi.org/10.7554/eLife.48081.019





