Inhibitory muscarinic acetylcholine receptors enhance aversive olfactory learning in adult Drosophila
Figures
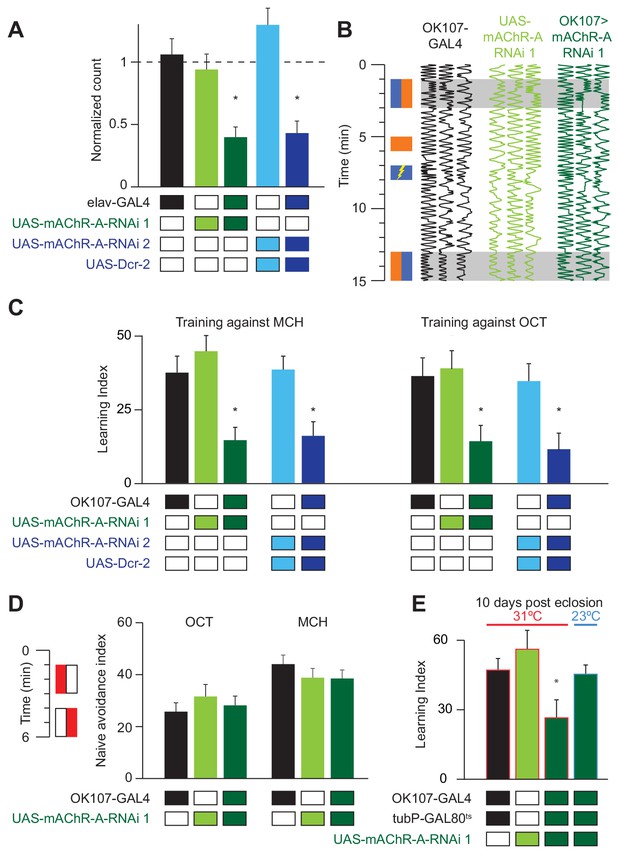
mAChR-A is required in the MB for short-term aversive olfactory learning and memory but not for naive behavior.
(A) qRT-PCR of mAChR-A with mAChR-A RNAi driven by elav-GAL4. The housekeeping gene eEF1α2 (eukaryotic translation elongation factor 1 alpha 2, CG1873) was used for normalization. Knockdown flies have ~40% of the control levels of mAChR-A mRNA (mean ± SEM; number of biological replicates (left to right): 6, 7, 7, 4, 4, each with three technical replicates; *p<0.05; Kruskal-Wallis test with Dunn’s multiple comparisons test and Welch ANOVA test with Dunnett’s T3 multiple comparisons test). For detailed statistical analysis see Supplementary file 1. (B) Each trace shows the movement of an individual fly during the training protocol, with fly position in the chamber (horizontal dimension) plotted against time (vertical dimension). Colored rectangles illustrate which odor is presented on each side of the chamber during training and testing. Flies were conditioned against MCH (blue rectangles; see Materials and methods). (C) Learning scores in flies with mAChR-A RNAi driven by OK107-GAL4. mAChR-A knockdown reduced learning scores compared to controls (mean ± SEM, n (left to right): 69, 69, 70, 71, 71, 47, 48, 53, 58, 51 *p<0.05; Kruskal-Wallis test with Dunn’s multiple comparisons test). (D) mAChR-A knockdown flies show normal olfactory avoidance to OCT and MCH compared to their genotypic controls (mean ± SEM, n (left to right): 68, 67, 58, 63, 91, 67, p=0.82 for OCT, p=0.64 for MCH; Kruskal-Wallis test). Colored rectangles show stimulus protocol as in (B); red for odor (MCH or OCT), white for air. (E) Learning scores in flies with mAChR-A RNAi 1 driven by OK107-GAL4 with GAL80ts repression. Flies raised at 23°C and heated to 31°C as adults (red outlines) had impaired learning compared to controls. Control flies kept at 23°C throughout (blue outline), thus blocking mAChR-A RNAi expression, showed no learning defects (mean ± SEM, n (left to right): 51, 41, 58, 51, **p<0.05, Kruskal-Wallis test with Dunn’s multiple comparisons test). For detailed statistical analysis see Supplementary file 1.
-
Figure 1—source data 1
Source data for Figure 1A.
- https://doi.org/10.7554/eLife.48264.006
-
Figure 1—source data 2
Source data for Figure 1C–E.
- https://doi.org/10.7554/eLife.48264.007
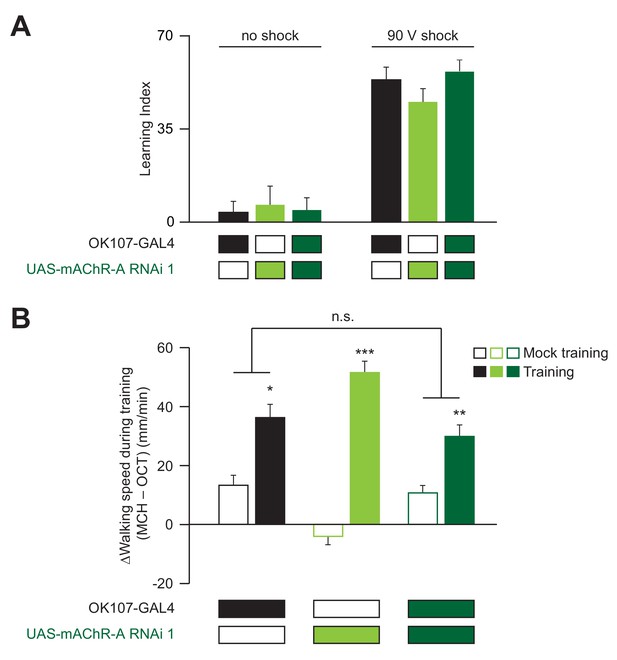
Controls and additional learning data.
(A) Flies were subjected to the same protocol as in Figure 1 but with no, or stronger, electric shock. With no electric shock, the flies do not change their odor preference and have a learning index which is not statistically different from 0 (n (left to right): 79, 73, 71; p>0.3, one-sample t-test). When flies were conditioned against MCH using 90 V electric shock instead of 50 V (as in the main Figures; see Materials and methods), driving mAChR-A RNAi in KCs using OK107-GAL4 did not affect learning compared to controls (mean ± SEM, n (left to right): 52, 46, 51, p>0.13, Kruskal-Wallis test). For detailed statistical analysis see Supplementary file 1. (B) Sensitivity to shock (extent to which flies walk faster while being shocked) is not affected by knocking down mAChR-A in KCs. Shown here is walking speed during training (time = 5–6 and 7–8 min in Figure 1B), taking the difference between speed during MCH (CS+) and speed during OCT (CS–). In mock training, the difference is close to zero, but during training, when MCH is paired with shock, flies walk much faster in MCH (*p<0.05, **p<0.01, ***p<0.001, Mann-Whitney test with Bonferroni correction, comparing training vs. mock training). The effect of shock is not significantly different between OK107 alone and OK107 >mAChR A-RNAi flies (n.s.: p=0.44 for interaction between genotype and training vs. mock training, 2-way ANOVA). n (left to right): 72, 100, 80, 80, 140, 160.
-
Figure 1—figure supplement 1—source data 1
Source data for Figure 1—figure supplement 1.
- https://doi.org/10.7554/eLife.48264.005
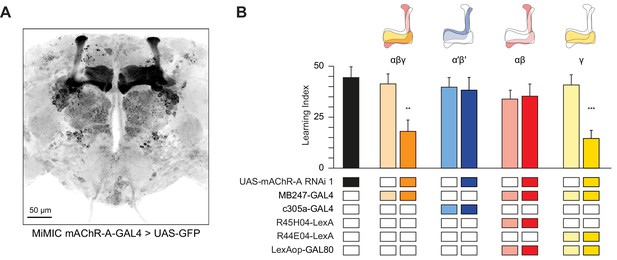
mAChR-A is required for short-term aversive olfactory learning and memory in γ KCs.
(A) Maximum intensity projection of 70 confocal sections (2 µm) through the central brain of a fly carrying MiMIC-mAChR-A-GAL4 and 20xUAS-6xGFP transgenes. MB αβ and γ lobes are clearly observed. No GFP expression is observed in α′β′ lobes. (B) mAChR-A RNAi 1 was targeted to different subpopulations of KCs. Learning scores were reduced compared to controls when mAChR-A RNAi 1 was expressed in αβ and γ KCs or γ KCs alone, but not when mAChR-A RNAi 1 was expressed in αβ or α′β′ KCs. (mean ± SEM, n (left to right): 69, 41, 70, 76, 69, 66, 71, 50, 68, **p<0.01, ***p<0.001, Kruskal-Wallis test with Dunn’s multiple comparisons test). For detailed statistical analysis see Supplementary file 1. The data for the UAS-mAChR-A RNAi 1 control are duplicated from Figure 1.
-
Figure 2—source data 1
Source data for Figure 2.
- https://doi.org/10.7554/eLife.48264.011
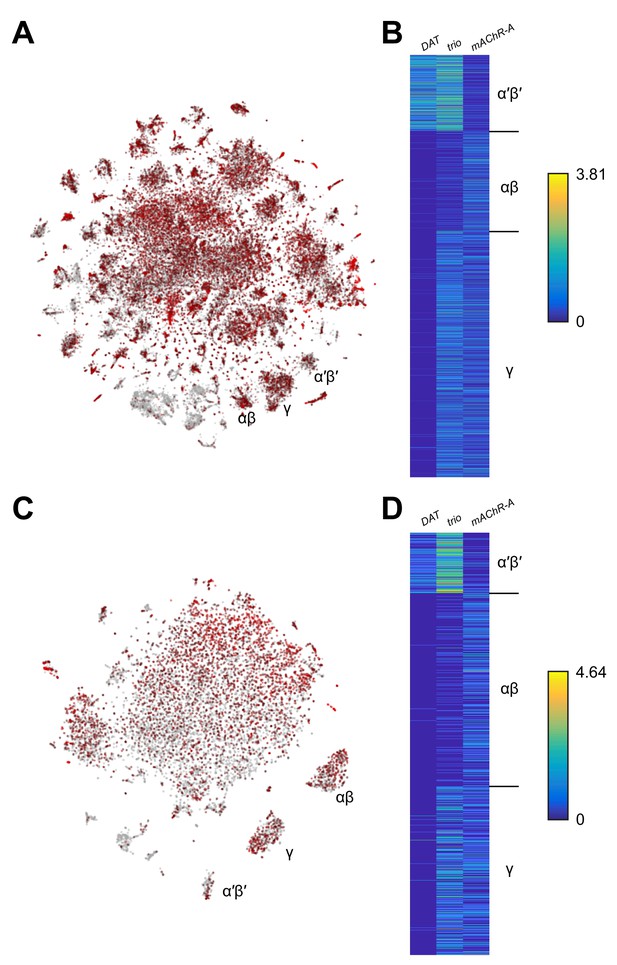
Expression of mAChR-A from single-cell transcriptome profiling.
(A) Data from Davie et al. (2018). 56,902 Drosophila brain cells arranged according to their single-cell transcriptome profiles, along the top two principal components using t-SNE. Red coloring indicates expression of mAChR-A. KC subtype clusters are labeled as identified in Davie et al. (2018). (B) Expression of DAT (marker for α′β′ KCs), trio (marker for α′β′ and γ KCs), and mAChR-A for cells identified as α′β′, αβ and γ KCs in Davie et al. (2018). mAChR-A expression is higher in αβ and γ KCs compared to α′β′ KCs. (C) As in A but with data from Croset et al. (2018) (10,286 Drosophila brain cells). (D) As in B but with data from Croset et al. (2018). Images screenshotted and raw data downloaded from SCope (http://scope.aertslab.org) on 24 June 2018.
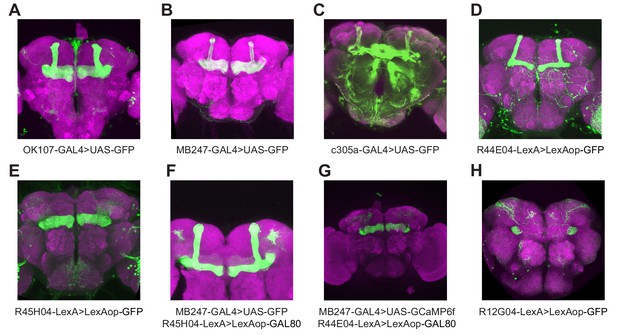
Expression patterns of GAL4 and LexA driver lines used in this study.
GFP expression was driven by the named GAL4 or LexA driver lines and the general neuropil was stained with an antibody to NC82 (magenta). Images are maximum-intensity Z-projections of confocal stacks. Panels A-D, G show only the planes of the mushroom body lobes and peduncle to more clearly show which lobes are labeled. (A) OK107-GAL4 labels all KCs. (B) MB247-GAL4 labels αβ and γ KCs. (C) c305a-GAL4 labels α′β′ KCs. (D) R44E04-LexA labels αβ KCs. (E) R45H04-LexA strongly labels γ KCs. (F) Silencing MB247-GAL4 expression in γ KCs by using R45H04-LexA to drive lexAop-GAL80 in γ KCs results in fairly specific expression in αβ KCs. (G) Silencing MB247-GAL4 expression in αβ KCs by using R44E04 to drive lexAop-GAL80 in αβ KCs results in fairly specific expression in γ KCs. (H) R12G04-GAL4 labels MBON-γ1pedc>α/β, aka MB-MVP2.
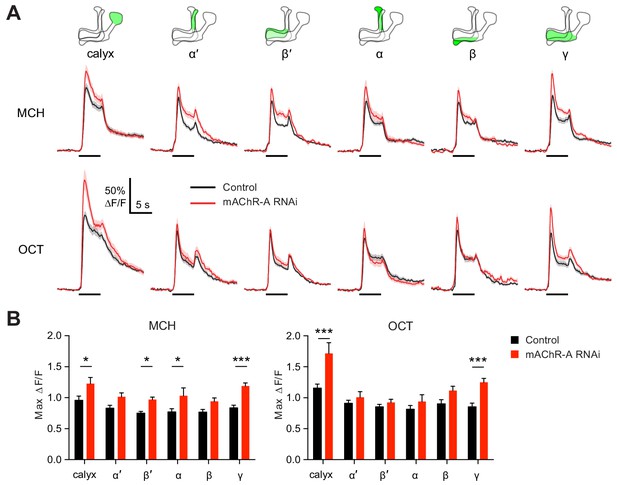
mAChR-A knockdown increases odor responses in γ KCs.
Odor responses to MCH and OCT were measured in control (OK107-GAL4 > GCaMP6f, Dcr-2) and knockdown (OK107-GAL4 > GCaMP6f, Dcr-2, mAChR-A-RNAi 2) flies. (A) ∆F/F of GCaMP6f signal in different areas of the MB in control (black) and knockdown (red) flies, during presentation of odor pulses (horizontal lines). Data are mean (solid line) ± SEM (shaded area). Diagrams illustrate which region of the MB was analyzed. (B) Peak response of the traces presented in A (mean ± SEM). n given as number of hemispheres (number of flies) for control and knockdown flies, respectively: calyx, 23 (13), 17 (10); α and α’, 24 (13), 20 (10); β, β’ and γ, 27 (14), 22 (11). *p<0.05, ***p<0.001, two-way ANOVA with Holm-Sidak multiple comparisons test). For detailed statistical analysis see Supplementary file 1.
-
Figure 3—source data 1
Source data for Figure 3.
- https://doi.org/10.7554/eLife.48264.014
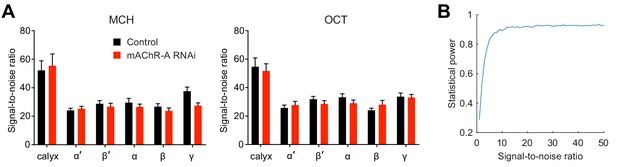
Statistical power is not affected by inter-lobe differences in signal-to-noise ratio (SNR).
(A) SNR in the baseline GCaMP6f signal differs among regions of the mushroom body. SNR was measured as the reciprocal of the standard deviation of ∆F/F during the 2 s immediately preceding odor onset (the period used to calculate baseline fluorescence, or F0). SNR is mean signal divided by standard deviation; here the standard deviation of ∆F/F equals (the standard deviation of F) divided by F0, which is the mean signal during the pre-stimulus period. (B) Statistical power to detect the effect size of the difference in γ lobe odor response between control and mAChR-A knockdown flies (Cohen’s d = 1.3 for OCT, Figure 3B), for different SNRs. Statistical power did not differ for SNRs in the range observed in (A) (SNR = 20–50). Materials and method: We simulated two groups of 20 random samples (n = 20 was the smallest sample size out of the αβ and α′β′ lobes) where the effect size of the difference between the two groups was 1.3. Each sample had a ‘ground truth’ value, from which we sampled three ‘time points’ that were subject to noise with SNR from 1 to 50 (we sampled three time points because the peak of the odor response almost always occurred between 1 and 2 s after odor onset, and our frame rate was ~3 Hz). The maximum of these three time points was taken as the measured ‘peak odor response’. We ran 1000 simulations, ran t-tests on the simulated data, and counted how many gave a p-value<0.0125 (a Holm-Bonferroni correction for the four mushroom body regions that did not consistently show significant differences between control and mAChR-A knockdown flies) – this fraction is the statistical power for detecting a difference in the non-γ lobes with effect size 1.3.
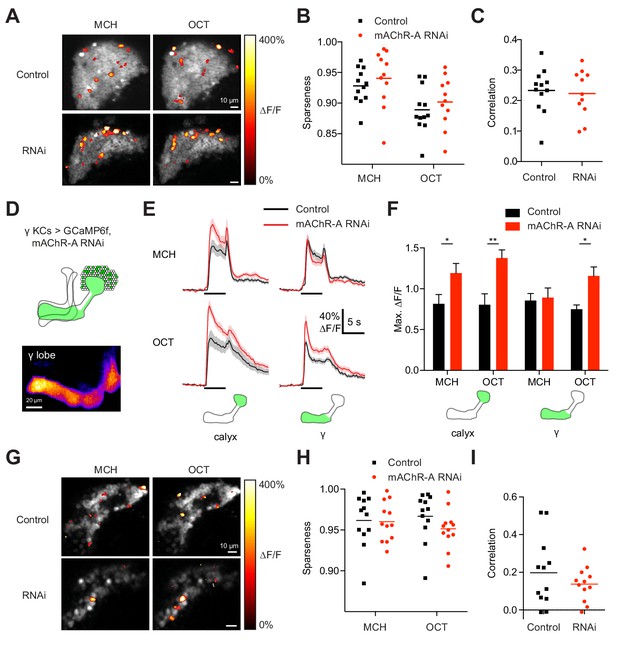
mAChR-A knockdown does not affect KC odor identity coding.
(A) Example activity maps (single optical sections from a z-stack) of KC odor responses to MCH and OCT in control (OK107-GAL4 > GCaMP6f, Dcr-2) and mAChR-A knockdown (OK107-GAL4 > GCaMP6f, Dcr-2, mAChR-A-RNAi 2) flies where all KCs are imaged. False-coloring indicates ∆F/F of the odor response, overlaid on grayscale baseline GCaMP6f signal. Scale bar, 10 µm. For detailed statistical analysis see Supplementary file 1. (B) Sparseness of pan-KC population responses is not affected by mAChR-A knockdown (p=0.38, two-way repeated-measures ANOVA). (C) Correlation between pan-KC population responses to MCH and OCT is not affected by mAChR-A knockdown (p=0.75, t-test). (D) Upper: diagram of γ KCs (green). Lower: False-colored average-intensity Z-projection of the horizontal lobe in a control fly imaged from a dorsal view in panel E (mb247-GAL4 > GCaMP6f, R44E04-LexA > GAL80), averaged over 10 s before the odor stimulus. R44E04-LexA > GAL80 almost completely suppresses β lobe expression. Scale bar, 20 µm. (E) Knocking down mAChR-A only in γ KCs increases γ KC odor responses. Shown here are odor responses in the calyx and γ lobe of control (mb247-GAL4 > GCaMP6f, R44E04-LexA > GAL80) and knockdown (mb247-GAL4 > GCaMP6f, mAChR-A-RNAi 1, R44E04-LexA > GAL80) flies. (F) Peak response of the traces presented in D (mean ± SEM.) n given as number of hemispheres (number of flies): 11 (6) for control, 12 (6) for knockdown. *p<0.05, **p<0.01, 2-way repeated-measures ANOVA with Holm-Sidak multiple comparisons test. (G) Example activity maps (single optical sections from a z-stack) of γ KC odor responses to MCH and OCT in control (mb247-GAL4 > GCaMP6f, R44E04-LexA > GAL80) and knockdown (mb247-GAL4 > GCaMP6f, mAChR-A-RNAi 1, R44E04-LexA > GAL80) flies. Note the gaps in baseline GCaMP6f signal due to lack of αβ and α′β′ KCs labeled. Scale bar, 10 µm (H) Sparseness of γ KC population responses is not affected by mAChR-A knockdown (p=0.76, two-way repeated-measures ANOVA). (I) Correlation between γ KC population responses to MCH and OCT is not affected by mAChR-A knockdown (p=0.32, t-test).
-
Figure 4—source data 1
Source data for Figure 4.
- https://doi.org/10.7554/eLife.48264.016
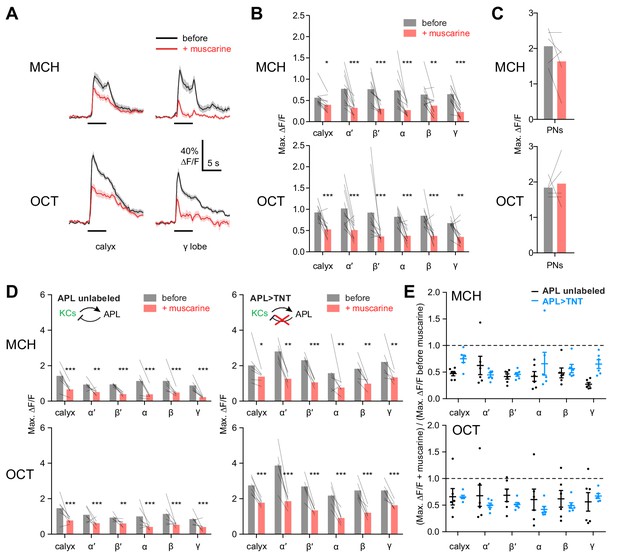
KC odor responses are decreased by muscarine.
(A) Odor responses in the calyx and γ lobe of OK107-GAL4 > GCaMP6f flies, before (black) and after (red) adding 10 µM muscarine in the bath. Data are mean (solid line) ± SEM (shaded area); horizontal lines indicate the odor pulse. Traces for all lobes are shown in Figure 5—figure supplement 1. For detailed statistical analysis see Supplementary file 1. (B) Peak ∆F/F during the odor pulse before and after muscarine. n = 11 hemispheres from 6 flies. *p<0.05, **p<0.01, ***p<0.001 by two-way repeated measures ANOVA with Holm-Sidak multiple comparisons test. (C) Odor responses in PN axons in the calyx are not affected by 10 µM muscarine, in GH146-GAL4 > GCaMP6f flies (p>0.49, two-way repeated measures ANOVA, n = 5 flies). (D) Peak ∆F/F during the odor pulse before and after muscarine in control hemispheres where APL was unlabeled (left, n = 6 hemispheres from 6 flies) and hemispheres where APL expressed tetanus toxin (TNT) (right, n = 6 hemispheres from 5 flies). *p<0.05, **p<0.01, ***p<0.001 by two-way repeated measures ANOVA with Holm-Sidak multiple comparisons test. (E) (Response (peak ∆F/F during the odor pulse) after muscarine) / (response before muscarine), using data from (D). No significant differences were observed (p>0.05, two-way repeated measures ANOVA with Holm-Sidak multiple comparisons test).
-
Figure 5—source data 1
Source data for Figure 5.
- https://doi.org/10.7554/eLife.48264.019
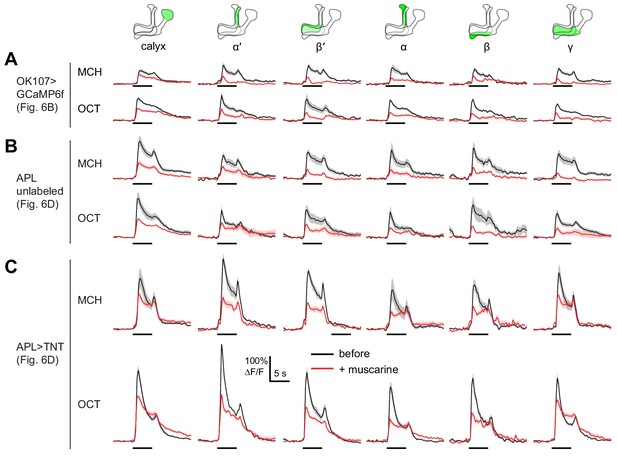
KC odor responses are decreased by muscarine — all traces.
Extended data for Figure 5. Odor responses in OK107-GAL4 > GCaMP6f flies (A), control APL unlabeled hemispheres (B), and APL >TNT hemispheres (C), before (black) and after (red) adding 10 µM muscarine in the bath. Data are mean (solid line) ± SEM (shaded area); diagrams illustrate which region of the MB was analyzed; horizontal lines indicate the odor pulse. These are the traces for the summary data shown in Figure 5B,D.
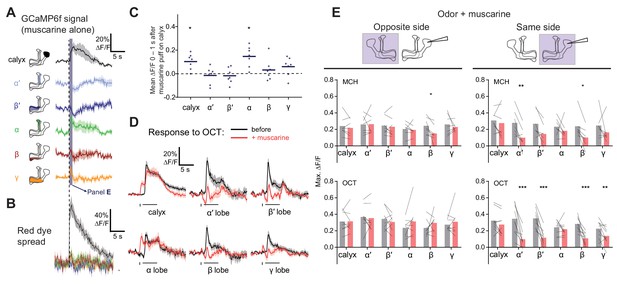
Local muscarine application to the calyx inhibits KC odor responses.
(A) Left: Schematic of MB, showing color scheme for the different regions where responses are quantified. Right: Average ∆F/F GCaMP6f signal in different areas of the MB of OK107 > GCaMP6f flies in response to a 10 ms pulse of 20 mM muscarine on the calyx. Data are mean (solid line) ± SEM (shaded area). Dashed vertical line shows the timing of muscarine application. Shaded bar indicates time window used to quantify responses in panel C. n = 7 hemispheres (5 flies). (B) ∆F/F traces of red dye indicator, showing which MB regions the muscarine spread to. The traces follow the same color scheme and visuals as shown in panel A. (C) Scatter plot showing average ∆F/F of GCaMP6f signal of the different MB regions at time 0–1 s 10 ms pulse of 20 mM muscarine on the calyx, quantified from traces shown in (A). n as in (A). *p<0.05, one-sample t-test (different from 0), Bonferroni correction for multiple comparisons. (D) Average ∆F/F GCaMP6f signal of different areas of the MB during odor pulses of OCT (horizontal bar), before (black) and after (red) muscarine application on the calyx, 1 s before the odor pulse (vertical bar). Data are mean (solid line) ± SEM (shaded area). n: 7 hemispheres (5 flies). See Figure 6—figure supplement 1 for all traces. (E) Line-bar plots showing paired peak ∆F/F GCaMP6f responses of the different MB regions during 5 s odor pulses of MCH or OCT, before (gray) and after (pink) muscarine application to the calyx, in the hemisphere where the muscarine was applied (same side, right) or the opposite (opposite side, left). Muscarine was applied 1 s before the odor pulse. Bars show mean value. n given as number of hemispheres (number of flies): Same side MCH 7 (6), OCT 9 (8), opposite side MCH 7 (5), OCT 8 (5). *p<0.05, **p<0.01, ***p<0.001 by two-way repeated measures ANOVA with Holm-Sidak multiple comparisons test.
-
Figure 6—source data 1
Source data for Figure 6.
- https://doi.org/10.7554/eLife.48264.022
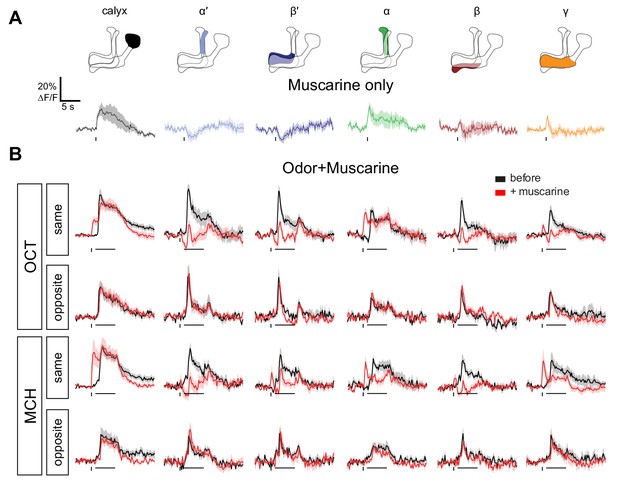
Local muscarine application to the calyx inhibits KC odor responses — all traces.
Average ∆F/F GCaMP6f traces of the different MB regions of OK107 >GCaMP6f flies that only received the muscarine pulse (A) or received an odor pulse (MCH or OCT) before (black) or after (red) 10 ms pulse of 20 mM muscarine (B,C). Panel A is duplicated from Figure 6A; panel B is the traces corresponding to the Figure 6E. Muscarine was applied in the calyx, 1 s before the odor pulse where applicable. Traces are from the same side or the opposite side that muscarine was applied. Data are mean (solid line) ± SEM (shaded area). Horizontal bars indicate odor pulse timing and duration. Vertical bars indicate timing of muscarine pulse. n, by number of hemispheres (number of flies): same side MCH 6 (4), OCT 7 (5), opposite side MCH 5 (3), OCT 5 (3), muscarine alone 7 (5).
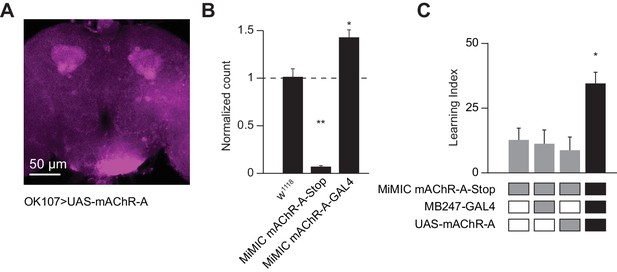
Dendritic function of mAChR-A suffices to rescue learning in mAChR-A mutants.
(A) mAChR-A-FLAG overexpressed in KCs by OK107-GAL4 appears in the calyx but not the lobes of the mushroom body. (B) Flies homozygous for the MiMIC mAChR-A-stop allele (which contains a stop cassette as part of the Minos gene-trap cassette in the 5’UTR) have virtually no mAChR-A mRNA. In contrast, flies with the MiMIC mAChR-A-GAL4 allele do not have reduced mAChR-A mRNA levels, because the stop cassette was replaced with GAL4 (indeed, their mAChR-A levels are slightly higher than the control). (mean ± SEM; n = 4 each with three technical replicates; **p=0.0001; Welch ANOVA test with Dunnett’s T3 multiple comparisons test). For detailed statistical analysis see Supplementary file 1. (C) Homozygous MiMIC mAChR-A-stop flies are defective in olfactory aversive learning, but learning is rescued by driving mAChR-A-FLAG in αβ and γ KCs by mb247-GAL4. n (left to right): 49, 70, 56, 47, *p<0.05, Kruskal-Wallis test with Dunn’s multiple comparisons test). For detailed statistical analysis see Supplementary file 1.
-
Figure 7—source data 1
Source data for Figure 7B.
- https://doi.org/10.7554/eLife.48264.025
-
Figure 7—source data 2
Source data for Figure 7C.
- https://doi.org/10.7554/eLife.48264.026
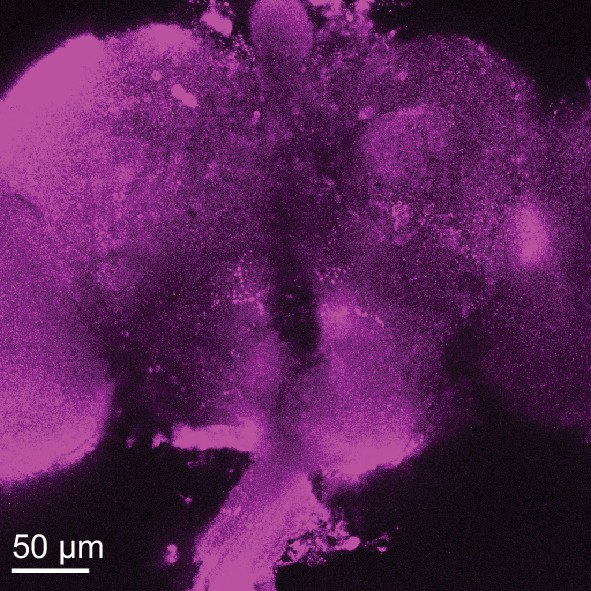
Localization of mb247-GAL4 > mAChR-A-FLAG.
Anti-FLAG immunostaining shows signal only in the calyx in flies expressing mAChR-A-FLAG under the control of mb247-GAL4 in a homozygous MiMIC mAChR-A-stop hypomorphic background.
The signal is less clear than in Figure 7A most likely because OK107-GAL4 is a stronger driver than mb247-GAL4.
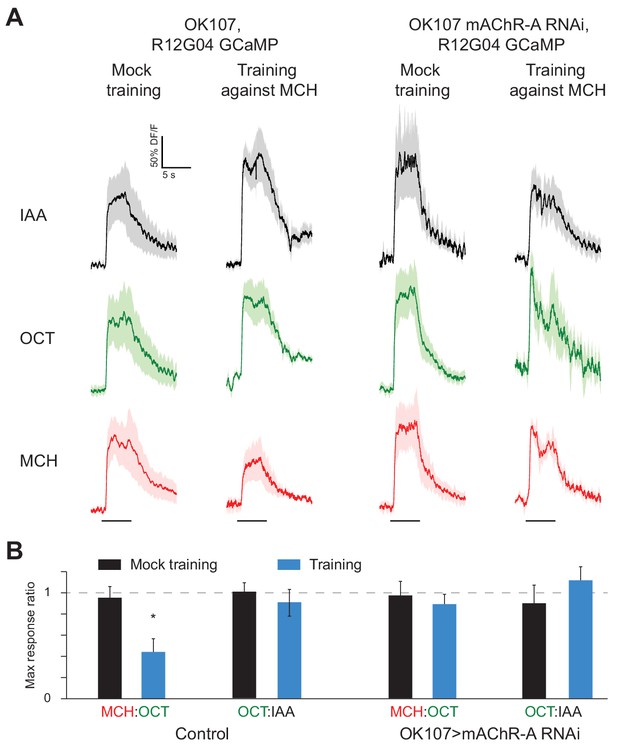
mAChR-A knockdown prevents aversive conditioning from decreasing the response to the trained odor in MB-MVP2.
(A) Odor responses in MB-MVP2 to isoamyl acetate (IAA, not presented during training), OCT (not shocked during training) and MCH (shocked during training), in control (OK107-GAL4, R12G04-LexA > GCaMP6f, mb247-dsRed) and knockdown (OK107-GAL4 > mAChR A-RNAi 1, R12G04-LexA > GCaMP6f, mb247-dsRed) flies, with mock training (no shock) or training against MCH. Traces show mean (solid line) ± SEM (shaded area). (B) MCH:OCT or OCT:IAA ratios of peak ∆F/F values from (A). n = 5. *p<0.05, Mann-Whitney test. Power analysis shows that n = 5 would suffice to detect an effect as strong as the difference between training and mock training in the MCH:OCT ratio, with power 0.9. See Figure 8—figure supplement 1 for absolute ∆F/F values.
-
Figure 8—source data 1
Source data for Figure 8 and Figure 8—figure supplement 1.
- https://doi.org/10.7554/eLife.48264.029
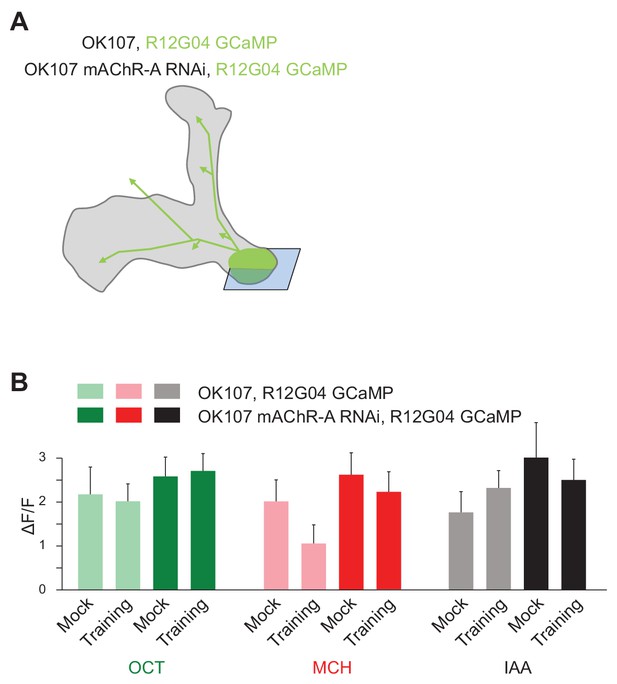
Diagram and additional data for Figure 8 (mAChR-A knockdown prevents learning-associated depression of odor responses in MVP2).
(A) Diagram of genotype: mAChR-A RNAi 1 was expressed in KCs with OK107 (gray), while GCaMP6f was expressed in MB-MVP2 with R12G04-LexA (green). The imaging plane is shown in blue. (B) Absolute ∆F/F values from MB-MVP2 corresponding to the ratios shown in Figure 8B. Odors and genotypes as in Figure 8B. No general depression was observed following RNAi expression. (mean ± SEM; n = 5, p>0.05 for all mock vs. trained comparisons, Mann-Whitney tests). The difference between mock vs. trained for MCH in control flies is not statistically significant because of variability in overall responsiveness to odors between flies. When MCH responses are normalized to OCT responses as in Figure 8B, the difference is statistically significant.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Drosophila melanogaster) | mAChR-A | FLYB: FBgn0000037 | Also known as: mAChR, mAcR-60C, DM1, Acr60C, CG4356 | |
| Genetic reagent (D. melanogaster) | MiMIC mAChR-A-stop | (Venken et al., 2011) PMID 21985007 | BDSC:59216 | mAChR-AMI13848 |
| Genetic reagent (D. melanogaster) | UAS-GCaMP6f (attP40) | (Chen et al., 2013) PMID 23868258 | BDSC:42747 | |
| Genetic reagent (D. melanogaster) | UAS-GCaMP6f (VK00005) | (Chen et al., 2013) PMID 23868258 | BDSC:52869 | |
| Genetic reagent (D. melanogaster) | lexAop-GCaMP6f | (Barnstedt et al., 2016) PMID 26948892 | Gift from S. Waddell | |
| Genetic reagent (D. melanogaster) | UAS-mAChR-A RNAi 1 | Bloomington Drosophila Stock Center | BDSC:27571 | TRiP.JF02725 |
| Genetic reagent (D. melanogaster) | UAS-mAChR-A RNAi 2 | Vienna Drosophila Resource Center | VDRC:101407 | |
| Genetic reagent (D. melanogaster) | UAS-Dcr-2 | Bloomington Drosophila Stock Center | BDSC:24651 | |
| Genetic reagent (D. melanogaster) | lexAop-GAL80 | Bloomington Drosophila Stock Center | BDSC:32216 | |
| Genetic reagent (D. melanogaster) | tub-GAL80ts | (McGuire et al., 2003) PMID 14657498 | BDSC:7108 | |
| Genetic reagent (D. melanogaster) | mb247-dsRed | (Riemensperger et al., 2005) PMID 16271874 | FLYB:FBtp0022384 | |
| Genetic reagent (D. melanogaster) | GH146-GAL4 | (Stocker et al., 1997) PMID 9110257 | BDSC:30026 | |
| Genetic reagent (D. melanogaster) | OK107-GAL4 | (Connolly et al., 1996) PMID 8953046 | BDSC:854 | |
| Genetic reagent (D. melanogaster) | c305a-GAL4 | (Krashes et al., 2007) PMID 17196534 | BDSC:30829 | |
| Genetic reagent (D. melanogaster) | mb247-GAL4 | (Zars et al., 2000) PMID 10784450 | BDSC:50742 | |
| Genetic reagent (D. melanogaster) | R44E04-LexA | (Jenett et al., 2012) PMID 23063364 | BDSC:52736 | Gift from A. Thum |
| Genetic reagent (D. melanogaster) | R45H04-LexA | (Bräcker et al., 2013) PMID 23770186 | FLYB:FBti0155893 | Gift from A. Thum |
| Genetic reagent (D. melanogaster) | R12G04-LexA | (Jenett et al., 2012) PMID 23063364 | BDSC:52448 | |
| Genetic reagent (D. melanogaster) | elav-GAL4 | (Lin and Goodman, 1994) PMID 7917288 | BDSC:458 | |
| Genetic reagent (D. melanogaster) | NP2631-GAL4 | (Lin et al., 2014; Tanaka et al., 2008) PMID 24561998, 18395827 | Kyoto Stock Center 104266 | |
| Genetic reagent (D. melanogaster) | GH146-FLP | (Hong et al., 2009; Lin et al., 2014) PMID 19915565,24561998 | FLYB:FBtp0053491 | |
| Genetic reagent (D. melanogaster) | tub-FRT-GAL80-FRT | (Gordon and Scott, 2009; Lin et al., 2014) PMID 19217375, 24561998 | BDSC:38880 | |
| Genetic reagent (D. melanogaster) | UAS-TNT | (Lin et al., 2014; Sweeney et al., 1995) PMID 24561998, 7857643 | FLYB:FBtp0001264 | |
| Genetic reagent (D. melanogaster) | UAS-mCherry-CAAX | (Kakihara et al., 2008;Lin et al., 2014) PMID 18083504, 24561998 | FLYB:FBtp0041366 | |
| Genetic reagent (D. melanogaster) | mb247-LexA | (Lin et al., 2014; Pitman et al., 2011) PMID 24561998 | FLYB:FBtp0070099 | |
| Genetic reagent (D. melanogaster) | 20xUAS-6xGFP | (Shearin et al., 2014) PMID 24451596 | BDSC:52266 | |
| Genetic reagent (D. melanogaster) | UAS-mCD8-GFP | (Lee et al., 1999) PMID 10457015 | BDSC:5130 | |
| Antibody | nc82 (mouse monoclonal) | Developmental Studies Hybridoma Bank | nc82 | (1:50, supernatant or 1:200, concentrate) |
| Antibody | FLAG (mouse monoclonal M2) | Sigma-Aldrich | F3165 | (1:250) |
| Antibody | Goat anti-mouse secondary Alexa 647 | Abcam | ab150115 | (1:500) |
| Antibody | Goat anti-mouse secondary Alexa 546 | Thermo Fisher | A11018 | (1:1000) |
Additional files
-
Supplementary file 1
Details of statistical analysis.
- https://doi.org/10.7554/eLife.48264.030
-
Supplementary file 2
Detailed genotypes used in this study.
- https://doi.org/10.7554/eLife.48264.031
-
Transparent reporting form
- https://doi.org/10.7554/eLife.48264.032





