Direct glia-to-neuron transdifferentiation gives rise to a pair of male-specific neurons that ensure nimble male mating
Figures
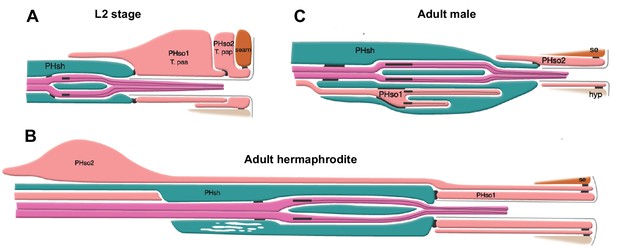
The phasmid sensillum.
Diagram of the phasmid sensillum in either sex at the L2 larval stage (A), in adult hermaphrodites (B) and in adult males (C). The socket-glial cells (PHso1 and PHso2) are coloured in light pink; the sheath glial cells (PHsh) in green; and the ciliated dendrites of the phasmid sensory neurons, in dark pink. The adherens junctions are depicted as black lines between cells. Axonemes and cilia are marked as black bars and black lines inside the dendrite tips. Each phasmid opens to the exterior on the extreme right (posterior), where grey lines mark the cuticle borders of the phasmid pore and fan. Hypodermis (hyp), seam (se). Diagram has been modified from and is used with permission from http://www.wormatlas.org.
© 2010, Wormatlas. Figure 1 is modified with permission from http://www.wormatlas.org/. It is not covered by the CC-BY 4.0 licence and further reproduction of this figure would need permission from the copyright holder.
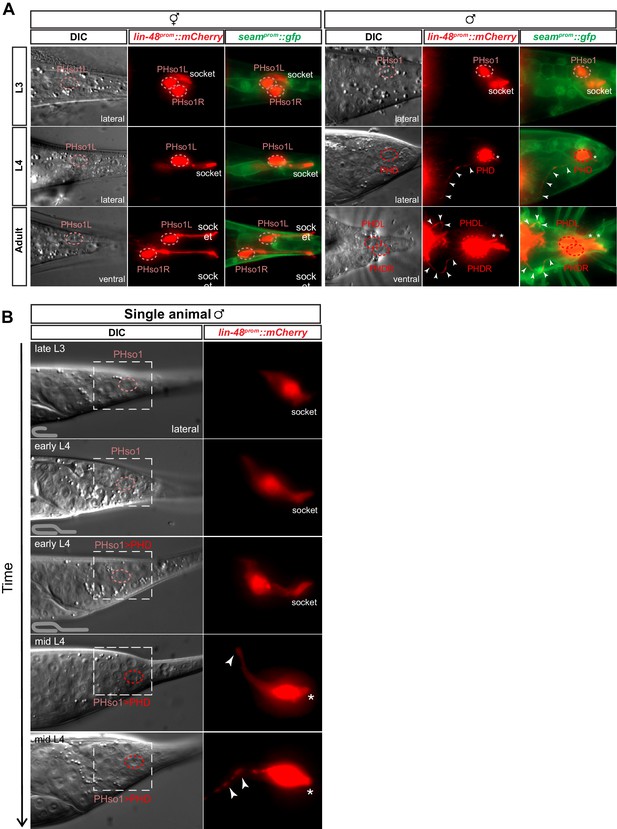
The sex-shared PHso1 cells undergo glia-to-neuron morphological changes in males.
(A) Expression of lin-48prom::mCherry and the seamprom::gfp (wrt-2prom::gfp) reporter transgenes in PHso1 of hermaphrodites (left panel) and males (right panel) at the third (L3) and fourth (L4) larval stages and in adults. The images show the morphological transformation of male PHso1 into the PHD neuron during sexual maturation. Arrowheads label the axonal process extending from the PHD into the pre-anal ganglion. Asterisks indicate the dendritic process of PHD. (B) DIC and fluorescent images of a time-lapse of PHso1-to-PHD remodelling in an individual male (see Materials and methods). The top two time-points show the late L3 stage after the gonad has looped back and early L4 after the gonad has crossed over itself (see cartoon at the bottom left side of the left panel). The subsequent time-points range from early-to-mid-L4, when the vas deferens has joined with the cloaca, to late mid-L4, when tail-tip retraction is almost complete. The dashed boxes on the DIC images indicate the position of the fluorescent images. Arrowheads indicate the nascent axon.
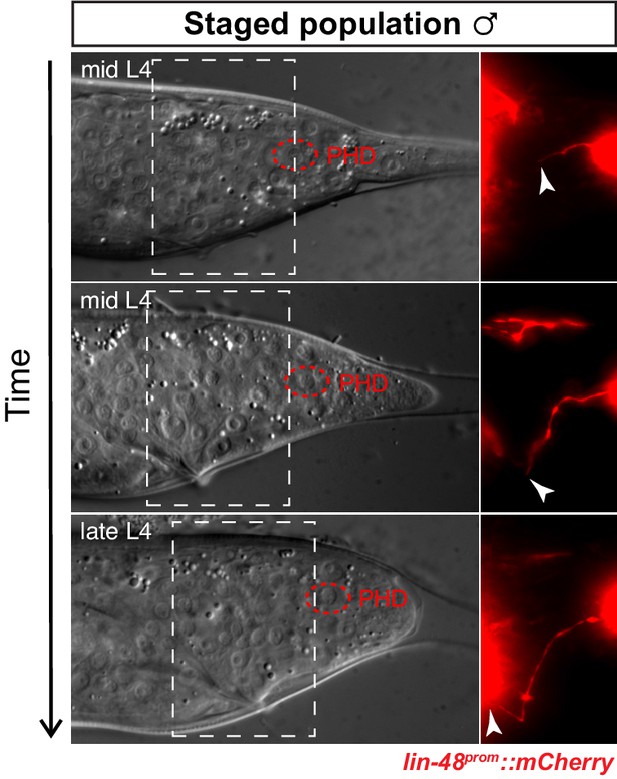
PHD axon outgrowth.
DIC and fluorescent images of L4 male tails from a staged population. The outgrowth of the anterior axon-like process of the PHso1/PHD cell is followed using lin-48::mCherry expression until late L4, when it reaches the pre-anal ganglion. The dashed boxes on the DIC images indicate the position of the fluorescent images. Arrowheads indicate the growing tip of the nascent axon.
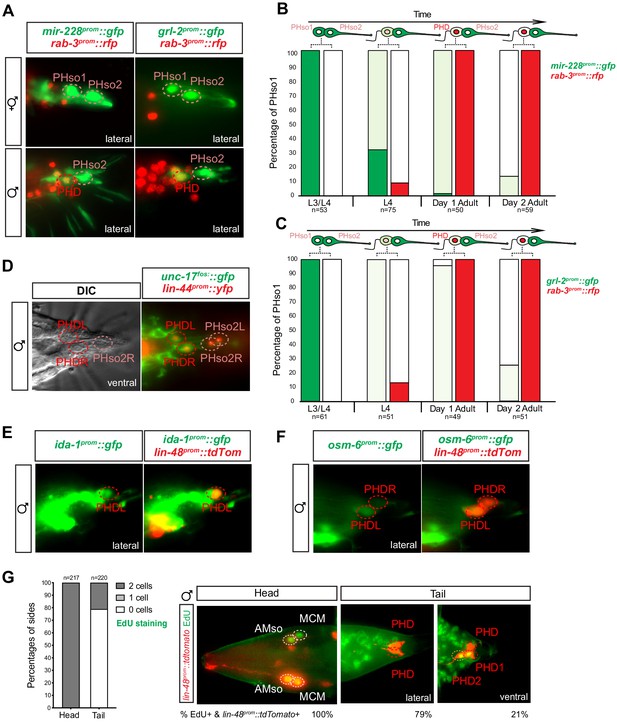
The sex-shared PHso1 cells undergo glia-to-neuron molecular changes in males occasionally accompanied by a division.
(A) Expression of the glial marker reporter transgenes mir-228prom::gfp and grl-2prom::gfp and pan-neuronal marker rab-3prom::rfp in the phasmid socket cells (PHso1 and PHso2) and the PHD neuron in adult male animals. (B) Bar chart showing the percentage of PHso1/PHD cells expressing the pan-glial marker reporter transgene mir-228prom::gfp and the pan-neuronal marker reporter transgene rab-3prom::rfp scored concomitantly in males at different stages of development and at adulthood. Intensity of the mir-228prom::gfp reporter transgene in the PHso1/PHD cells was assessed by eye in comparison with PHso2: dark green indicates PHso1/PHD = PHso2, light green indicates PHso1/PHD < PHso2 and white is non-detectable in PHso1. (C) As B but for the subtype-specific glial marker grl-2prom::gfp scored concomitantly with rab-3prom::rfp. (D) Expression of the acetylcholine vesicle uploader reporter transgene unc-17prom::gfp in the PHD neurons of adult males. The lin-44prom::yfp reporter transgene has been coloured red. (E) Expression of an osm-6prom::gfp reporter transgene in the PHD neurons of adult males, which are co-labelled with a lin-48prom::tdTomato transgene. (F) Expression of an ida-1prom::gfp reporter transgene in the PHD neurons of adult males, which are co-labelled with a lin-48prom::tdTomato transgene. (G) EdU staining to assess PHso1 division. Left panel: quantification of EdU labelling in cells per side in adult males. The AMso division that gives rise to AMso and MCM cells was scored as a positive control. Right panel: representative images of EdU DNA labelling (green) present in the nuclei of the AMso socket cell and MCM neuron (head), and absent in the PHD neuron (tail), unless two cells per side are observed (PHD1 and PHD2). All cells scored were labelled with a lin-48prom::tdTomato transgene.
-
Figure 3—source data 1
Scoring data of glial and neuronal fluorescent reporters and EdU labelling in the PHD neurons of wildtype animals.
- https://cdn.elifesciences.org/articles/48361/elife-48361-fig3-data1-v1.xlsx
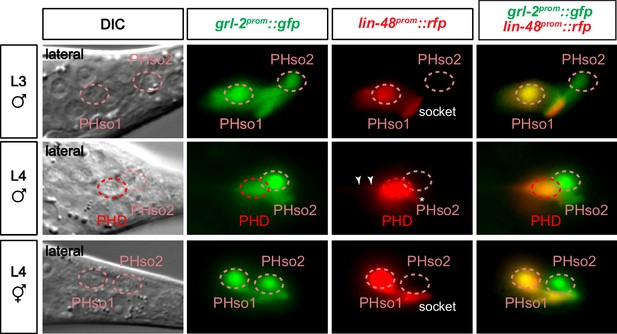
Expression of glial markers is downregulated in the PHso1 of males.
Expression of the glial marker reporter transgene grl-2prom::gfp and the PHso1 marker reporter transgene lin-48prom::tdTomato in the phasmid socket cells (PHso1 and PHso2) and the PHD neuron in L3 males and L4 males and hermaphrodites. Arrowheads label the process extending from the PHD into the pre-anal ganglion. Asterisks indicate the dendritic process of PHD.
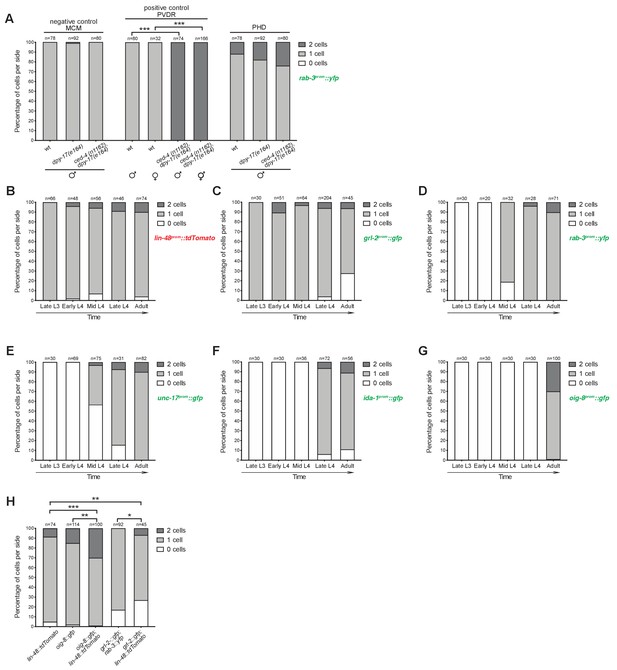
PHso1 divides at low frequency in a background-dependent manner.
(A) Bar chart showing the percentage of PHD neurons per side that express the pan-neuronal marker rab-3prom::yfp transgene (otIs291) in mutant animals for the apoptosis-inducing gene ced-4 (strain: ced-4(n1162) dpy-17(e164)). The lin-48prom::tdTomato reporter transgene was used to identify the PHD neurons. Scorings for control single dpy-17(e164) mutants are included, as are a negative (MCM lineage; Sammut et al., 2015) and positive (PVDR lineage; Sulston and Horvitz, 1977) control for cell death. Cell death in this mutant background is not sex-specific as demonstrated by PVDR scorings in both male and hermaphrodite animals. Male animals from late L4 to adult stages were scored. No statistical difference is observed between any of the groups using Fisher’s exact test. Of note, wildtype animals and ced-4 dpy-17 animals are homozygous siblings obtained from the same cross. Control dpy-17 animals, however, were built from an independent strain. (B-G) Bar charts showing the percentage of cells per side expressing a battery of reporter transgenes before, during and after PHso1 remodelling. Male animals were subdivided into five categories on the basis of gonad migration and tail morphogenesis: late L3 – from the stage at which the gonad begins its posterior migration to the point at which the gonad crosses itself (~28–32 hr); early L4 – from the stage at which the gonad crosses itself to full extension towards the tail (~32–36 hr); mid-L4 – from the start to 50% tail morphogenesis (i.e. complete tail-tip retraction and beginning of ray precursor cell fusion into the tail seam syncytium) (~36–41 hr); late L4 – from 50% to complete tail morphogenesis (i.e. fully developed fan, rays and syncytium;~41–45 hr); adult – from completion of tail-tip retraction to 1-day adult. (B) Quantification of the lin-48prom::tdTomato reporter transgene which was used to label in red PHso1 in larvae and PHD in adults. Graphs C-G represent the number of lin-48prom::tdTomato expressing cells that concomitantly express different green reporter transgenes. (C) Quantification of the glia subtype marker reporter transgene grl-2prom::gfp. Note that grl-2prom::gfp is lost in most animals after the remodelling of PHso1 into the PHD neuron (white bar). (D) Quantification of the pan-neuronal marker reporter transgene rab-3prom::yfp (synaptic vesicle associated Ras GTPase). Note that neuronal reporters are turned on as PHso1 remodels into the PHD neuron. (E) Quantification of the neuron subtype marker reporter transgene unc-17prom::gfp (acetylcholine vesicle uploader). (F) Quantification of the neuron subtype marker reporter transgene ida-1prom::gfp (phogrin orthologue for dense-core vesicle secretion). (G) Quantification of the neuron subtype marker reporter transgene oig-8prom::gfp (sensory-neuron related IG domain containing protein). (H) Quantification of PHD1 and PHD2 expressing the oig-8prom::gfp (drpIs4) and the grl-2prom::gfp (drpIs1) reporter transgenes per side, with and without the lin-48prom::tdTomato (drpIs3) transgene in the background. As grl-2prom::gfp is expressed in PHD and PHso2, to identify PHD unambiguously, rab-3prom::rfp (otIs356) was included in the background and only double positive cells were scored as PHD. The first lin-48prom::tdTomato is repeated from B for comparison. Fisher’s exact test was used to compare all categories between genotypes and only statistically significant differences from the ‘2 cell’ phenotype are indicated (*p≥0.05, **p≥0.01, ***p≥0.001).
-
Figure 3—figure supplement 2—source data 1
Scoring data of PHD background-dependent division.
- https://cdn.elifesciences.org/articles/48361/elife-48361-fig3-figsupp2-data1-v1.xlsx
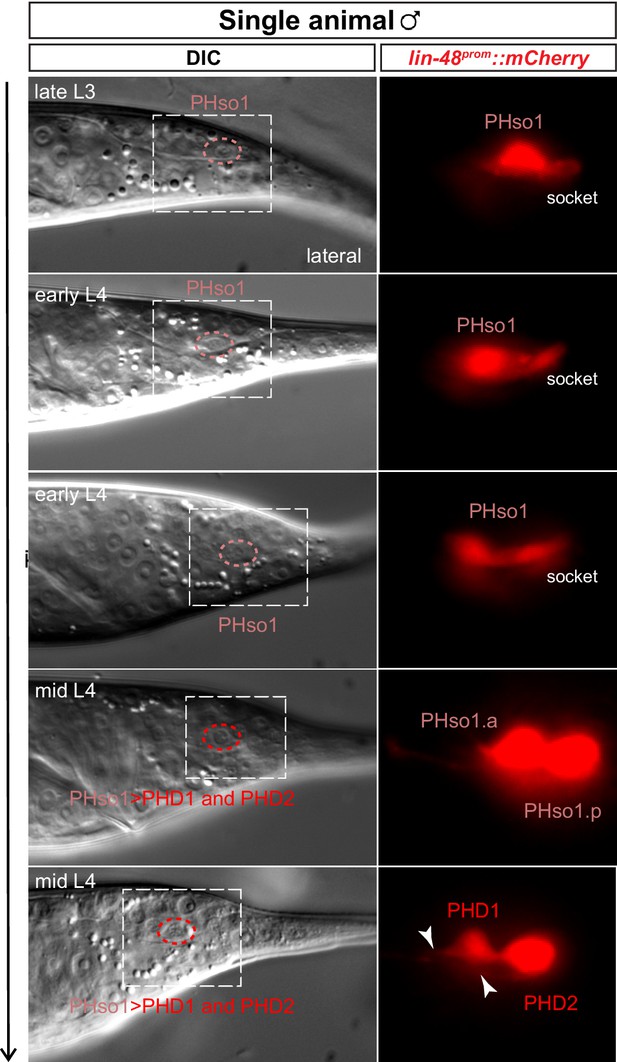
Live division of PHso1 in a single-animal time-lapse.
DIC and fluorescent images of a time-lapse of PHso1-to-PHD remodelling in an individual male (see Materials and methods). The first time-point shows the late-L3 stage after the gonad has looped back. The subsequent time-points, progress through the L4 stage, until mid-late L4, when tail-tip retraction is underway. The dashed boxes on the DIC images indicates the position of the fluorescent images. Arrowheads indicate nascent axon. Images show the right PHso1 with a wildtype morphology and socket structure. In early-to-mid-L4 the PHso1 divides, socket morphology is no longer visible and two cells are observed, which appear to send to a process anteriorly. After the division, DIC images focus on the nucleus of the anterior daughter of the right PHso1. The left PHso1 in this animal does not undergo division and directly assumes the PHD morphology (not shown) as in Figure 2B.
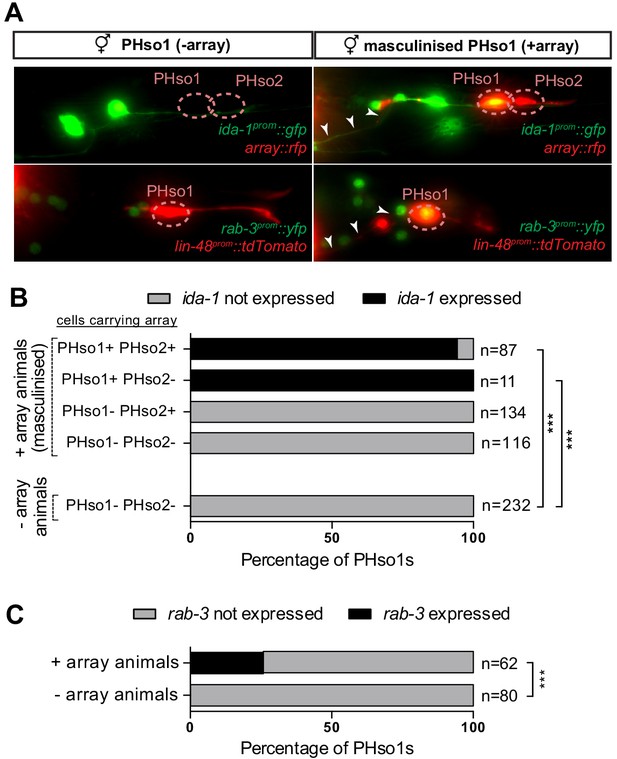
PHso1-to-PHD plasticity is intrinsically regulated.
(A) Expression of the ida-1prom::gfp and rab-3prom::yfp reporter transgenes in adult hermaphrodites carrying the masculinising array grl-2prom::fem-3::mCherry in PHso1 (right panel) and in non-array-carrying hermaphrodites (left panel). (B) Bar chart showing the percentage of PHso1 and PHso2 cells expressing the ida-1prom::gfp reporter transgenes in adult hermaphrodites carrying or not carrying the masculinising grl-2prom::fem-3::mCherry array. Of note, the grl-2 promoter fragment is also expressed in the AMso, excretory pore and excretory duct cells in the head (Hao et al., 2006). Fisher’s exact test was used to compare all categories between genotypes and only statistically significant differences from the non-array carrying animals are indicated (*p≤0.05, **p≤0.01, ***p≤0.001). (C) Bar chart showing the percentage of PHso1 cells expressing the rab-3prom::yfp reporter transgene in adult hermaphrodites carrying or not carrying the masculinising grl-2prom::fem-3::mCherry array. Fisher’s exact test was used to compare all categories between genotypes and only statistically significant differences from non-array carrying animals are indicated (*p≤0.05, **p≤0.01, ***p≤0.001).
-
Figure 4—source data 1
Scoring data of PHso1 masculinised cells in hermaphrodite worms.
- https://cdn.elifesciences.org/articles/48361/elife-48361-fig4-data1-v1.xlsx
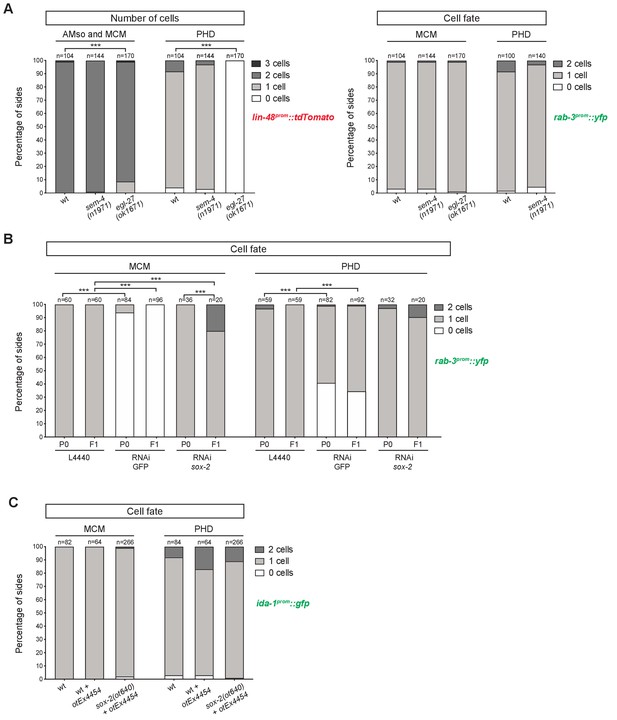
Factors required for Y-to-PDA transdifferentiation are largely dispensable for PHso1-to-PHD and AMso-to-AMso+MCM transdifferentiation.
(A) Bar chart showing the percentage of AMso, MCM, and PHD cells expressing the lin-48prom::tdTomato transgene (drpIs3; left panel) and the pan-neuronal marker rab-3prom::yfp reporter transgene (otIs291; right panel), in sem-4(n1971) putative null and egl-27(ok1670) strong loss-of-function mutant animals. The presence and morphology of cells was assessed by lin-48prom::tdTomato that is expressed, in the head, in the MCM mother (the AMso) and retained in the AMso and MCM daughters after the division (two cells per side). In the tail, lin-48prom::tdTomato is expressed in PHso1 before the cell remodelling and after, in the PHD neuron (one cell per side unless PHD1 and PHD2 are observed). Cells per side were scored in male animals from late L4 to adult stages. Neuronal identity was assessed by rab-3prom::yfp. For the PHD neuron rab-3prom::yfp was scored concomitantly with lin-48prom::tdTomato due to the high number of rab-3prom::yfp-expressing neurons in the tail. Fisher’s exact test was used to compare all categories between genotypes and only statistically significant differences from the wildtype phenotype are indicated (*p≤0.05, **p≤0.01, ***p≤0.001). Of note, 4/7 cells lacking rab-3prom::yfp expression in the tail of sem-4 mutants retained a socket morphology, which is never observed in the cells lacking the reporter in the control strain. This could suggest a block to the initiation of transdifferentiation in PHso1 or that the severe morphological defects of male tails in these mutant animals impair the remodelling process. (B) Bar chart showing the percentage of MCM and PHD neurons expressing rab-3prom::yfp after RNAi-knockdown of sox-2. L4440 empty vector was used as a negative control and GFP RNAi-knockdown as a positive control. Fisher’s exact test was used to compare all categories between genotypes and statistically significant differences from the wildtype phenotype are indicated (*p≤0.05, **p≤0.01, ***p≤0.001). (C) Bar chart showing the percentage of MCM and PHD neurons expressing the ida-1prom::gfp neuron subtype marker in sox-2(ot460) null mutant animals rescued for lethality with a sox-2 fosmid-based extrachromosomal array: (otEx4454[sox-2(fosmid)::mCherry + elt-2prom::DsRed]). A mixed population of of sox-2 mutant (mosaic or non-rescued) and wildtype (mutant-rescued) cells were scored. No statistical difference is observed between any of the groups using Fisher’s exact test.
-
Figure 5—source data 1
Scoring data of glial and neuronal fluorescent reporters in the PHD neurons of sem-4, egl-27, and sox-2 mutant animals.
- https://cdn.elifesciences.org/articles/48361/elife-48361-fig5-data1-v1.xlsx
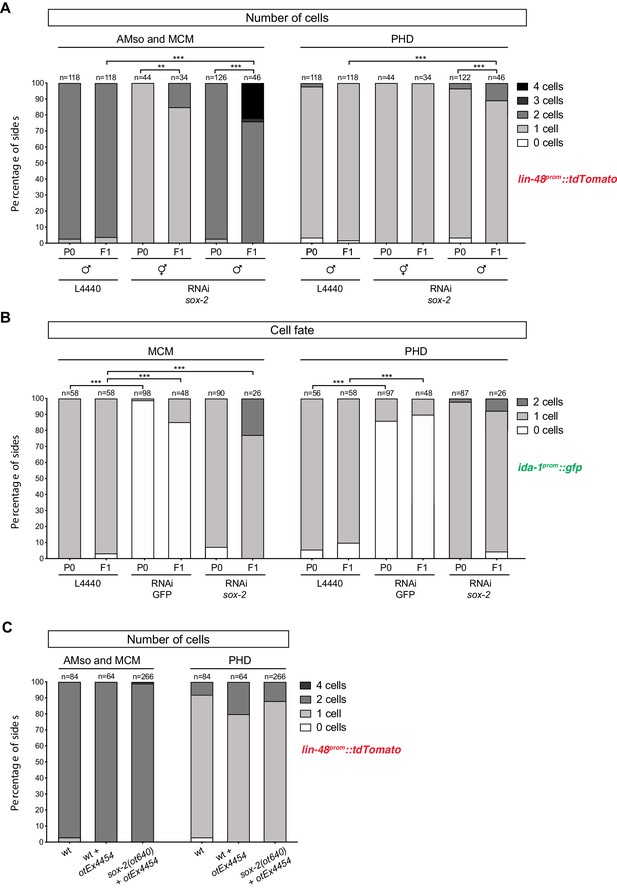
Factors required for Y-to-PDA transdifferentiation are largely dispensable for PHso1-to-PHD and AMso-to-AMso+ MCM transdifferentiation – additional data.
As in Figure 5, cells per side were scored in animals from late-L4 to adult stages. Fisher’s exact test was used to compare all categories between genotypes and only statistically significant differences from the wildtype phenotype are indicated (*p≥0.05, **p≥0.01, ***p≥0.001). (A) Bar chart showing the percentage of AMso, MCM, and PHD cells expressing the lin-48prom::tdTomato reporter transgene (drpIs3) after RNAi-knockdown of sox-2 in male animals. In hermaphrodites AMso was scored, as it never divides to produce the MCM neuron. Of note, scored lin-48::tdTomato-positive cells correspond to the same animals scored in Figure 5B and 5 SB. (B) Bar chart showing the percentage of MCM and PHD neurons expressing the ida-1prom::gfp reporter transgene (inIs179) after RNAi knock-down of sox-2. Ectopic AMso cells are observed in sox-2 depleted F1 animals but not in sox-2 depleted P0 animals. (C) Bar chart showing the percentage of AMso, MCM and PHD cells expressing the lin-48prom::tdTomato reporter transgene (drpIs3) in sox-2(ot460) null mutant animals rescued from lethality with a sox-2 fosmid-based extrachromosomal array (otEx4454[sox-2(fosmid)::mCherry + elt-2p::DsRed]). A mixed population of of sox-2 mutant (mosaic or non-rescued) and ‘wildtype’ (mutant-rescued) cells were scored.
-
Figure 5—figure supplement 1—source data 1
Additional scoring data of glial and neuronal fluorescent reporters in the PHD neurons of sem-4, egl-27, and sox-2 mutant animals.
- https://cdn.elifesciences.org/articles/48361/elife-48361-fig5-figsupp1-data1-v1.xlsx
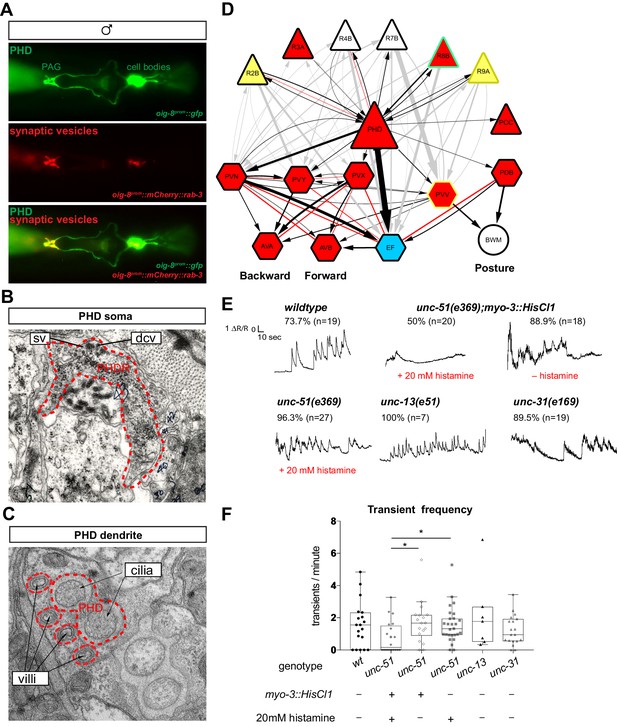
The PHDs are putative proprioceptive neurons of male-specific copulation circuits.
(A) Expression of the oig-8prom::mCherry::rab-3 and oig-8prom::gfp reporter in the PHD neurons of adult males. The synapses made by the PHDs in the pre-anal ganglion (PAG) can be observed (ventral view). (B) Electron micrographs of the soma of a PHD neuron of an adult male. sv, synaptic vesicle; dcv, dense-core vesicles; PHDR, right PHD neuron. (C) As B for a PHD dendrite. (D) Diagram depicting the connectivity of the PHD neurons with their main pre-synaptic inputs (ray neurons) and post-synaptic targets. The connections between the ray neurons (RnA/B) and their post-synaptic targets independently of PHD are indicated in grey. Arrows and red lines indicate chemical and electrical synaptic connections, respectively. The thickness of the arrows is proportional to the anatomical strength of their connections (# serial sections). Neurons are colour-coded according to their neurotransmitter: red, cholinergic; yellow, glutamatergic; dark yellow, dopaminergic; blue, GABAergic; green, serotonergic; white, orphan. Note that some neurons (R8B, R9A, PVV) express more than one neurotransmitter. (E) Example traces showing PHD activity as normalised GCaMP/RFP fluorescence ratio in restrained animals. Traces are shown for a wildtype male, an unc-51(e359) mutant with and without a histamine-inducible silencing transgene in muscle (myo-3prom::HisCl1), a mutant in synaptic transmission (unc-13), and a mutant in dense-core vesicle exocytosis (unc-31, CADPS/CAPS). The proportion of traces where calcium peaks were identified is indicated for each genotype and treatment. n = number of neurons imaged. (F) Plots of frequency values of calcium transients per neuron. Dots represent individual neurons imaged. Tukey box-and-whisker plots indicate the interquartile ranges and median. *p<0.05; One-way ANOVA with multiple comparisons. Two groups were compared: unc-51 genotypes and treatments and another group including wt, unc-13, and unc-31. Only statistically significant comparisons are indicated.
-
Figure 6—source data 1
Value of calcium transients elicited by PHD in different genetic backgrounds.
- https://cdn.elifesciences.org/articles/48361/elife-48361-fig6-data1-v1.xlsx
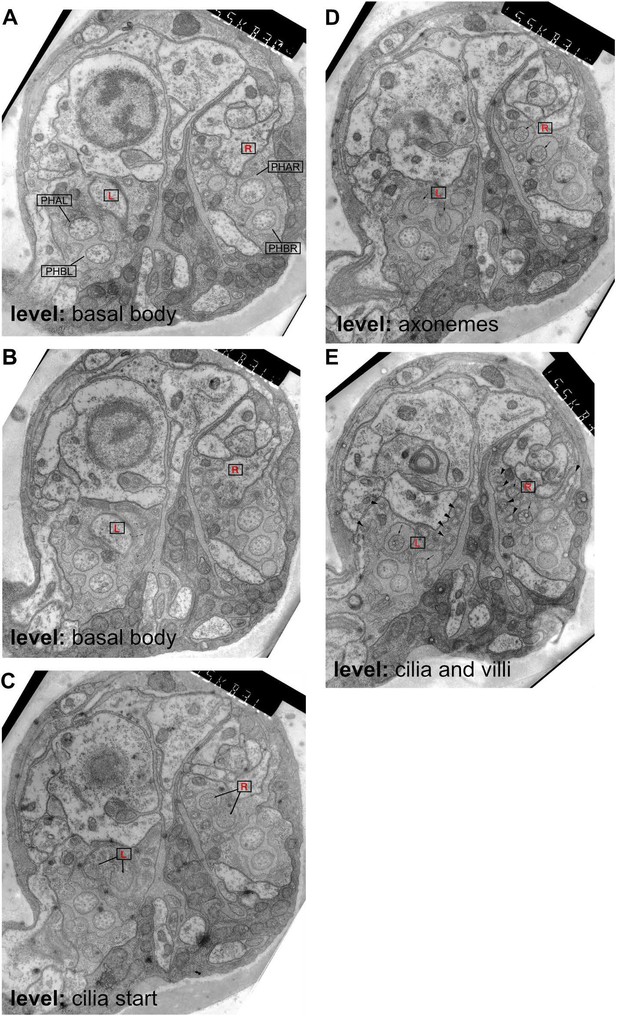
Ultrastructure of the male phasmid sensilla.
Transmission electron microscopy images from serial transverse thin sections of an adult male tail, from anterior (A) to posterior (E). Dorsal is up. The level at which the sections are taken is labelled on the figure according to the cilia structures visible in PHD (compare to Figure 6). In all sections, the left and right PHD neurons are indicated with a boxed red R(ight) or L(eft). In A, the other phasmid neurons are also labelled and their cilia can be observed in each panel. (A) The basal body of the cilia is visible in PHDL and PHDR. (B) The PHD cilia are first visible in this section. (C) The PHD axonemes are visible in this section. (D) The finger-like villi are labelled in this section with arrowheads (but are also visible in all sections), and can be traced in serial sections to the basal body of PHD. Their appearance is identical to the more numerous villi that extend from the AFD cilium in the amphid (Ward et al., 1975).
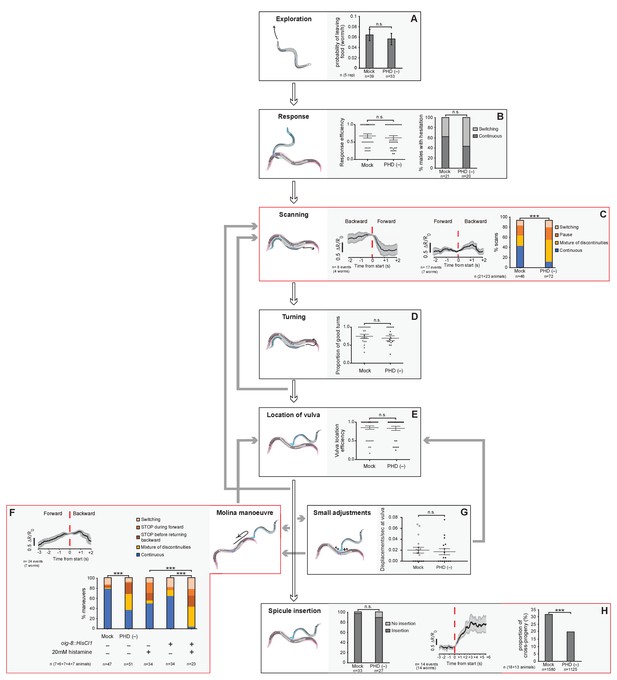
The PHD neurons are required for coordinated backward locomotion and effective intromission during mating.
Diagram depicting the steps of reproductive behaviours controlled by the male tail circuits. The steps affected by PHD ablation are highlighted in red. Intact (mock) and ablated males carried either an oig-8prom::gfp or an unc-17prom::gfp transgene to identify the PHD neurons for ablation. In F, males carrying an oig-8prom::HisCl1 transgene were used to silence PHD neurons acutely. White arrows indicate the transitions between the mating sequence. Grey arrows indicate the corrective transitions that males perform when they fail to attain the subsequent goal. The corrective transitions that occur upon failure of spicule insertion attempts (between location of vulva and spicule insertion) are always preceded by a displacement from the vulva (not depicted). Behavioural analysis in intact and PHD-ablated males are shown for each step. Calcium imaging in PHD neurons is shown for steps C, F, and H. The black trace shows PHD activity as normalised GCaMP/RFP fluorescence ratio changes averaged for several events and phase locked (red dotted line) to the switch in the direction of locomotion (C and F) or to the start of spicule insertion (H). The grey shadow shows S.E.M. (A) Male exploratory behaviour measured as PL values (probability of leaving food per worm per hour). n, number of males tested. Maximum likelihood statistical analysis was used to compare PL values. n.s., no statistically significant difference, p≥0.05; error bars, S.E.M. (B) Response efficiency, measured as 1/number of contacts with a mate before responding; and hesitation, measured as a switch in direction of locomotion during response. (C) Scanning locomotion during vulva search. Categories: switching (change in the direction of locomotion from backward to forward); pause (stopping during backward scanning); mixture (scans with switches and pauses); continuous (uninterrupted backward movement along the mate’s body). p<0.001 (χ2 test of continuous and discontinuous scans); n = number of events. (D) Turning, measured as proportion of good turns per male. (E) Vulva location efficiency measured as 1/number of vulva encounters before stopping. (F) Proportion of continuous and discontinuous Molina manoeuvres. Categories: switching (a brief change in the direction of movement either during forward locomotion away from the vulva or during backward locomotion returning to the vulva); STOP during forward (stopping during forward movement away from the vulva and then continuing in the same direction); STOP before returning backward (stopping at the transition between forward movement away from the vulva and returning backwards to the vulva); mixture (manoeuvres that displayed more than one of the discontinuities described above); continuous (smooth movement forward away from the vulva and return backwards to the vulva without stopping or switching in between). p<0.001 (χ2 test of continuous and discontinuous manoeuvres); n = number of events. (G) Number of displacements away from the vulva per unit of time spent at vulva. (H) Left bar chart, proportion of males able to insert their spicules; n.s., no statistically significant difference, p≥0.05 (χ2 test); n = animals tested. Right bar chart, sperm transfer efficiency measured as percentage of cross-progeny; p<0.001 (χ2 test); n = total progeny. For B, D, E, and G, bar and dots represent mean and individual animal values, respectively; error bars, S.E.M. n.s., no statistically significant difference, p≥0.05 (Mann-Whitney U test). Worm cartoons were modified with permission from original drawings by Rene García.
© 2018, Genetics Society of America. Figure 7 contains modified versions of the male and hermaphrodite worm cartoons from Barr et al., 2018 used with permission. They are not covered by the CC-BY 4.0 licence and further reproduction of this panel would need permission from the copyright holder.
-
Figure 7—source data 1
Analysis of mate searching, fertility, and male-mating behaviours and measurements of PHD neuronal activity during mating in wildtype and PHD-ablated animals.
- https://cdn.elifesciences.org/articles/48361/elife-48361-fig7-data1-v1.xlsx
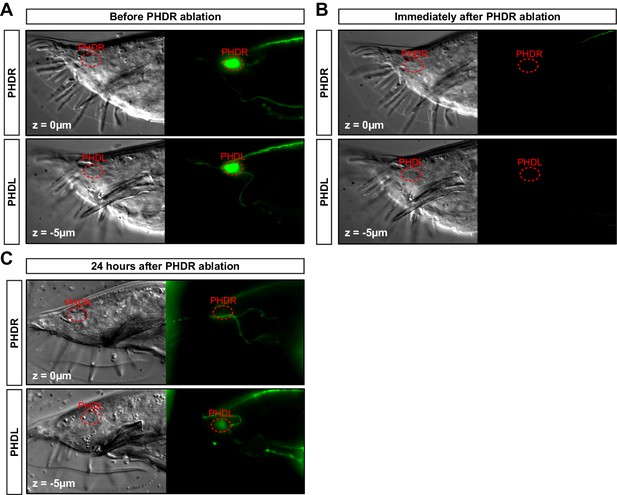
PHD neuron ablation control.
(A) DIC and fluorescent images of a one-day adult male expressing the unc-17prom::gfp transgene in both PHDR and PHDL neurons in the tail, before the laser ablation. (B) Same animal after the PHDR neuron has been ablated. The laser photobleaches all GFP signal in the region. (C) Same animal after 24 hr after the ablation. The GFP signal from the transgene reappears in the non-ablated PHDL and in other structures of the male tail but is specifically lost in the ablated PHDR. Of note, the GFP signal coming from the PHDL is dimmer due to the older age of the animal (2 day adult).
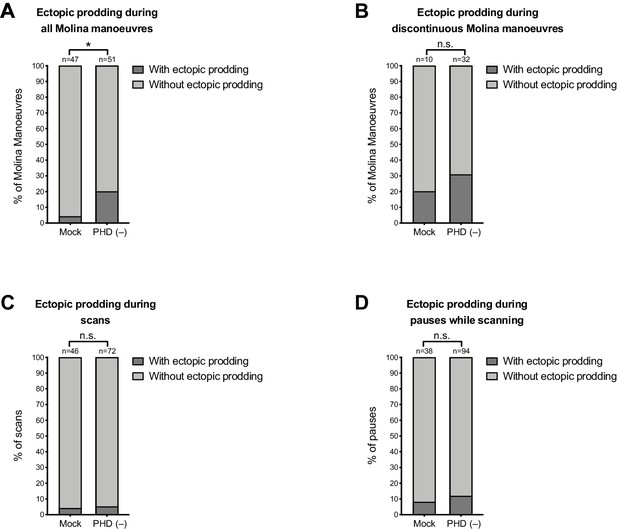
Some ectopic prodding occurs during discontinuous Molina manoeuvres and during pauses while scanning.
Plots show proportion of events with ectopic prodding in mock and PHD-ablated males. n = events: (A) all Molina manoeuvres; (B) discontinuous Molina manoeuvres; (C) all scans; (D) all pauses while scanning. For A, p<0.05 (χ2 test). For B, C, and D, p≥0.05 (χ2 test).
-
Figure 7—figure supplement 2—source data 1
Scoring of ectopic prodding events during mating in wildtype and PHD-ablated animals.
- https://cdn.elifesciences.org/articles/48361/elife-48361-fig7-figsupp2-data1-v1.xlsx
Videos
Imaging of neuronal activity in PHD neurons with GCaMP6f (left channel) and RFP (right channel) in restrained animals: wildtype male.
Animals are expressing an oig-8prom::GCaMP6f::sl2::rfp transgene. Videos play at 100 fps (recorded at 20 fps).
Imaging of neuronal activity in PHD neurons with GCaMP6f (left channel) and RFP (right channel) in restrained animals: unc-51(e359) male expressing a histamine-inducible silencing transgene in muscle (myo-3prom::HisCl1::mCherry) and treated with 20mM histamine.
Animals are expressing an oig-8prom::GCaMP6f::sl2::rfp transgene. Videos play at 100 fps (recorded at 20 fps).
Imaging of neuronal activity in PHD neurons with GCaMP6f (left channel) and RFP (right channel) in restrained animals: unc-51(e359) male expressing a histamine-inducible silencing transgene in muscle (myo-3prom::HisCl1::mCherry) and treated with 20mM histamine.
Animals are expressing an oig-8prom::GCaMP6f::sl2::rfp transgene. Videos play at 100 fps (recorded at 20 fps).
Imaging of neuronal activity in PHD neurons with GCaMP6f (left channel) and RFP (right channel) in restrained animals: unc-51(e359) male treated with 20mM histamine.
Animals are expressing an oig-8prom::GCaMP6f::sl2::rfp transgene. Videos play at 100 fps (recorded at 20 fps).
Males performing Molina manoeuvres during mating: wildtype male performing a Molina manoeuvre during mating with a paralysed unc-51(e359) hermaphrodite.
Videos are played at 40 fps (sped up x2).
Males performing Molina manoeuvres during mating: wildtype male performing a Molina manoeuvre during mating with a wildtype hermaphrodite.
Videos are played at 40 fps (sped up x2).
Males performing Molina manoeuvres during mating: PHD-ablated male performing a defective, discontinuous Molina manoeuvre.
Videos are played at 40 fps (sped up x2).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (C. elegans) | C. elegans: Strain N2 | Caenorhabditis Genetics Center | WormBase: N2 | |
| Genetic reagent (C. elegans) | AW827 | Dr Alison Woollard laboratory | him-5(e1490), heIs63 [wrt-2p::GFP::PH + wrt-2p::GFP::H2B + lin-48p::mCherry] V | |
| Genetic reagent (C. elegans) | CHL56 | This study | drpIs3 [lin-48p::tdTomato] I; him-5(e1490) V | |
| Genetic reagent (C. elegans) | BAR37 | This study | nsIs198 [mir-228p::GFP + lin-15(+)]; otIs356 [rab-3p::2xNLS::TagRFP], him-5(e1490) V | |
| Genetic reagent (C. elegans) | CHL32 | This study | dpy-5(e907) I; otIs356 [rab-3p::2xNLS::TagRFP], him-5 (e1490) V; drpIs1 [grl-2p::GFP, dpy-5(+)] X | |
| Genetic reagent (C. elegans) | OH13083 | Pereira et al. Elife. 2015; Caenorhabditis Genetics Center | him-5(e1490) V; otIs576 [unc-17fos::GFP + lin-44::YFP] | |
| Genetic reagent (C. elegans) | CHL36 | This study | drpIs3 [lin-48p::tdTomato] I; inIs179 [ida-1p::GFP] II; him-8(e1489) IV | |
| genetic reagent (C. elegans) | CHL57 | This study | drpIs3 [lin-48::tdTomato] I; mnIs17 [osm-6p::GFP + unc-36(+)]; him-5(e1490) V | |
| Genetic reagent (C. elegans) | CHL58 | This study | drpIs3 [lin-48::tdTomato] I; otIs291 [rab-3p::2xNLS::YFP + rol-6(su1006)], him-5(e1490) V | |
| Genetic reagent (C. elegans) | CHL59 | This study | drpIs3 [lin-48p::tdTomato] I; dpy-17(e164) III; otIs291 [rab-3p::2xNLS::YFP + rol-6(su1006)], him-5(e1490) V | |
| Genetic reagent (C. elegans) | CHL60 | This study | drpIs3 [lin-48p::tdTomato] I; ced-4(n1162), dpy-17(e164) III; otIs291 [rab-3p::2xNLS::YFP + rol-6(su1006)], him-5(e1490) V | |
| Genetic reagent (C. elegans) | CHL61 | This study | drpIs3 [lin-48p::tdTomato] I; him-5(e1490) V; drpIs1 [grl-2p::GFP, dpy-5(+)] X | |
| Genetic reagent (C. elegans) | CHL63 | This study | drpIs3 [lin-48p::tdTomato] I; him-5(e1490) V; vsIs48 [unc-17p::GFP] X | |
| Genetic reagent (C. elegans) | CHL64 | This study | drpIs3 [lin-48p::tdTomato] I; inIs179[ida-1p::GFP] II; him-5(e1490) V | |
| Genetic reagent (C. elegans) | CHL67 | This study | drpIs3 [lin-48p::tdTomato] I; drpIs4 [oig-8p::GFP + pha-1(+)]; him-5(e1490) V | |
| Genetic reagent (C. elegans) | CHL65 | This study | drpIs4 [oig-8p::GFP + pha-1(+)]; him-5(e1490) V | |
| Genetic reagent (C. elegans) | BAR77 | This study | oleEx24 [grl-2(1 kb)::fem-3::SL2::mCherry (8 ng)+ elt-2p::GFP (40 ng)]; inIs179 [ida-1p::GFP] II; him-8(e1489) IV | |
| Genetic reagent (C. elegans) | CHL62 | This study | oleEx18 [grl-2(1 kb)::fem-3::SL2:: mCherry(20 ng) + elt-2p::GFP (40 ng)]; drpIs3 [lin-48p::tdTomato] I; otIs291 [rab-3p::2xNLS::YFP + rol-6(su1006)], him-5(e1490) V | |
| Genetic reagent (C. elegans) | CHL68 | This study | sem-4(n19719), drpIs3 [lin-48p::tdTomato] I; otIs291 [rab-3p::2xNLS::YFP + rol-6(su1006)], him-5(e1490) V | |
| Genetic reagent (C. elegans) | CHL69 | This study | drpIs3 [lin-48p::tdTomato] I; egl-27(ok1670) II; otIs291 [rab-3p::2xNLS::YFP + rol-6(su1006)], him-5(e1490) V | |
| Genetic reagent (C. elegans) | CHL70 | This study | drpIs3 [lin-48p::tdTomato] I; otIs291 [rab-3p::2xNLS::YFP + rol-6(su1006)], him-5(e1490) V; nre-1(hd20), lin-15B(hd126) X | |
| Genetic reagent (C. elegans) | CHL71 | This study | drpIs3 [lin-48p::tdTomato] I; inIs179 [ida-1p::GFP] II; him-8(e1489) IV Generated during sox-2 mutant crosses. Distinct background to CHL36. | |
| Genetic reagent (C. elegans) | CHL72 | This study | drpIs3 [lin-48p::tdTomato] I; inIs179 [ida-1p::GFP] II; him-8(e1489) IV; otEx4454 [sox-2(fosmid)::mCherry + elt-2p::DsRed] | |
| Genetic reagent (C. elegans) | CHL73 | This study | drpIs3 [lin-48p::tdTomato] I; inIs179 [ida-1p::GFP] II; him-8(e1489) IV; sox-2(ot640[unc-119(+)]) X; otEx4454 [sox-2(fosmid)::mCherry + elt-2p::DsRed] | |
| Genetic reagent (C. elegans) | CHL74 | This study | drpIs3 [lin-48p::tdTomato] I; inIs179 [ida-1p::GFP] II; him-8(e1489) IV; nre-1(hd20), lin-15B(hd126) X | |
| Genetic reagent (C. elegans) | EM1370 | Dr Scott Emmons laboratory | bxEx271 [oig-8p::GFP + oig-8p::mCherry::rab-3]; him-5(e1490) V | |
| Genetic reagent (C. elegans) | EM1371 | Dr Scott Emmons laboratory | bxEx272 [oig-8p::GFP + oig-8p::mCherry::rab-3]; him-5(e1490) V | |
| Genetic reagent (C. elegans) | BAR90 | This study | him-5(e1490) V; oleEx27 [oig-8p::GCaMP6f::SL2::mRFP(30 ng/µl) + cc::GFP(30 ng/µl)] | |
| Genetic reagent (C. elegans) | BAR115 | This study | unc-51(e369), him-5(e1490) V; oleEx27 [oig-8p::GCaMP6f:: SL2::mRFP(30 ng/µl) + cc::GFP(30 ng/µl)] | |
| Genetic reagent (C. elegans) | BAR115 | This study | unc-51(e369), him-5(e1490) V; oleEx27 [oig-8p::GCaMP6f:: SL2::mRFP(30 ng/µl) + cc::GFP(30 ng/µl)]; oleEx38 [myo-3::HisCL1::SL2::Cherry(20 ng/µl)] | |
| Genetic reagent (C. elegans) | BAR95 | This study | unc-13 (e51) I; him-5 (e1490) V; oleEx27 [oig-8p::GCaMP6f:: SL2::mRFP(30 ng/µl) + cc::GFP(30 ng/µl)] | |
| Genetic reagent (C. elegans) | BAR106 | This study | unc-31(e169) IV; him-5(e1490) V; oleEx27 [oig-8p::GCaMP6f:: SL2::mRFP(30 ng/µl) + cc::GFP(30 ng/µl)] | |
| Genetic reagent (C. elegans) | EM1251 | Dr Scott W. Emmons laboratory | bxEx201[oig-8p::GFP + pha-1(+)]; him-5(e1490) V - line#1 | |
| Genetic reagent (C. elegans) | EM1253 | Dr Scott W. Emmons laboratory | bxEx201[oig-8p::GFP + pha-1(+)]; him-5(e1490) V - line#3 | |
| Genetic reagent (C. elegans) | BAR94 | This study | him-5(e1490) V; lite-1(ce314) X; oleEx30[oig-8p::GCaMP6f::SL2::mRFP(50 ng/µl)] | |
| Genetic reagent (C. elegans) | CB369 | Caenorhabditis Genetics Center | unc-51(e369) V | |
| Genetic reagent (C. elegans) | BAR160 | This study | him-5(e1490) V; oleEx53 [pAB6(oig-8p::HisCl1::SL2::RFP) (30 ng/uL)+unc-122p::GFP(25 ng/uL)] | |
Strain, strain background (E. coli) | Strain OP50 | Caenorhabditis Genetics Center | OP50 | |
| Strain, strain background (E. coli) | Strain MG1693 (Thy-) | E. coli stock centre | MG1693 | Strain for EdU staining experiments |
| Strain, strain background (E. coli) | Strain: HT115 (DE3) | Caenorhabditis Genetics Center | HT115 | |
| Tecombinant DNA reagent | Plasmid: pPD95.75 | Addgene | #1494 | See: DNA constructs and transgenic strains Used to create oig-8prom::GFP and for PCR fusions |
| Recombinant DNA reagent | Plasmid: oig-8prom::gfp | This study | See: DNA constructs and transgenic strains 964 bp upstream of the oig-8 ATG | |
| Recombinant DNA reagent | Plasmid: pkd- 2prom::mCherry::rab-3 | Dr María I. Lázaro-Peña | See: DNA constructs and transgenic strains Replaced promoter by oig-8prom | |
| Recombinant DNA reagent | Plasmid: oig- 8prom::mCherry::rab-3 | This study | See: DNA constructs and transgenic strains Created from pkd-2prom::mCherry::rab-3 | |
| Recombinant DNA reagent | Plasmid pLR306 | Dr Luis Rene García Laboratory | See: DNA constructs and transgenic strains Gateway plasmid used to amplify GCaMP6f::sl2::rfp | |
| Recombinant DNA reagent | Plasmid: oig- 8prom::GCaMP6f::SL2::RFP | This study | See: DNA constructs and transgenic strains Created by PCR fusion (see primers) | |
| Recombinant DNA reagent | Plasmid: pNP471 | Dr Cori Bargmann Laboratory | See: DNA constructs and transgenic strains Used to amplify HisCl1::sl2::mCherry | |
| Recombinant DNA reagent | Plasmid: oig- 8prom::HisCl1::sl2::rfp | This study | See: DNA constructs and transgenic strains Created by Gibson | |
| Recombinant DNA reagent | Plasmid: myo- 3prom::HisCl1::sl2::mCherry | This study | See: DNA constructs and transgenic strains Created by PCR fusion (see primers) | |
| Recombinant DNA reagent | Plasmid: lin- 48prom::tdTomato | Dr Mike Boxem Laboratory | See: DNA constructs and transgenic strains 6.8 kb upstream of lin-48 ATG | |
| Recombinant DNA reagent | Plasmid pPD129.36 (L4440) | Dr Andrew Fire Laboratory, Addgene | #1654 | Control plasmid for RNAi experiments |
| Recombinant DNA reagent | Plasmid: sox-2 RNAi clone | Source BioScience | Silence endogenous sox-2 gene | |
| Sequence-based reagent | primer_oig-8 fusion_F | This study | PCR fusion primers | See: DNA constructs and transgenic strains gggagtgacctatgcaaacc |
| Sequence-based reagent | primer_oig-8 fusion_R | This study | PCR fusion primers | See: DNA constructs and transgenic strains CGACGTGATGAGTCGACCAT tgttttacctgaaatctttt |
| Sequence-based reagent | primer_myo-3 fusion_F | This study | PCR fusion primers | See: DNA constructs and transgenic strains cgtgccatagttttacattcc |
| Sequence-based reagent | primer_myo-3 fusion_R | This study | PCR fusion primers | See: DNA constructs and transgenic strains gctagttgggctttgcatGCttctagatggatctagtggtc |
| Sequence-based reagent | primer_ced-4 (n1162)_F | This study | PCR genotyping primers | See: C. elegans strains gatgctctgcgaaatcgaatgc |
| Sequence-based reagent | primer_ced-4 (n1162)_R | This study | PCR genotyping primers | See: C. elegans strains tcacctaaatcacacatctcgtcg |
| Sequence-based reagent | primer_dpy-17 (e164)_F | This study | PCR genotyping primers | See: C. elegans strains aggaggaagcccaatcaacc |
| Sequence-based reagent | primer_dpy-17 (e164)_R | This study | PCR genotyping primers | See: C. elegans strains cagttggtccttcttctccagc |
| Sequence-based reagent | primer_sem-4 (n1971)_F | This study | PCR genotyping primers | See: C. elegans strains aaggtgatgcgatgatgtctcc |
| Sequence-based reagent | primer_sem-4 (n1971)_R | This study | PCR genotyping primers | See: C. elegans strains taatgatcggcttgggtgtgg |
| Sequence-based reagent | primer_egl-27 (ok1679)_F ext | This study | PCR genotyping primers | See: C. elegans strains tcatcgtttccagtctcttcagg |
| Sequence-based reagent | primer_egl-27 (ok1679)_R ext | This study | PCR genotyping primers | See: C. elegans strains cgctggttatcaaatgacgcc |
| Sequence-based reagent | primer_egl-27 (ok1679)_F int | This study | PCR genotyping primers | See: C. elegans strains agacaccagaagctacgaaacc |
| Sequence-based reagent | primer_egl-27 (ok1679)_R int | This study | PCR genotyping primers | See: C. elegans strains gtttgcatcacggtcttcacg |
| Sequence-based reagent | primer_nre-1(hd20) lin-15B(hd126)_F | This study | PCR genotyping primers | See: C. elegans strains catgagagctgcgctgaggc |
| Sequence-based reagent | primer_nre-1(hd20) lin-15B(hd126)_R | This study | PCR genotyping primers | See: C. elegans strains ggttcgggctcgcggtagtc |
| Chemical compound, drug | Paraformaldehyde | Thermo Fisher Scientific | #28908 | Used at 4% |
| Chemical compound, drug | β-mercapto-ethanol | Sigma Aldrich | M6250 | Used at 5% |
| Chemical compound, drug | Collagenase type IV | Sigma Aldrich | C-5138 | Used at 1 mg/ml |
| Chemical compound, drug | Vectashield antifade | Vector Laboratories | H-1900 | |
| Chemical compound, drug | Isopropyl-β-D- thiogalactopyranoside (IPTG) | Generon | GEN-S-02122 | Used at 0.6 mM in plates for RNAi knock-down |
| Chemical compound, drug | Histamine | Sigma | H7125 | Used at 20 mM in plates for behavioural assays |
| Chemical compound, drug | Wormglu | GluStitch, Delta, Canada | ||
| Antibody | Anti-RFP pAb (Rabbit polyclonal antibody) | MBL International | PM005 | (1:500) |
| Antibody | Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 555 (Donkey polyclonal antibody) | Molecular probes | A-31572 | (1:200) |
| Commercial assay or kit | Click-IT EdU Alexa Fluor 594 | Invitrogen | C10339 | |
| Software, algorithm | ImageJ | ImageJ (http://imagej.nih.gov/ij/) | RRID:SCR_003070 | |
| Software, algorithm | GraphPad Prism | GraphPad Prism (graphpad.com) | RRID:SCR_002798 | |
| Software, algorithm | Affinity Designer | https://affinity.serif.com/ en-us/designer/ | RRID:SCR_016952 |
Additional files
-
Supplementary file 1
PHD connectivity.
A table indicating the quantification of pre-synaptic inputs and post-synaptic outputs of the PHD neurons as derived from the electron microscopy serial reconstruction (see Materials and methods). The number of synapses and number of sections over which synapses are observed for each input/output neuron are indicated.
- https://cdn.elifesciences.org/articles/48361/elife-48361-supp1-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/48361/elife-48361-transrepform-v1.docx





