Ultrastructural heterogeneity of layer 4 excitatory synaptic boutons in the adult human temporal lobe neocortex
Figures
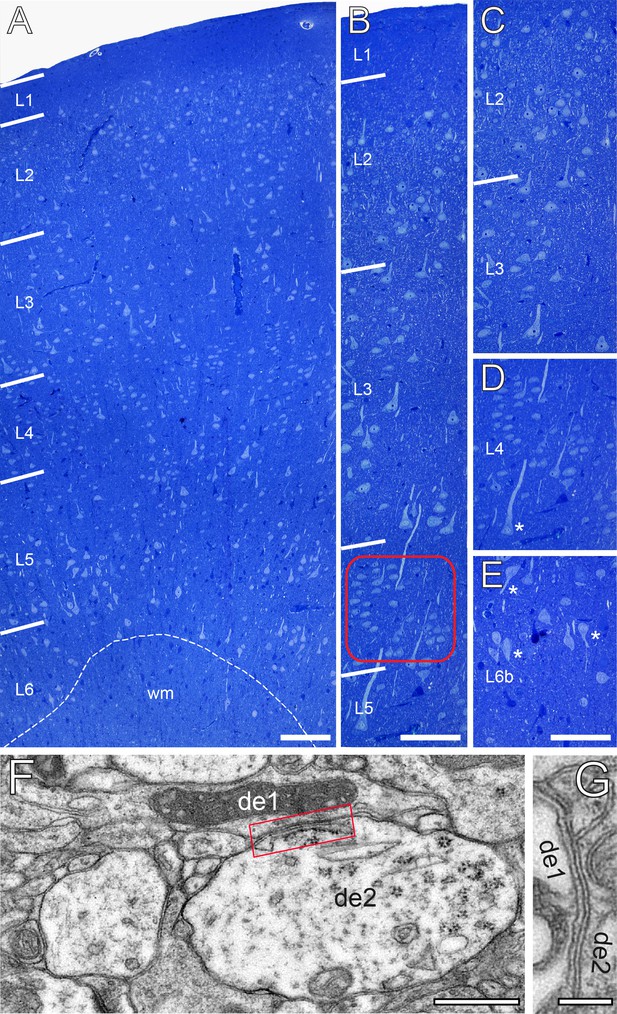
The cytoarchitecture of the human TLN.
(A) Low-power light micrograph of a methylene blue stained semithin section through the gyrus temporalis inferior (baso-lateral region) in humans, showing the typical six-layered organization of a granular neocortex. Dashed line indicates the gray/white matter border. (B) High-power light micrograph through the same section shown in (A). Note the different density of neurons in L2 and L3. Red frame: region of interest in L4. (C) High-power light micrograph through layers 2 and 3. (D) Cluster-like arrangement of neurons in L4. Note the large L5 pyramidal cell marked by asterisk. (E) Neuronal organization of L6b containing several inverted pyramidal cells marked by asterisks. Scale bars (A–E) 100 µm. (F, G) Two examples of GAP-junctions between dendrites (de1, de2) one in (F, red frame) and the second in (G) at higher magnification. Scale bars 0.5 µm in F 0.2 µm in G.
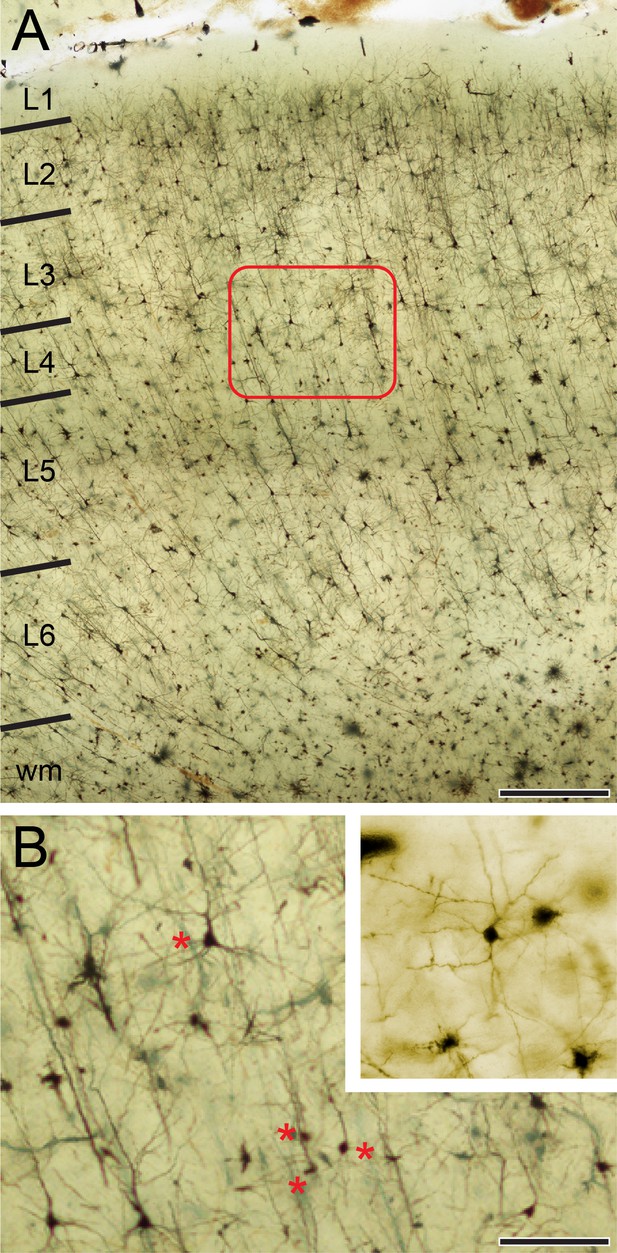
Golgi-Cox impregnation of the TLN.
(A) Overview through the six layers of the TLN. Scale bar 500 µm. (B) High-power micrograph of the framed area in A. Asterisks indicate spiny stellate cells and star pyramidal neurons characteristic of L4. Scale bar 100 µm. Inset: Representative example of a spiny stellate cell in L4.
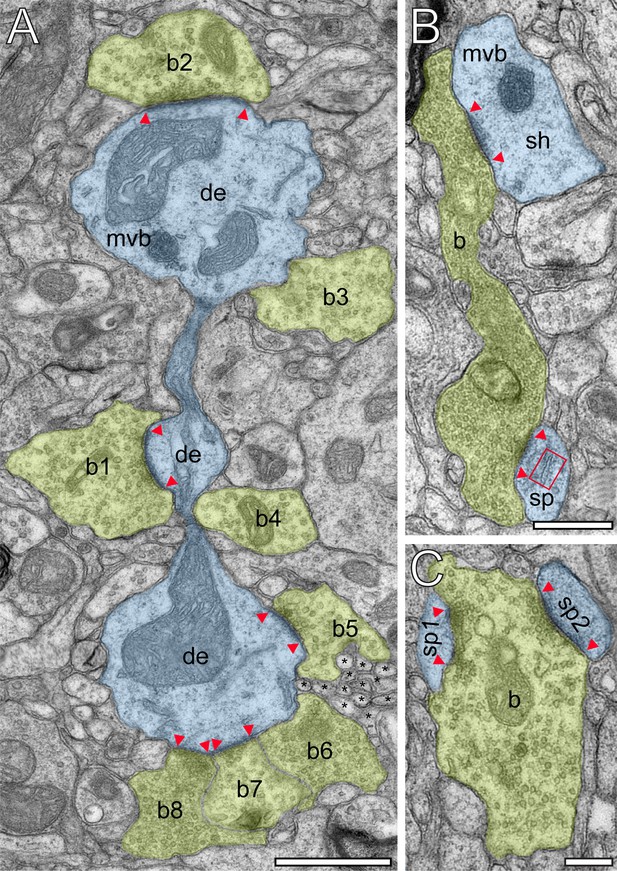
Innervation patterns of excitatory L4 SBs and target specificity.
(A) Dense innervation of endterminal SBs (b1-b8; transparent yellow) terminating on a dendrite (de; transparent blue) of ~5 µm length. Note the presence of a cluster of unmyelinated axons (asterisks) isolating SB b5 from SB b6. Scale bar 1 µm. (B) En passant SB (b) innervating a dendritic shaft (sh) and a spine (sp) identified by a spine apparatus (framed area). (C) Large endterminal SB (b) innervating two small spines (sp1, sp2). Scale bars in (B) and (C) 0.5 µm. Note the presence of multivesicular bodies (mvb) in (A) and (B). In all images, the AZs are marked by red arrowheads. In (B) and (C) same color code as in (A).
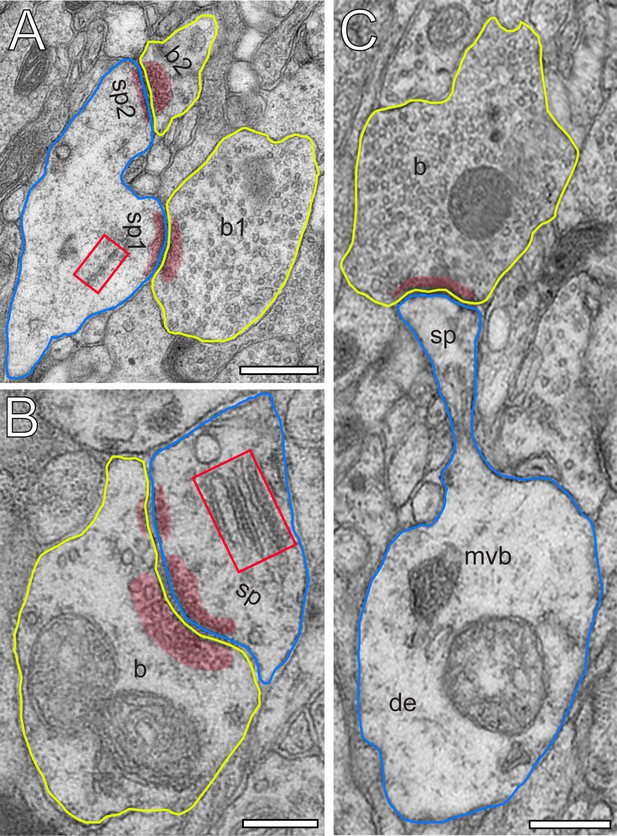
Excitatory L4 SBs innervate different types of spines.
(A) Two SBs (b1, b2; yellow contour) terminating on two stubby spines (sp1, sp2; blue contour). Scale bar 0.5 µm. (B) SB (b; yellow contour) on the head of a large mushroom spine (sp; blue contour). Note the presence of two mitochondria occupying a large fraction of the total volume of the SB. Here, SVs were found in closer proximity to the PreAZ. Scale bar 0.25 µm. (C) SB (b; yellow contour) terminating on the head of an elongated spine (sp; blue contour) emerging from a relatively large dendritic segment (de; blue contour) containing a multivesicular body (mvb). Scale bar 0.5 µm. Note the presence of a spine apparatus (framed area) in (A) and (B). All AZs are highlighted in transparent red.
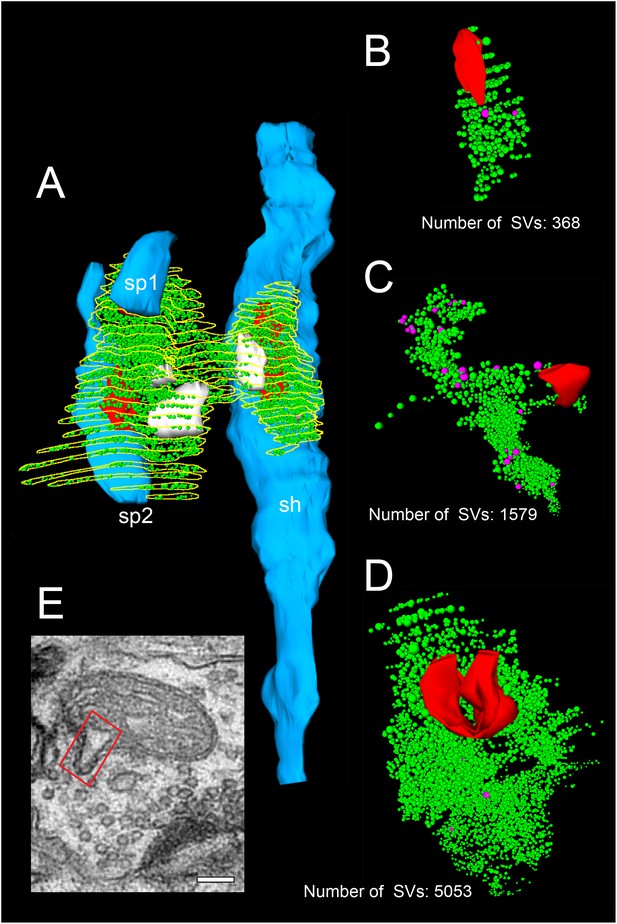
3D-volume reconstructions showing the high variability in the size of the total pool of SVs.
(A) 3D-volume reconstruction of a SB (yellow outline) innervating a dendritic shaft (sh; blue) and two spines (sp1, sp2; blue). Note the relatively large total pool of SVs (green dots); the size of the PreAZs (red) and the mitochondria (white) always associated to the pool of the SVs. (B–D) 3D-volume reconstructions of individual total pools of SVs (green dots) at either non-perforated (B, C) or perforated (D) AZs. Large DCVs (magenta) were frequently observed. (E) Example of a mitochondrial-derived vesicle (framed area) in the process of separation from the mitochondrion. Scale bar 0.1 µm.
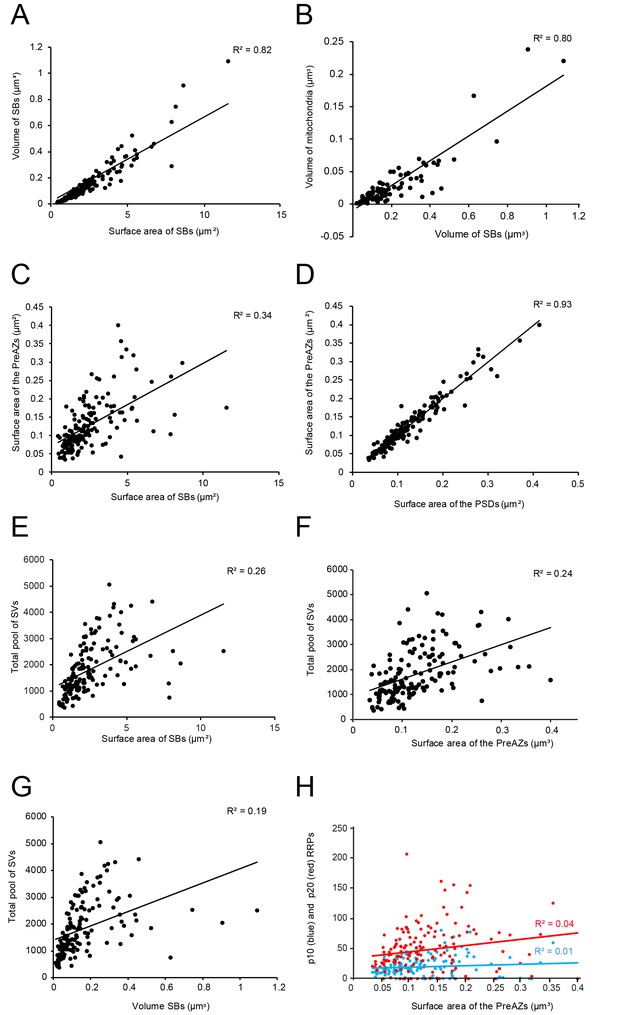
Correlations between various structural parameters of L4 SBs.
(A) The surface area vs. the volume of the SBs. (B) The volume of SBs vs. the volume of mitochondria. (C) The surface area of SBs vs. the surface area of the PreAZs. (D) The surface area of PSDs vs. the surface area of the PreAZs. (E) The surface area of SBs vs. the total pool of SVs. (F) The surface area of the PreAZs vs. the total pool of SVs. (G) The volume of SBs vs. the total pool of SVs. (H) The surface area of the PreAZs vs. p10 nm (blue dots) and p20 nm (red dots) RRPs, respectively. *Data points were fitted by linear regression and the R2 is given for each correlation.
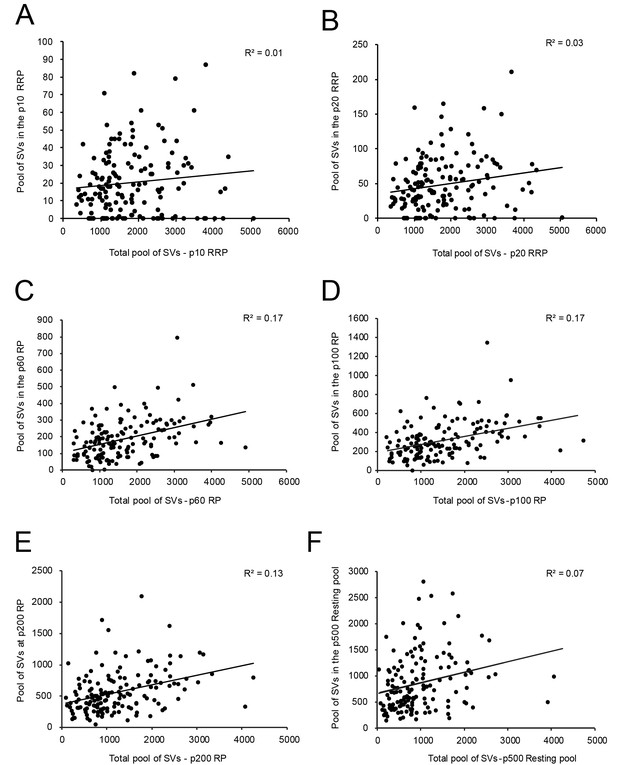
Correlations between various structural parameters of L4 SBs and SVs.
(A) The total pool of SVs vs. the RRP at p10 nm. (B) The total pool of SVs vs. the RRP at p20 nm. (C) The total pool of SVs vs. the RP at p60 nm. (D) The total pool of SVs vs. the RP at p100 nm. (E) The total pool of SVs vs. the RP at p200 nm. (F) The total pool of SVs vs. the resting pool at p500 nm.
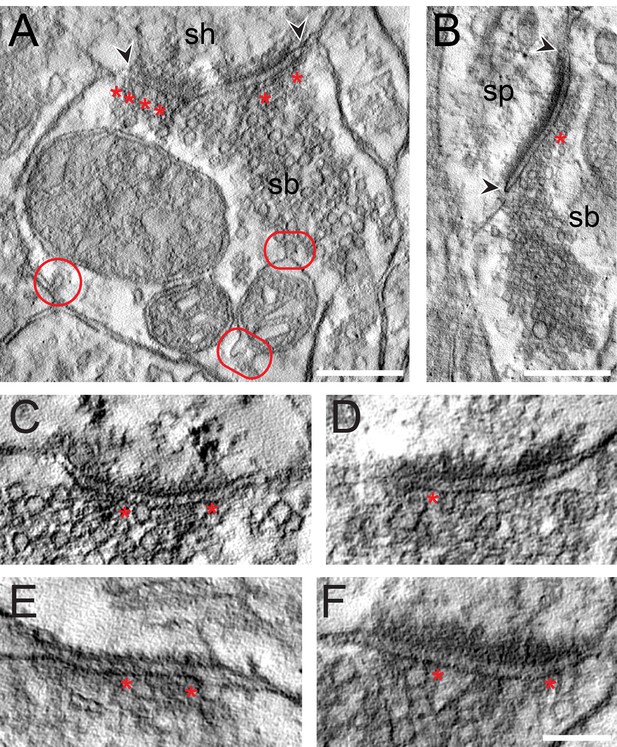
EM tomography of L4 SBs in the TLN.
(A) Example of a SB (sb) terminating on a dendritic shaft (sh) with a large, perforated AZ marked by arrowheads. Asterisks indicate ‘docked’ SVs. The two frames point to the MDVs and the circle to the clathrin-coated pit. Scale bar 0.25 µm. (B) Axo-spinous synapse (sb, sp) with a large non-perforated AZ (arrowheads) and an omega-shaped body (asterisk). Scale bar 0.25 µm. (C, D, E, F) Four examples of high-power images of AZs where ‘docked’ SVs or omega-shaped bodies were marked by asterisks. Scale bar 0.1 µm.
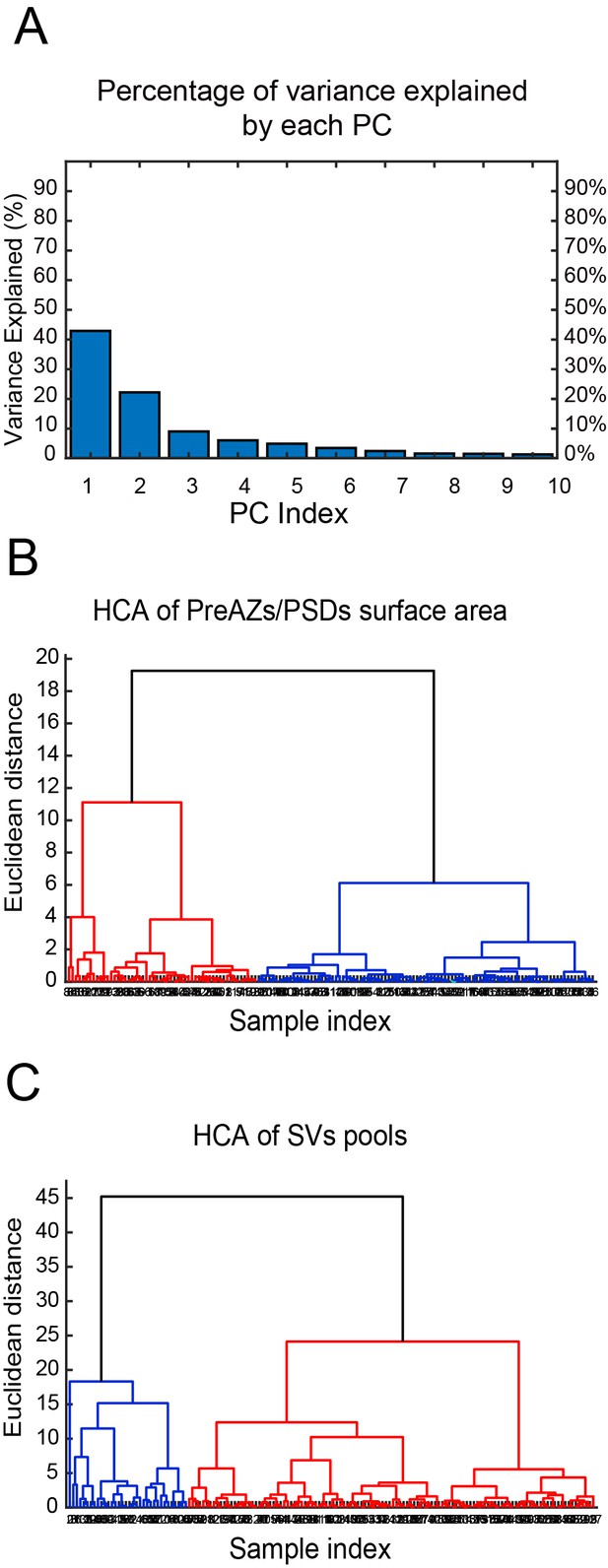
CA of PreAZs/PSDs surface area and SVs pools in excitatory L4 SBs in the human TLN.
(A) Bar histogram showing the PCs of all structural parameters analyzed. (B, C) Two dendrograms generated from the CA, identifying two groups (clusters) of L4 SBs according to the PreAZs/PSDs surface area (B) and the size of SV pools (C). The Euclidian height indicates the difference or dissimilarities between the clusters.
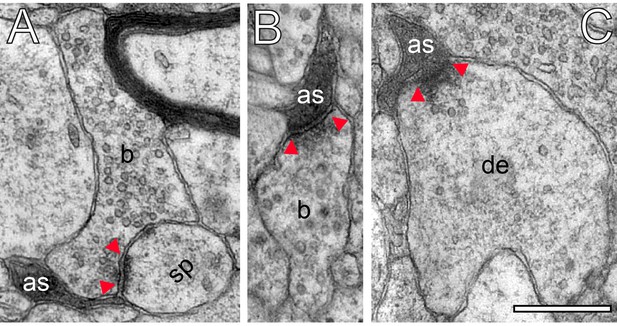
Astrocytic interactions.
(A) Fine astrocytic process (as) reaching as far as the synaptic cleft at the synaptic apposition zone between a spine (sp) and a SB (b). (B), (C) Direct synaptic contact established between an astrocytic finger (as) with a SB (b) in (B) and a putative dendrite (de) in (C). Note the presence of SVs in the dendrite in (C). Scale bars in (A–C) 0.5 µm. In all images, the AZs are indicated by red arrowheads in (A–C).
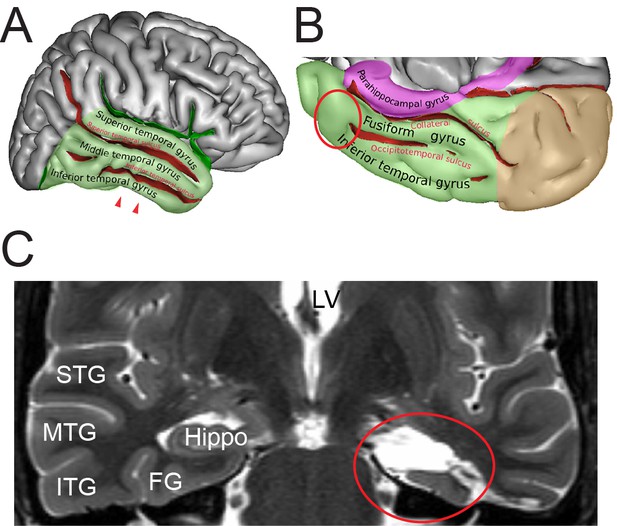
Identification of the region of interest in the human TLN.
(A) Lateral view of the human right cerebral hemisphere. The areas highlighted in transparent green represent the TL. The depth of the sulci are colored in red. The arrowheads indicate the region of interest (ROI) in the inferior temporal gyrus. (B) Midsagittal and oblique view of the human right cerebral hemisphere. Color code as in (A) Brown represents the occipital lobe and purple the parahippocampal region respectively. The red circle indicates the ROI. Figures (A) and (B) were retrieved and modified from https://en.wikipedia.org/wiki/Inferior_temporal_gyrus#/media/File:TempCapts.png under the following license: https://en.wikipedia.org/wiki/Creative_Commons. (C) Postoperative fMRI after the corresponding epilepsy surgery (selective amygdalohippocampectomy). The sampling site of the biopsy material is circled in red, representing the region between the inferior temporal gyrus and the fusiform gyrus. Abbreviations: FG: fusiform gyrus, Hippo: hippocampus, ITG: inferior temporal gyrus, LV: lateral ventricle, MTG: middle temporal gyrus, STG: superior temporal gyrus.
© Sebastian, 2011. Panels A and B retrieved and modified from https://en.wikipedia.org/wiki/Inferior_temporal_gyrus#/media/File:TempCapts.png under a Creative Commons Attribution-ShareAlike License (CC BY-SA 3.0)
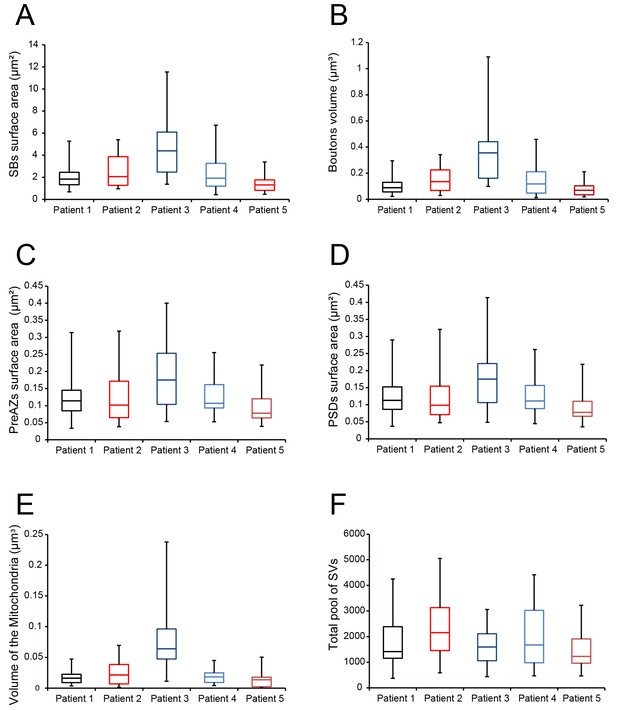
Boxplots of various structural parameters.
Data distributions for each patient are indicated by the medians (horizontal bars), IQRs (framed areas), minimum and maximum (vertical lines) for the distribution of: (A) Surface area of SBs; (B) Volume of SBs; (C) PreAZ surface area; (D) PSD surface area; (E) Volume of mitochondria; (F) total pool of SVs. Note that most structural parameters are not significantly different.
Videos
EM tomography of L4 SBs in the human TLN.
Two SBs terminating on dendritic spines, both with a large non-perforated AZ occupying half of the pre- and postsynaptic apposition zone with a relatively large pool of SVs, one of which is clustered around the AZ (lower right corner). Note, also the SB establishing a synaptic contact with a dendritic shaft (upper left corner). All three SBs contain a single but large mitochondrion. Scale bar 0.25 µm.
FIB-SEM sequential video through L4 of the human TLN.
Note the dynamic changes in the shape and size of dendritic and synaptic structures through the z-stack (250 single images).
Tables
Density of synaptic contacts in L4 of the TLN.
| *Patients | Hu_1 ♀ | Hu_2 ♀ | Hu_3 ♀ | Average ±SD ♀ | Hu_4 ♂ | Hu_5 ♂ | Hu_6 ♂ | Average ±SD ♂ | Total average ±SD |
|---|---|---|---|---|---|---|---|---|---|
| Total density of synaptic contacts/mm3 | 4.1*106 | 1.3*106 | 6.0*106 | 3.80*106 ± 2.36*106 | 0.8*106 | 0.5*106 | 1.5*106 | 0.93*106 ±0.51*106 | 2.37*106 ± 2.19*106 |
| Excitatory SBs on spines (%) | 60 | 60 | 73.08 | 64.36 ± 7.55 | 100 | 100 | 66.66 | 88.89 ± 19.25 | 76.62 ± 18.75 |
| Excitatory SBs on shafts (%) | 33.33 | 40 | 15.38 | 29.57 ± 12.73 | 0 | 0 | 33.33 | 11.11 ± 19.24 | 20.34 ± 17.75 |
| Inhibitory SBs on spines (%) | 6.66 | 0 | 0 | 2.22 ± 3.84 | 0 | 0 | 0 | 0 ± 0 | 1.33 ± 2.98 |
| Inhibitory SBs on shafts (%) | 0 | 0 | 7.69 | 2.56 ± 4.44 | 0 | 0 | 0 | 0 ± 0 | 1.54 ± 3.44 |
-
*Patients identity is anonymous due to the new European protection of data privacy (EU) 2016/679.
Comparative quantitative analysis of various synaptic parameters in L4 and L5 of the TLN.
| Mean ± SD | Median | IQR | CV | Skewness | Variance | ||
|---|---|---|---|---|---|---|---|
| Synaptic boutons | Layers | ||||||
| Surface area (µm²) | L4 | 2.50 ± 1.78 | 2.05 | 1.67 | 0.72 | 1.97 | 3.24 |
| L5*** | 6.09 ± 0.92 | 6.05 | 0.87 | 0.15 | 4.49 | 23.04 | |
| Volume (µm³) | L4 | 0.16 ± 0.16 | 0.11 | 0.12 | 1.01 | 2.89 | 0.03 |
| L5*** | 0.63 ± 0.18 | 0.63 | 0.21 | 0.29 | 2.05 | 0.46 | |
| Active zones | |||||||
| PreAZ surface area (µm²) | L4 | 0.13 ± 0.07 | 0.11 | 0.08 | 0.54 | 1.35 | 0.005 |
| L5*** | 0.23 ± 0.05 | 0.22 | 0.07 | 0.22 | 1.86 | 0.03 | |
| PSD surface area (µm²) | L4 | 0.13 ± 0.07 | 0.11 | 0.08 | 0.53 | 1.44 | 0.005 |
| L5*** | 0.29 ± 0.15 | 0.23 | 0.16 | 0.45 | 2.77 | 0.06 | |
| Cleft width (nm) | |||||||
| Lateral | L4 | 14.11 ± 0.69 | 14.43 | 1.19 | 0.05 | 0.74 | 8.86 |
| L5*** | 17.25 ± 2.39 | 17.51 | 3.74 | 0.13 | 1.14 | 20.73 | |
| Central | L4 | 16.47 ± 1.85 | 15.72 | 3.26 | 0.11 | 0.80 | 17.09 |
| L5*** | 19.05 ± 2.94 | 18.85 | 2.95 | 0.15 | 1.82 | 30.84 | |
| Mitochondria | |||||||
| Volume (µm³) | L4 | 0.03 ± 0.04 | 0.02 | 0.02 | 1.04 | 3.71# | 0.001 |
| L5*** | 0.12 ± 0.09 | 0.07 | 0.16 | 0.87 | 8.22 | 50.30 | |
| % to the total volume | L4 n.s. | 13.11 ± 6.20 | 12.78 | 9.25 | 0.47 | 0.17 | 38.47 |
| L5 | 12.04 ± 1.20 | 11.89 | 2.18 | 0.10 | 0.57 | 23.04 | |
| Synaptic vesicles | |||||||
| Total number | L4*** | 1820.64 ± 980.34 | 1544.5 | 1119.5 | 0.54 | 0.91 | 961066.59 |
| L5 | 1518.52 ± 303.18 | 1347.21 | 541.98 | 0.19 | 2.39 | 1655452.24 | |
| Diameter (nm) | L4 | 19.80 ± 5.63 | 18.00 | 0.28 | 3.41 | 2.10 | 31.69 |
| L5*** | 36.69 ± 1.71 | 37.02 | 3.26 | 0.04 | −2.07 | 153.21 | |
| Volume (µm³) | L4 | 0.01 ± 0.01 | 0.01 | 0.01 | 1.28 | 3.95# | 0.0002 |
| L5*** | 0.05 ± 0.02 | 0.05 | 0.03 | 0.4 | 3.60# | 0.002 | |
| Pool size of SVs | |||||||
| Putative RRP at p10 nm | L4*** | 20.20 ± 18.58 | 17 | 27.25 | 0.92 | 1.11 | 345.04 |
| L5 | 5.42 ± 4.09 | 4.93 | 6.29 | 0.75 | 2.17 | 39.93 | |
| Putative RRP at p20 nm | L4*** | 48.59 ± 39.02 | 41 | 53 | 0.80 | 1.17 | 1523.14 |
| L5 | 15.21 ± 9.02 | 13.55 | 16.34 | 0.59 | 2.06 | 206.69 | |
| Putative RP 60–200 nm | L4*** | 382.1 ± 248.23 | 313 | 376.79 | 0.65 | 1.41 | 61617.55 |
| L5 | 181.86 ± 27.05 | 180.89 | 47.42 | 0.15 | 1.25 | 11469.97 | |
| Putative resting pool > 200 nm | L4 | 1251.82 ± 471.17 | 541 | 471.17 | 0.38 | 1.70 | 87678.29 |
| L5 n.s. | 1264.07 ± 301.77 | 1150.76 | 540.39 | 0.24 | 0.66 | 72853.49 | |
-
Summary of various structural parameter measurements provided from the detailed 3D-volume reconstructions of SBs in L4 (present study) and L5 (Yakoubi et al., 2019) of the human TLN. Mean ± SD, Median, Interquartile Range, CVs, Skewness and Variance were given for each parameter in all patients investigated. #: Values with a skew >3 indicating non-normal distributions. Abbreviations: p10 nm, p20 nm: perimeter 10 and 20 nm from AZ (see Materials and methods), n.s: not significant, ***p≤0.001.
Summary of patient data
| Patient identity | Gender | Age (years) | Age at epilepsy onset (years) | Histopathological result | Antiepileptic drugs (pre-op) | Reconstructed SBs |
|---|---|---|---|---|---|---|
| Hu_1 | Female | 36 | 4 | GGL | LTG, LEV | 53 |
| Hu_2 | Female | 25 | 12 | AHS | LTG | 25 |
| Hu_3 | Female | 25 | 23 | GGL | Zebinix, LEV | 25 |
| Hu_4 | Male | 33 | 5 | Gliosis | LEV, CBZ | 25 |
| Hu_5 | Male | 63 | 24 | AHS | LEV, LTG, CBZ | 22 |
| Hu_6 | Male | 49 | 36 | AHS | Vimpat, ZNS | - |
-
AHS: Ammon's horn sclerosis; GGL: Ganglioglioma; LEV: Levetiracetam; LTG: Lamotrigine; ZNS: Zonisamide; CBZ: Carbamazepine.
Additional files
-
Source code 1
Matlab code for CA of all synaptic parameters investigated.
- https://cdn.elifesciences.org/articles/48373/elife-48373-code1-v2.m
-
Source code 2
Matlab code for CA of PreAZs and PSDs.
- https://cdn.elifesciences.org/articles/48373/elife-48373-code2-v2.m
-
Source code 3
Matlab code for CA of pools of SVs.
- https://cdn.elifesciences.org/articles/48373/elife-48373-code3-v2.m
-
Source data 1
Original data for synaptic density measurements.
- https://cdn.elifesciences.org/articles/48373/elife-48373-data1-v2.docx
-
Source data 2
Original data of all synaptic parameters analyzed.
- https://cdn.elifesciences.org/articles/48373/elife-48373-data2-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/48373/elife-48373-transrepform-v2.pdf





