In vivo functional diversity of midbrain dopamine neurons within identified axonal projections
Figures
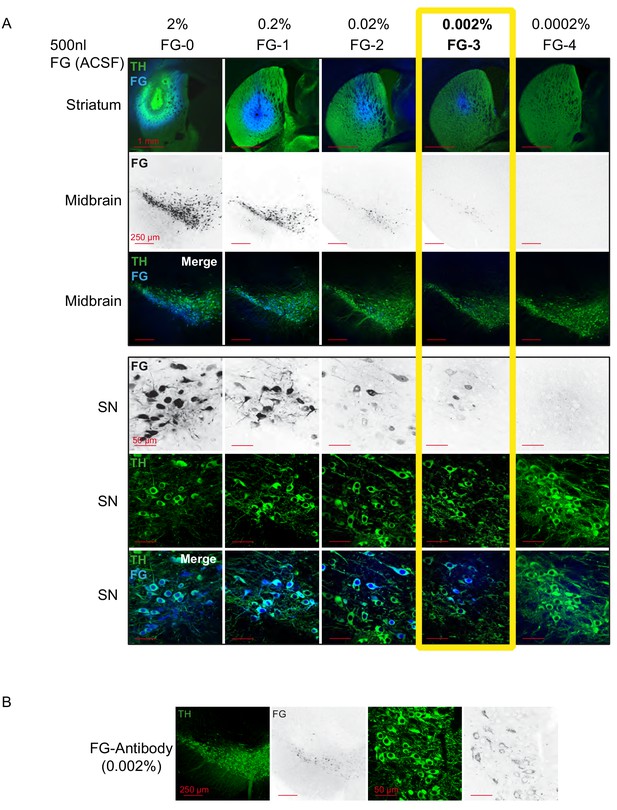
1000-fold reduction of conventional FG-concentrations leads to intact FG-labelled DA neurons in vivo.
(A) Confocal images of a dilution series of Fluorogold/ACSF injections. (Top panel, 1st row) merged images of injections sites (4x magnification) in dorsal striatum with TH in green and FG in blue. (2nd and 3rd row) 10x images of retrogradely traced neurons (FG black in the 2nd row and blue in the 3rd row) in the midbrain (TH green) at bregma −3.16 mm (10x magnification). (Bottom panel) Images of retrogradely traced SN DA neurons at higher magnification (60x; FG black in the 1st row and blue in the 3rd row, TH green). Note that 0.002% FG constitutes the lowest detectable concentration. (B) Use of FG-Antibody facilitates identification of FG-labelled neurons after 0.002% FG/ACSF infusion. Note the increased detectability in comparison to the intrinsic FG signal after 0.002% infusion of FG (Figure 1A, column four).
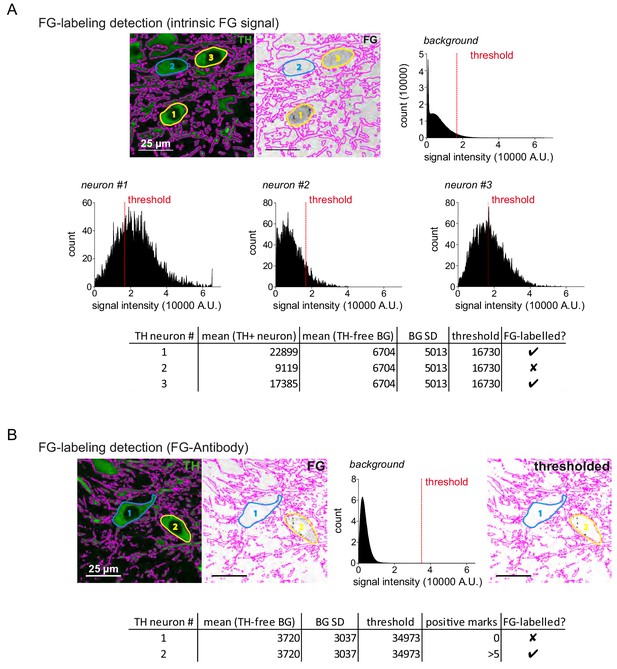
FG-labelling detection method.
(A) Detection method of intrinsic FG signals. The left image shows the TH-mask (TH in green), the right image shows the FG-channel (FG in black), the TH-mask is overlaid on both images in magenta, the four intensity histograms depict the background and single neuron FG-intensity distributions with a dotted line at the threshold of labelling detection. Threshold for detection was set to Thr = mean background intensity + 2*SD of the mean background. TH-negative nuclei were subtracted from the background. (B) Detection method of immune-FG-signal. The left image shows the TH-mask (TH in green), the middle image shows the FG-channel (FG in black), the intensity histogram displays the FG-intensity distribution of the TH-free background, the right image shows the thresholded FG-channel, the TH-mask is overlaid on all images in magenta. Here, threshold for detection was set to Thr = mean background intensity + 10*SD of the mean background.
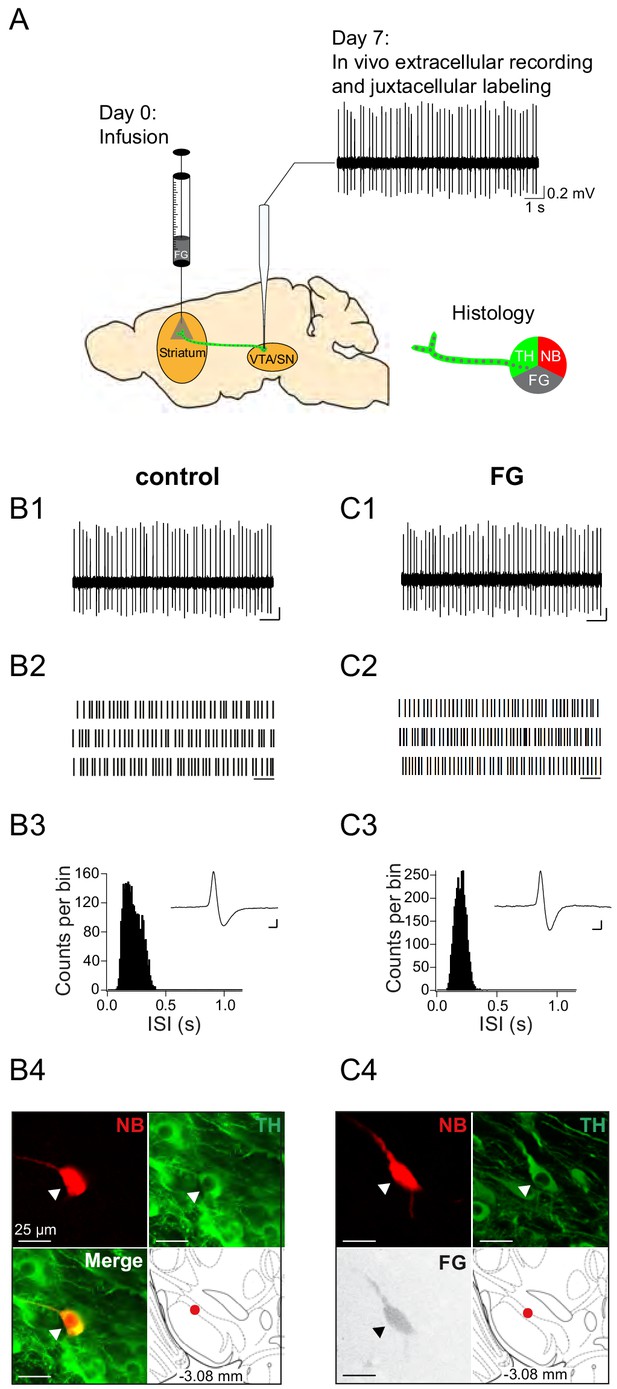
Experimental design and electrophysiological data of control neuron and FG-labelled SN DA neuron.
(A) Experimental design for FG-labelled DA dataset. Control DA neurons were recorded in untreated mice (only ‘Day 7’). (B1, C1) spontaneous in vivo extracellular single-unit activities of a representative control DA lSN neuron (B) and a representative FG-labelled DLS-projecting DA lSN neuron (C), shown as 10 s of original recording traces (scale bar: 0.2 mV, 1 s). (B2, C2) 30 s raster plots (scale bar: 1 s). (B3, C3) ISI-distributions. Inset, averaged AP waveform showing biphasic extracellular action potentials in high resolution (scale bar: 0.2 mV, 1 ms). (B4, C4) Confocal images of extracellularly recorded and juxtacellularly labelled neurons, the location of the neurons in the lSN at bregma −3.08 mm is shown in the bottom right image. Note the similarities in mean frequency, regularity, firing pattern, AP waveform and anatomical location.
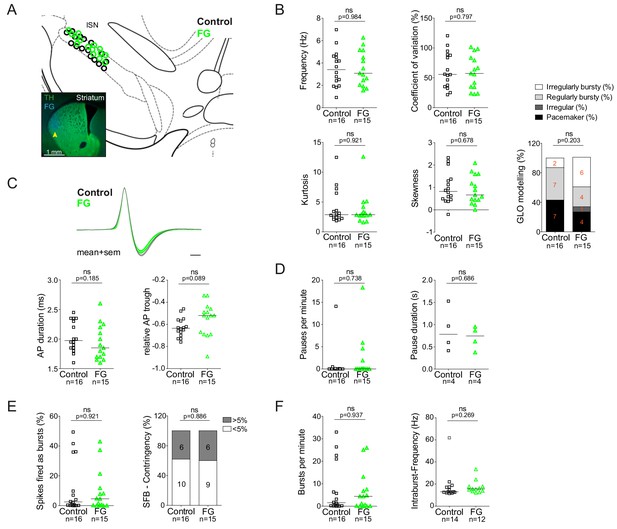
Retrograde tracing with highly-diluted (0.002%) fluorogold prevents perturbation of in vivo electrophysiological properties of identified nigrostriatal dopamine neurons.
(A) Anatomical mapping of all extracellularly recorded and juxtacellularly labelled neurons (projected to bregma −3.16 mm; control in black, FG-labelled in green). Note the anatomical overlap of recorded DA populations in the lSN. Inset, FG-injection site in DLS (FG in blue, TH in green). (B–F) Scatter dot-plots (line at median) showing no significant differences in firing frequency (Hz), coefficient of variation (%), kurtosis and skewness of the ISI-distributions, GLO-based firing pattern (all B), normalized AP waveform, AP duration (ms), relative AP trough (all C), pauses per minute, pause duration (s) (both D), SFB (%), SFB contingency (% of neurons > and < 5% SFB) (both E), bursts per minute, intraburst-frequency (Hz) (both F).
-
Figure 3—source data 1
Electrophysiological data.
All included electrophysiological parameters analyzed for recorded control and projection-defined DA neurons.
- https://doi.org/10.7554/eLife.48408.007
-
Figure 3—source data 2
Labelling detection.
Labelling detection parameters (labelling intensity of individual cells, backgrounds, Delta, standard deviation of backgrounds and number of positive FG-positive marks after thresholding) for FG-labelled DA neurons.
- https://doi.org/10.7554/eLife.48408.008
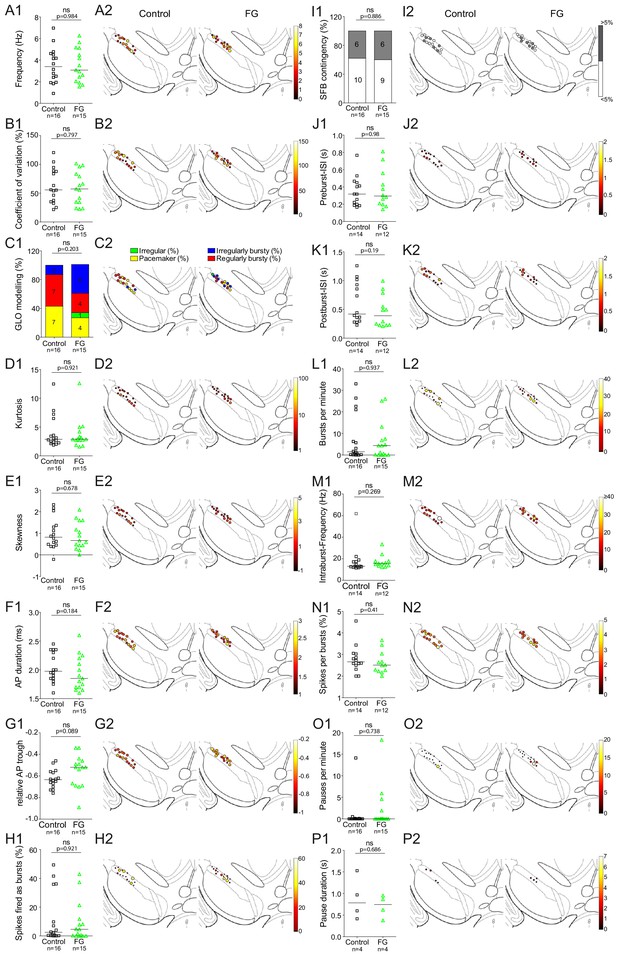
Comparison of in vivo firing properties of control and FG-labelled neurons.
(A1–P1) Scatter dot-plots (line at median) and bar graphs showing no significant differences in various parameters of firing. (A2–P2) Feature maps of respective parameters.
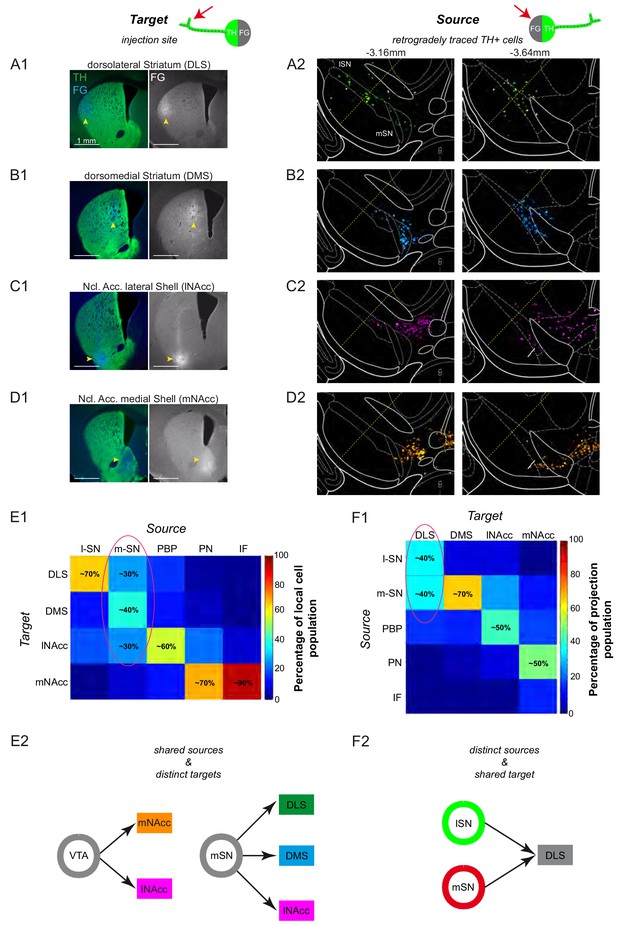
Dorsal and ventral striatal-projecting DA neurons intermix within the mSN.
(A1–D1) Injection sites for fluorescent tracing experiments injecting FG into the DLS (A1; bregma +0.86 mm), DMS (B1; bregma +0.74 mm), lNAcc (C1; bregma +0.86 mm), mNacc (D1; bregma +1.54 mm). (Left images) merged TH-(green) and FG-(blue) signals. (Right images) FG mono. (A2–D2) locations of retrogradely traced DA neurons (accumulated from n = 3 animals) at a rostral (−3.16 mm) and a caudal (−3.64 mm) midbrain section, shown as colored heat maps. DLS-projecting DA neurons (A2) are shown in green, DMS-projecting DA neurons (B2) in cyan, lNAcc-projecting DA neurons (C2) in magenta, mNAcc-projecting DA neurons (D2) in orange. The border between lSN and mSN is indicated with a yellow dotted line. Note that DLS-projecting DA neurons are located throughout the mediolateral extent of the SN (A2) and that the mSN is a substantial midbrain source area for three mayor projection-defined systems (A2–C2). (E1) Local cell population matrix (%, average of three animals). Note that the lSN, PBP, PN and IF are populated mainly by one projection-defined population each (lSN - DLS, PBP - lNAcc, PN and IF - mNAcc). However, the mSN is a region of overlap of DLS-, DMS- and lNAcc- projecting DA neurons. (F1) Projection population matrix (%, average of three animals). Note that the DLS receives dopaminergic input from both SN source areas (lSN and mSN). On the other side, DMS mainly receives input from the mSN, lNAcc from PBP, mNAcc from PN. (E2) Shared sources and distinct targets. The mSN is a region of overlap of three different projection target-defined systems (DLS, DMS and lNAcc). The VTA harbors both the mNAcc and lNAcc projection system. (F2) Distinct sources and shared target. The DLS receives dopaminergic input from two distinct midbrain source areas (lSN and mSN).
-
Figure 4—source data 1
Anatomical reconstruction.
(A1, B1) Mean (N = 3 animals for each projection) and accumulated numbers of FG-labelled and projection-defined DA neurons in distinct midbrain regions. (A2, B2) Percentages of mean and accumulated projection population. (A3, B3) Percentages of mean and accumulated local cell population.
- https://doi.org/10.7554/eLife.48408.014
-
Figure 4—source data 2
Anatomical reconstruction - DLS DLS-projecting cells counted in different regions and at distinct caudo-rostral levels.
- https://doi.org/10.7554/eLife.48408.015
-
Figure 4—source data 3
Anatomical reconstruction - DMS DMS-projecting cells counted in different regions and at distinct caudo-rostral levels.
- https://doi.org/10.7554/eLife.48408.016
-
Figure 4—source data 4
Anatomical reconstruction - lNAcc DLS-projecting cells counted in different regions and at distinct caudo-rostral levels.
- https://doi.org/10.7554/eLife.48408.017
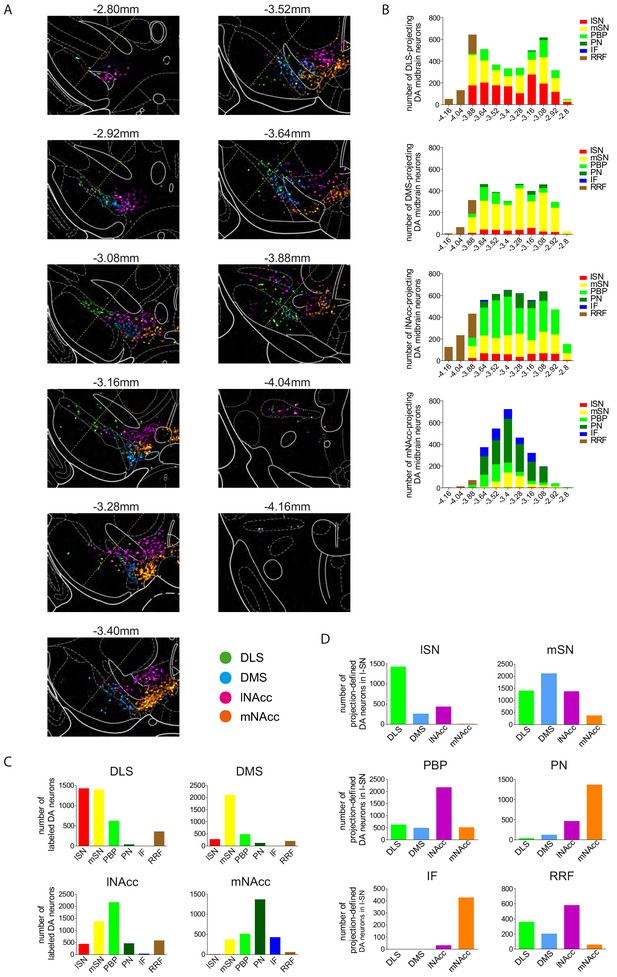
Anatomical segregation of different DA systems in the midbrain.
(A) Serial reconstruction of retrogradely traced DA neurons across the caudorostral extent of the midbrain reveals anatomical position of DA neurons projecting to DLS (green), DMS (cyan), lNAcc (magenta) and mNAcc (orange), shown as colored heat maps from rostral (bregma −2.80 mm) to caudal (bregma −4.16 mm). Note the high degree of overlap in the mSN. (B) Absolute distribution of retrogradely labelled DA neurons between the different caudorostral levels for each projection-defined system (numbers of neurons accumulated from three animals for each group), shown as stacked bar graphs with color-coding for distinct midbrain source areas. (C) Cumulative numbers of retrogradely labelled neurons (n = 3 animals) in distinct midbrain areas for each projection-defined DA system, shown as bar graphs. (D) Cumulative numbers of retrogradely traced populations in a distinct midbrain area (average of 3 animals ± SEM). Note the heterogeneous population in the mSN in contrast to more homogene populations in lSN, PBP, PN and IF. Data is given as mean ± SEM.
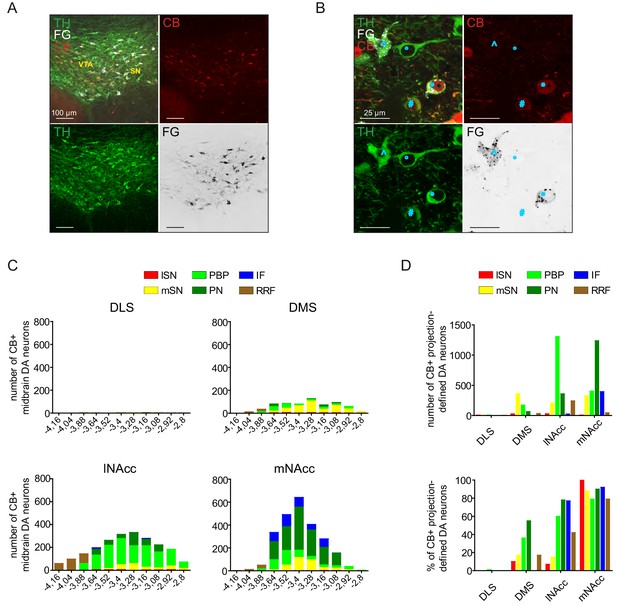
Calbindin expression of midbrain DA neurons does not co-segregate with specific axonal projections.
(A) Confocal 20x images of retrogradely FG-traced midbrain DA neurons (injection site: lNAcc) costained for TH (green), FG (white) and CB (red). Note the intermingling of CB-positive and CB-negative neurons in the VTA and mSN. (B) Higher magnification (60x, 2.5 zoom) confocal images of retrogradely traced DA neurons (same section as in (A)) costained for TH (green), FG (white) and CB (red). Note the variety of TH-, FG- and CB-costaining: The circumflex (^) shows a TH- and FG-positve but CB-negative neuron. The circle (°) displays a TH-positive but FG- and CB-negative neurons. The number sign (#) presents a TH- and CB-positive but FG-negative neuron. Finally, the star (*) represents a TH-, FG- and CB-positive neuron. (C) Accumulated numbers of retrogradely FG-labelled and CB-positive DA neurons (n = 3 animals) for each projection-defined DA system through the caudorostral axis, shown as stacked bar graphs with color-coding for distinct midbrain source areas. Note the gradient of CB-expression: from low to high: DLS-DMS-lNAcc-mNAcc. Also, note the gradient on the caudorostral axis. Most CB-expressing neurons are located in the intermediate midbrain (bregma −3.52,–3.4, −3.28 mm). CB-expression levels decrease at more caudal and more rostral midbrain areas. (D) Comparison of CB-expression levels of different projection-defined DA systems. (Top graph) Accumulated numbers of retrogradely FG-labelled CB-positive DA neurons for distinct color-coded midbrain areas. (Bottom graph) Percentages of CB-positive FG-labelled DA neurons out of all FG-labelled DA neurons in distinct color-coded midbrain areas. Note that CB-expression levels for DMS- and lNAcc-projecting neurons are different overall, but similar in the mSN.
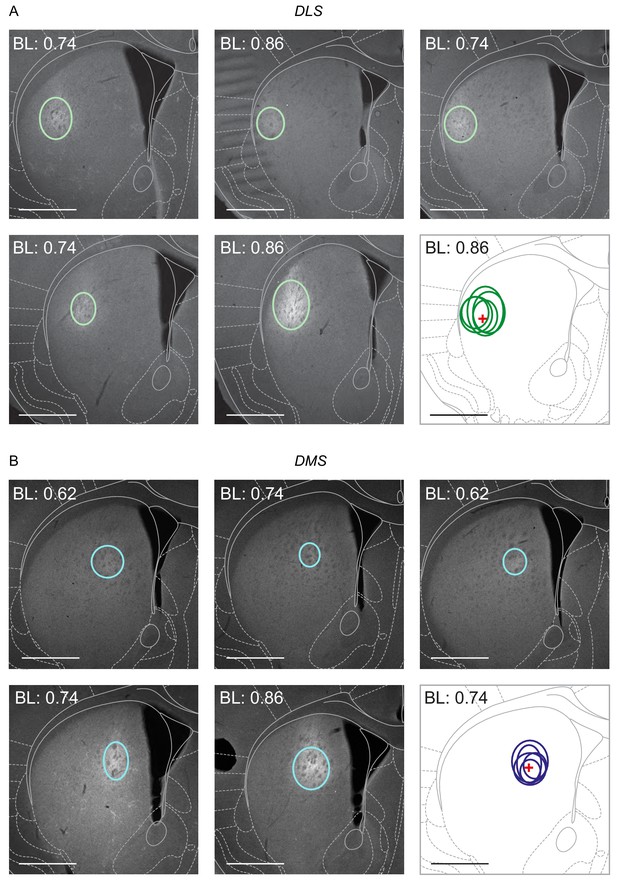
Striatal fluorogold infusion sites show high reliability across animals.
(A–D) Monochromatic (FG) images of infusion sites for DLS (A), DMS (B), lNAcc (C) and mNAcc (D), given as five examples, each outlining detectable FG with overlay of corresponding stereotactic maps. Red crosses indicate respective target coordinates for stereotactically-guided infusion. Scale bar = 1 mm.
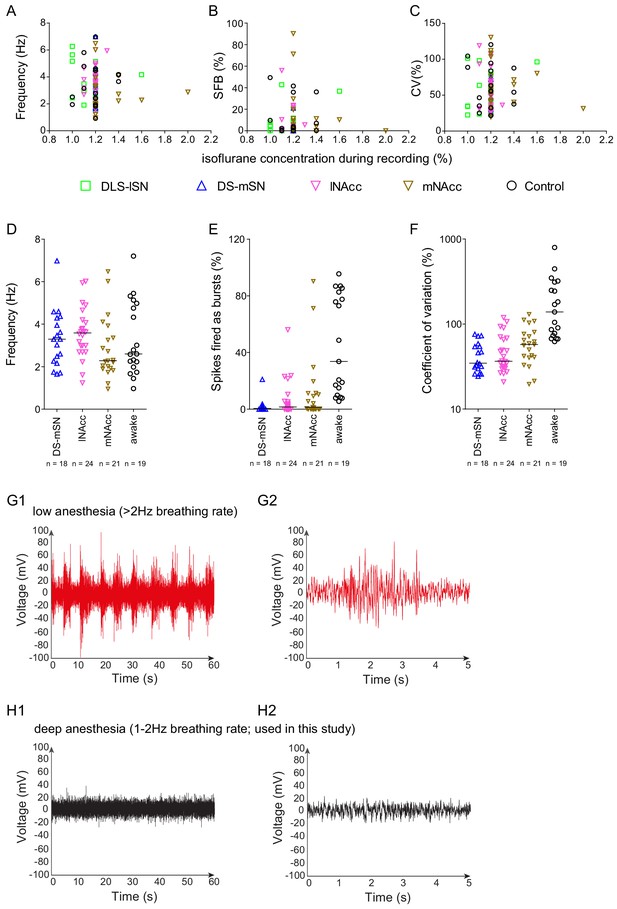
In vivo activities of midbrain DA neurons recorded under anesthesia do not correlate with isoflurane concentrations.
(A–C) Frequency (Hz), SFB (%) and CV (%) of individual neurons plotted against isoflurane concentrations during respective recordings. Note the absence of significant correlations between functional neuronal properties and isoflurane concentrations. (D–F) Scatter dot-plots (line at median) comparing mean frequency, spikes fired in bursts and coefficient of variation between distinct projection-defined mSN/VTA DA neurons recorded under isoflurane anesthesia and pharmacologically identified mSN/VTA DA neurons recorded in awake, behaving animals in the homecage (dataset from Duvarci et al., 2018). (G–H) Cortical LFP recordings during low (breathing rate >2 Hz) (G) and deep (breathing rate 1–2 Hz) (H) anesthesia (G1, H1 – one minute; G2, H2 – 5 s).
-
Figure 4—figure supplement 4—source data 1
Electrophysiological data – DA neurons, recorded in awake mice, home cage.
All included electrophysiological parameters for recorded DA neurons in awake mice during home cage activity. Data were collected in the context of a previous study (Duvarci et al., 2018).
- https://doi.org/10.7554/eLife.48408.018
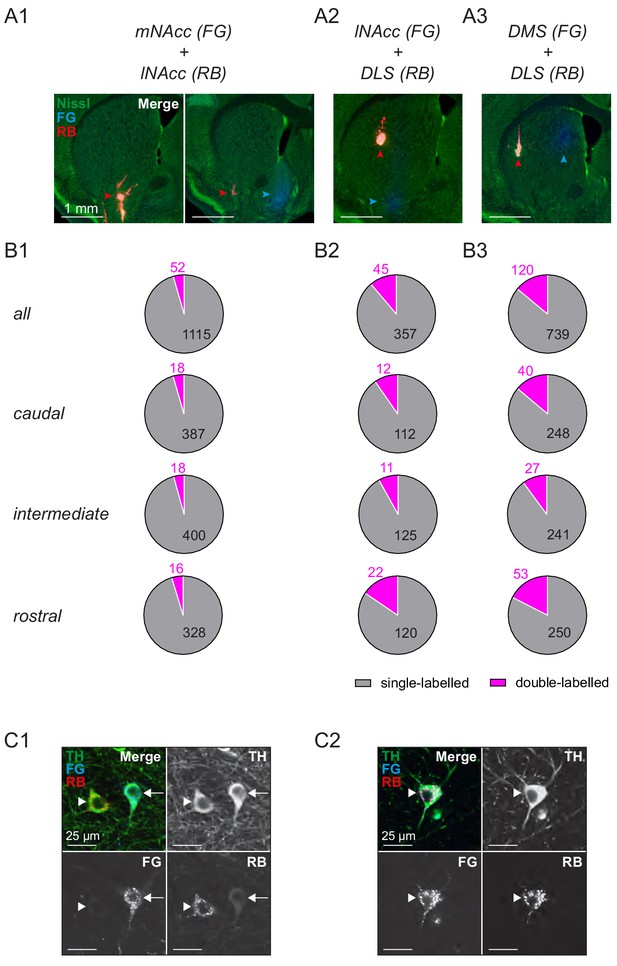
Projection-defined midbrain DA systems projecting to dorsal and ventral striatum are predominantly parallel and non-overlapping.
(A) Infusion sites in green nissl counterstained 100 μm sections of double retrograde tracing experiments using fluorogold (blue) for mNAcc (A1), lNAcc (A2) and DMS (A3) and red fluorescently labelled latex beads (red) for lNAcc (A1) and DLS (A2 and A3). Arrowheads indicate the respective infusion sites. (B) Distribution of single-labelled and double-labelled DA neurons for each double tracing experiment (single-labelled in grey, double-labelled in magenta). Numbers indicate accumulated total numbers of neurons counted (first row, N = 3 per group), as well as those in caudal sections (−3.64 mm from bregma; second row), intermediate sections (−3.28 mm from bregma; third row) and rostral sections (−3.08 mm from bregma; fourth row). (C) Confocal images of TH-positive (green) cells in the m-SN labelled with fluorogold (blue) for lNAcc neurons and red beads (red) for DLS neurons. (C1) Example of two neighboring single-labelled neurons. (C2) Example of a double labelled neuron.
-
Figure 5—source data 1
Double labelling Single-labelled and double-labelled.
DA neurons counted in caudal, intermediate and rostral sections (N = 3 animals for each comparison).
- https://doi.org/10.7554/eLife.48408.020
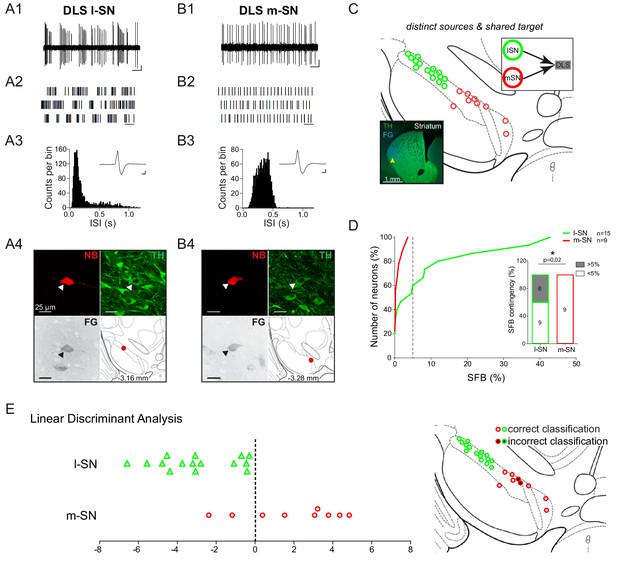
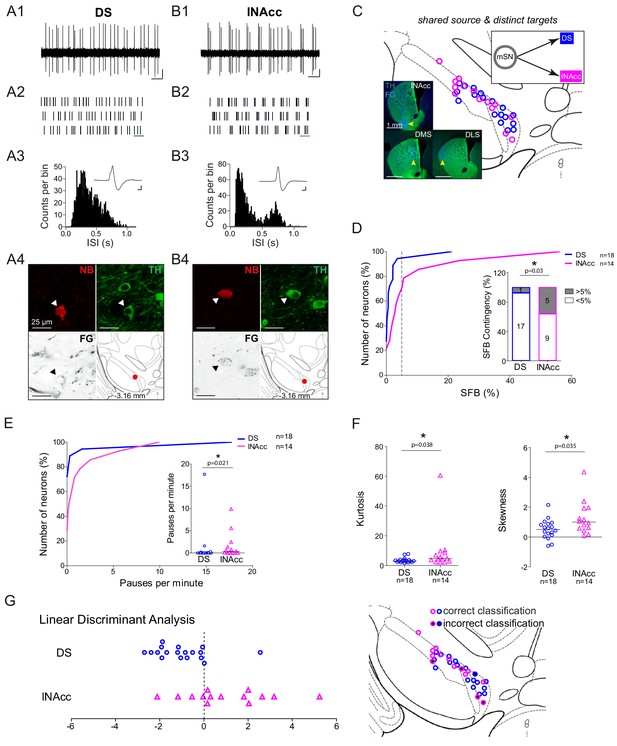
High in vivo burstiness in lateral compared to medial DLS-projecting SN DA neurons.
(A1, B1) Spontaneous in vivo extracellular single-unit activities of a representative DLS-projecting DA neuron located in the lSN (A1) and a representative DLS-projecting DA neuron located in the mSN (B1), shown as 10 s of original recording traces (scale bar: 0.2 mV, 1 s). Note the differences in burstiness. (A2, B2) 30 s raster plots (scale bar: 1 s). (A3, B3) ISI-distributions. Note the presence of ISIs below 80 ms and 160 ms indicating bursts in A3 in contrast to B3. Inset, averaged AP waveform showing biphasic extracellular action potentials in high resolution (scale bar: 0.2 mV, 1 ms). (A4, B4) Confocal images of retrogradely traced, extracellularly recorded and juxtacellularly labelled DA neurons, the location of the neurons is displayed in the bottom right images. (C) Anatomical mapping of all extracellularly recorded and juxtacellularly labelled neurons (projected to bregma −3.16 mm; DLS-lSN in green, DLS-mSN in red). Inset, FG-injection site in DLS (FG in blue, TH in green). (D) Cumulative SFB distribution histograms (dotted line at SFB = 5% threshold) and bar graphs of SFB contingencies (% of neurons > and < 5% SFB) showing significant differences in burstiness. Note that no DLS-projecting DA neurons located in the mSN displayed a SFB above 5%. (E) Linear discriminant analysis of DLS-projecting DA neurons located either in the mSN or lSN. (Right Picture) Mapping of correctly- vs incorrectly-classified DLS-projecting DA neurons located in mSN or lSN.
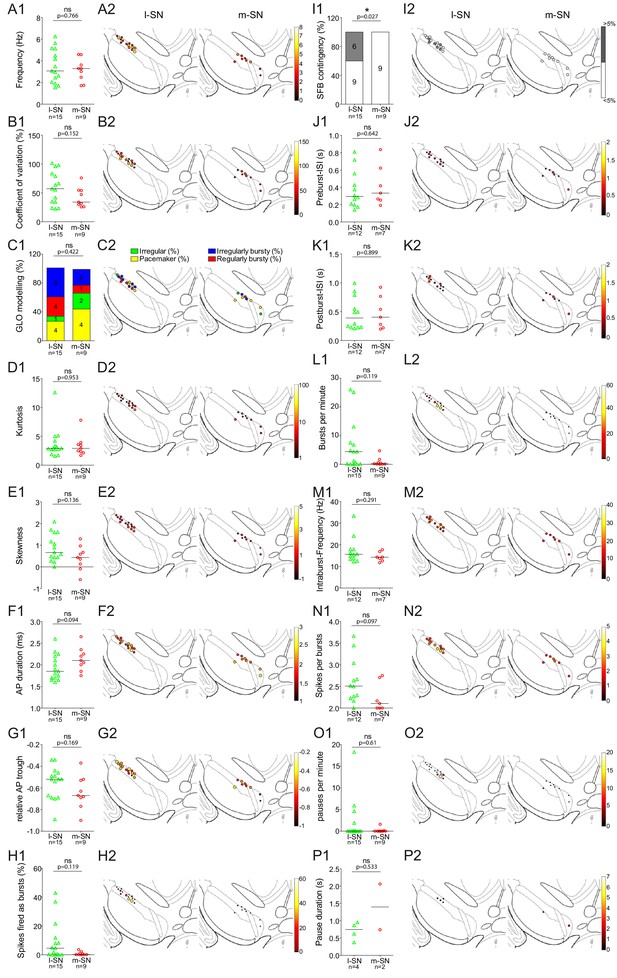
Comparison of in vivo firing properties of lSN and mSN DA neurons projecting to DLS.
(A1–P1) Scatter dot-plots (line at median) and bar graphs showing significant differences selectively in SFB contingency (% of >and < 5%). (A2–P2) Feature maps of respective parameters.
Functional segregation of in vivo electrophysiological properties mSN DA neurons with distinct axonal projections.
(A1, B1) Spontaneous in vivo extracellular single-unit activities of a representative DS-projecting DA neuron located in the mSN (A1) and a representative lNAcc-projecting DA neuron located in the mSN (B1), shown as 10 s of original recording traces (scale bar: 0.2 mV, 1 s). Note the differences in bursting. (A2, B2) 30 s raster plots (scale bar: 1 s). (A3, B3) ISI-distributions. Note the presence of ISIs below 80 ms and 160 ms indicating bursts in B3 in contrast to A3. Inset, averaged AP waveform showing biphasic extracellular action potentials in high resolution (scale bar: 0.2 mV, 1 ms). (A4, B4) Confocal images of retrogradely traced, extracellularly recorded and juxtacellularly labelled DA neurons, the location of the neurons is displayed in the bottom right images. (C) Anatomical mapping of all retrogradely traced, extracellularly recorded and juxtacellularly labelled neurons (projected to bregma −3.16 mm; DS-lSN in blue, lNAcc-mSN in magenta). Inset, FG-injection sites in DMS, DLS and lNAcc (FG in blue, TH in green). (D) Cumulative SFB distribution histograms (dotted line at SFB = 5% threshold) and bar graphs of SFB contingencies (% of neurons > and < 5% SFB) showing significant differences in burstiness. (E) Cumulative distribution histograms and bar graphs of pauses per minute showing significant differences. (F) Scatter dot-plots showing significant differences in kurtosis and skewness of the ISI-distributions. (G) Linear discriminant analysis of DA mSN neurons projecting either to DLS/DMS or lNAcc. 70.8% of neurons were classified correctly based on skewness of the ISI-distribution, AP duration, repolarization speed and the precision of spiking (log(σ2)) when randomly bisecting the data 1000 times. When the whole data set was used, 84.4% of neurons were classified correctly. (Right Picture) Mapping of correctly- vs incorrectly-classified DA mSN neurons projecting to DS or lNAcc.
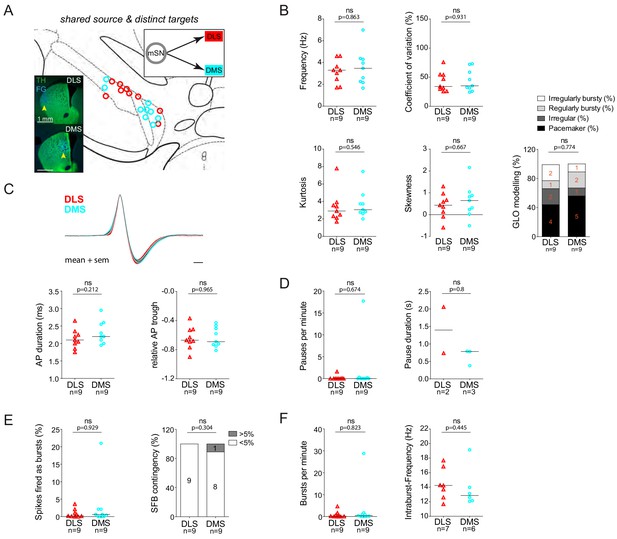
DLS and DMS-projecting DA neurons located in the mSN do not exhibit significantly different in vivo firing properties.
(A) Anatomical mapping of all extracellularly recorded and juxtacellularly labelled neurons (projected to bregma −3.16 mm; DLS in red, DMS in cyan). Inset, FG-injection sites in the DLS and DMS (FG in blue, TH in green). (B–F) Scatter dot-plots and bar graphs (line at median) showing no significant differences in firing frequency (Hz), coefficient of variation (%), kurtosis and skewness of the ISI-distributions, GLO-based firing pattern (all B), normalized AP waveform, AP duration (ms), relative AP trough (all C), pauses per minute, pause duration (s) (both D), SFB (%), SFB contingency (% of neurons > and < 5% SFB) (both E), bursts per minute, intraburst-frequency (Hz) (both F).
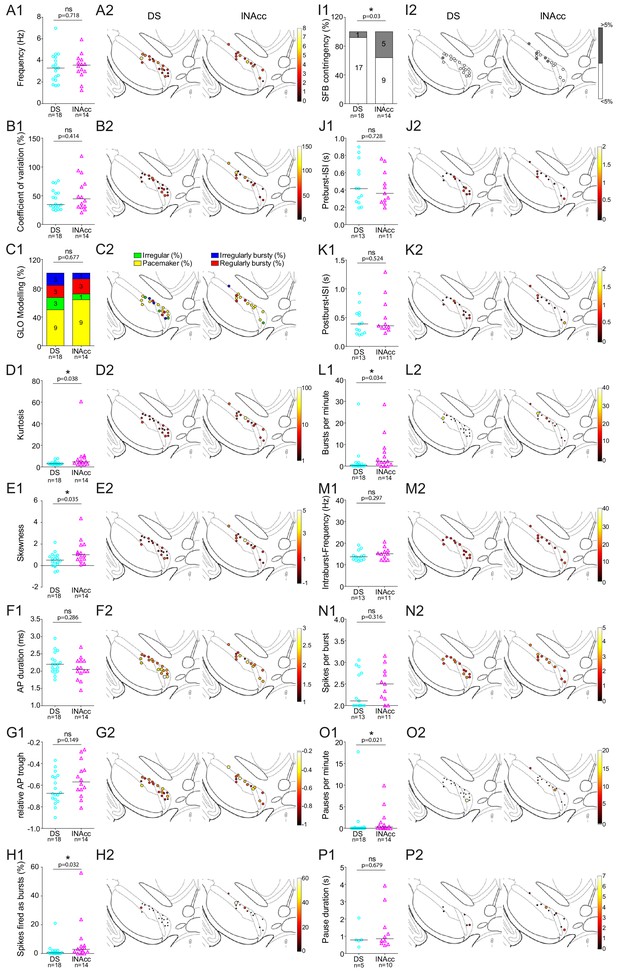
Comparison of in vivo firing properties of DS and lNAcc-projecting DA mSN neurons.
(A1–P1) Scatter dot-plots (line at median) and bar graphs showing significant differences in kurtosis, skewness, SFB (%), SFB contingency (% of >and < 5%), bursts per minute, pauses per minute. (A2–P2) Feature maps of respective parameters.
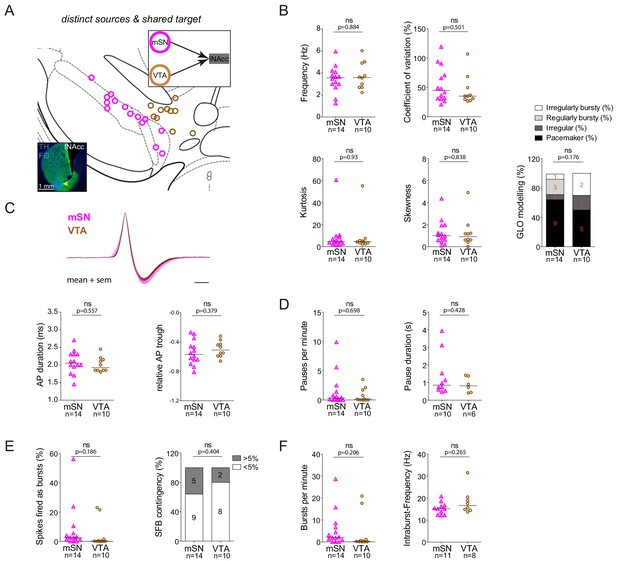
lNAcc-projecting DA neurons located either in mSN or VTA do not exhibit significantly different in vivo firing properties.
(A) Anatomical mapping of all extracellularly recorded and juxtacellularly labelled neurons (projected to bregma −3.16 mm; mSN in magenta, VTA in brown). Inset, FG-injection site in the DLS (FG in blue, TH in green). (B–F) Scatter dot-plots (line at median) and bar graphs showing no significant differences in firing frequency(Hz), coefficient of variation (%), kurtosis and skewness of the ISI-distributions, GLO-based firing pattern (all B), normalized AP waveform, AP duration (ms), relative AP trough (all C), pauses per minute, pause duration (s) (both D), SFB (%), SFB contingency (% of neurons > and < 5% SFB) (both E), bursts per minute, intraburst-frequency (Hz) (both F).
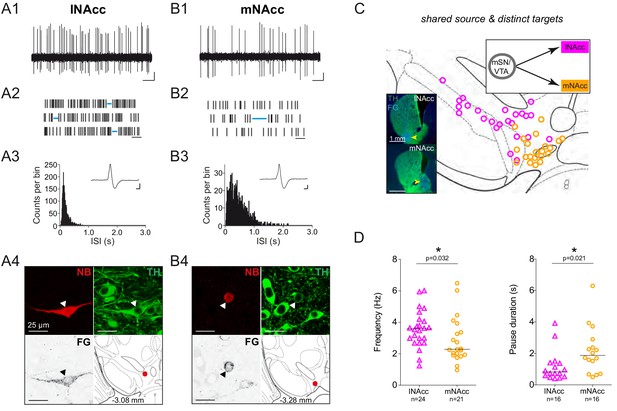
DA neurons projecting to medial or lateral shell of the nucleus accumbens display distinct in vivo firing rates.
(A1, B1) Spontaneous in vivo extracellular single-unit activities of a representative lNAcc-projecting DA neuron located in the mSN (A1) and a representative mNAcc-projecting DA neuron located in the VTA (B1), shown as 10 s of original recording traces (scale bar: 0.2 mV, 1 s). Note the differences in firing frequency. (A2, B2) 30 s raster plots (scale bar: 1 s). Blue lines indicate pauses. Note the differences in pause duration. (A3, B3) ISI-distributions. Note the differences in overall shape and maximal ISIs. Inset, averaged AP waveform showing biphasic extracellular action potentials in high resolution (scale bar: 0.2 mV, 1 ms). (A4, B4) Confocal images of retrogradely traced, extracellularly recorded and juxtacellularly labelled DA neurons, the location of the neurons is displayed in the bottom right images. (C) Anatomical mapping of all extracellularly recorded and juxtacellularly labelled neurons (projected to bregma −3.16 mm; lNAcc in magenta, mNAcc in orange). Insert, FG-injection sites in lNAcc and mNAcc (FG in blue, TH in green). (D) Scatter dot-plots (line at median) showing significant differences in firing frequency (Hz) and pause duration (s).
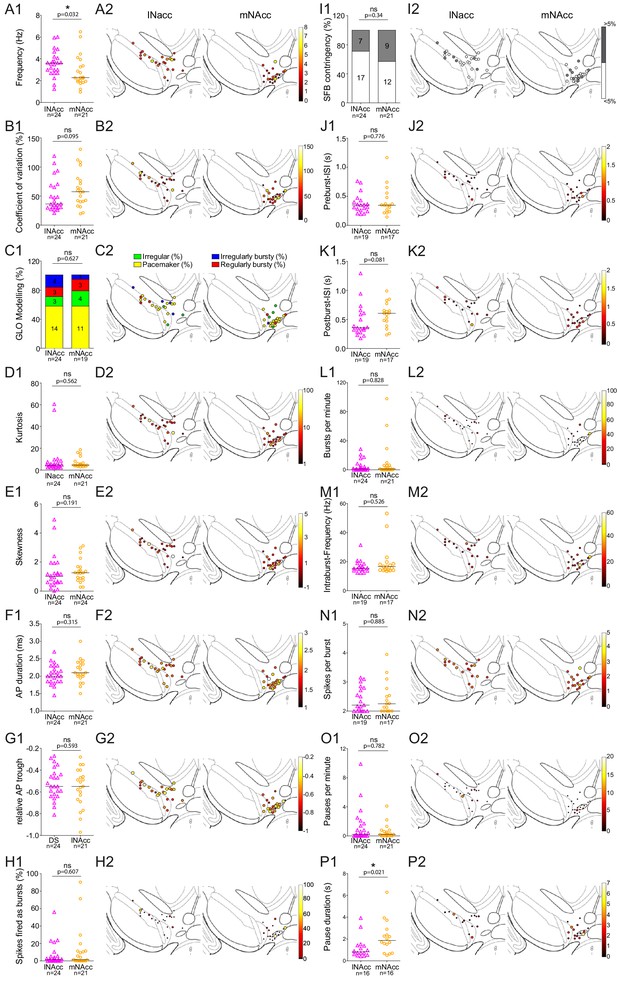
lNAcc and mNAcc-projecting midbrain DA neurons do not exhibit significantly different in vivo firing properties.
(A1–P1) Scatter dot-plots (line at median) and bar graphs showing significant differences in firing frequency (Hz) and pause duration (s) (A2–P2) Feature maps of respective parameters.
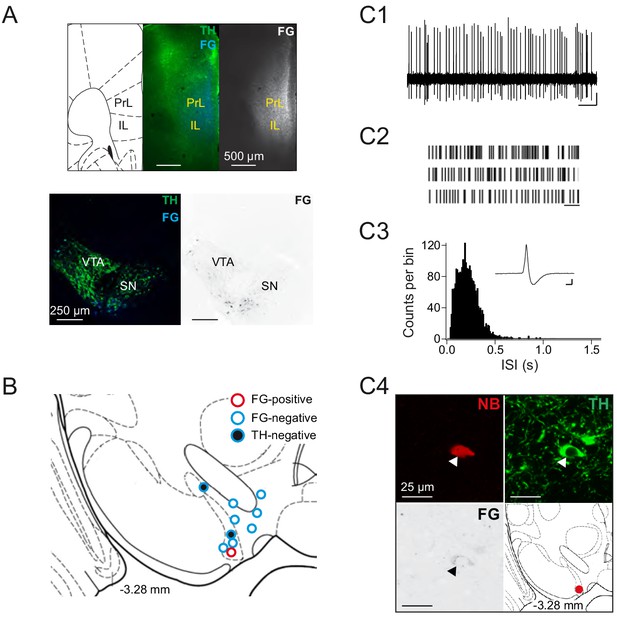
Low success rate in identifying mPFC-projecting DA neurons.
(A) (Top panel) FG-injection sites (0.002%, 2 × 1 μl, 50 nl/min) in PrL (bregma: 1.98 mm, lateral: 0.3 mm, ventral: 1.5 mm) and IL (bregma: 1.98 mm, lateral: 0.3 mm, ventral:2.1 mm) nucleus. (bottom panel) Retrogradely FG-labelled neurons in VTA and SN (B) Anatomical mapping of all extracellularly recorded and juxtacellularly labelled neurons in mPFC-traced mice (open red circle: TH+/FG+, open blue circle: TH+/FG-, filled blue circle: TH-) (C1–C4) In vivo extracellular activity of the single mPFC-projecting midbrain DA neuron recorded, shown as 10 s of original recording traces (C1; scale bar: 0.2 mV, 1 s) and 30 s raster plots (C2; scale bar: 1 s). (C3) ISI-distributions. Inset, averaged AP waveform showing biphasic extracellular action potential in high resolution (scale bar: 0.2 mV, 1 ms). (C4) Confocal images of extracellularly recorded and juxtacellularly labelled neuron, the location of the neuron at bregma −3.28 mm is shown in the bottom right image.
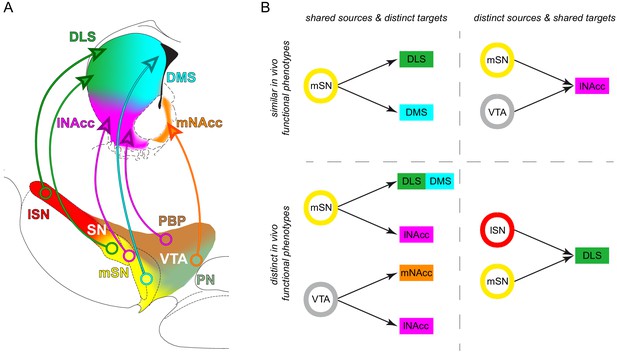
Midbrain dopamine neurons with distinct source-target topographies display functional in vivo firing differences.
(A) Diagram of the organization of distinct midbrain DA subtypes defined by their source-target topography. Single midbrain source areas and striatal target areas are colored individually (midbrain: lSN red, mSN yellow, PBP brown, PN light green; striatum: DLS green, DMS cyan, lNAcc magenta, mNAcc orange). Distinct subtypes are illustrated with a circle in the midbrain source area and an arrow to the projection target area. (B) Comparisons of in vivo activities of distinct source-target topography defined DA subtypes.
Additional files
-
Source code 1
Matlab scripts for generation of feature mapsCustom script used in order to create feature maps as in Figure 3—figure supplement 1, Figure 6—figure supplement 1, Figure 7—figure supplement 2 and Figure 8—figure supplement 1.
- https://doi.org/10.7554/eLife.48408.031
-
Transparent reporting form
- https://doi.org/10.7554/eLife.48408.032





