Stochastic cell-cycle entry and cell-state-dependent fate outputs of injury-reactivated tectal radial glia in zebrafish
Figures
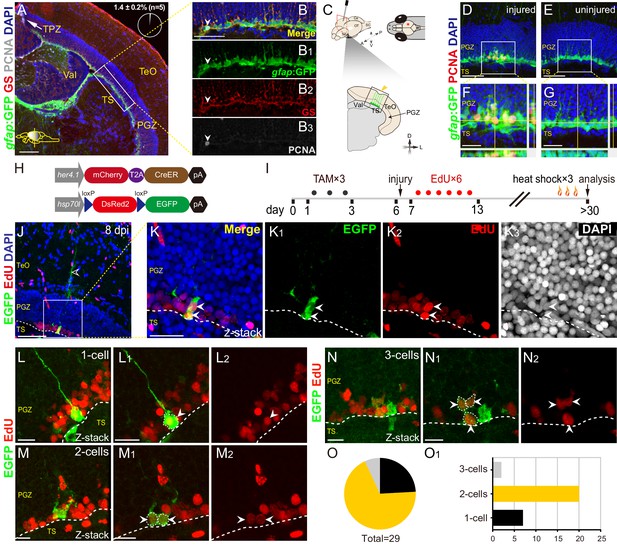
Injury reactivates dormant RG to proliferate and divide.
(A–B3) Tg(gfap:GFP) (green), GS (red) and PCNA (gray) immunofluorescences show that PCNA+ proliferative cells (gray cells) are restricted to the TPZ (white arrow in (A)) and very few radial glia (RG) (1.4 ± 0.2%, n = 5, mean ± SEM, gray cells, white arrowheads in (B–B3)) is PCNA+. (B–B3) The high-magnification images of the boxed area (white box) in (A). (C) Schematic representation of stab injury assay. A 30G needle is stabbed into the central-dorsal region of the right hemisphere of zebrafish optic tectum. The red asterisk and yellow arrowhead indicates the injury site. RG (green cells) at the bottom of PGZ underneath the injury site are analyzed. (D–G) Tg(gfap:GFP) (green) and PCNA (red) immunofluorescences show that injury induces the proliferation of RG (GFP+/PCNA+, yellow cells) underneath the injury site at 3 days post-injury (dpi). (F and G) The high-magnification images of boxed areas in (D) and (E), respectively. (H) The design of Cre-loxP transgenic fish lines used for clonal analysis of individual tectal RG. Fish expressing mCherryT2ACreERT2 controlled by the her4.1-promotor are crossed to red-to-green reporter fish controlled by the hsp70l promoter. In Tg(her4.1:mCherry-CreERT2×hsp70l:DsRed2(floxed)EGFP) double transgenic fish, EGFP expression is specifically induced in her4.1-expressing RG and their progeny by TAM applications and heat shocks. (I) Experimental time course of Cre-loxP-based clonal analysis experiments shown in (J–O1). Double transgenic fish are administrated with TAM for three consecutive days (black dots) before the injury. EdU is injected to the injured fish to label the newborn cells for six consecutive days (red dots). Fish (21 to 24 dpi) are heat-shocked to induce EGFP expression in recombined cells and their progeny. (J–K3) Representative RG-derived clone (EGFP+/EdU+, white arrows) underneath the injury site at 8 dpi. (K–K3) The high-magnification images of the boxed area in (J). Two EGFP+/EdU+ (white arrowheads) cells and an EGFP+ radial process (open white arrowhead in (J)) are found underneath the injury site in this clone. (L–N2) Representative 1 cell (L–L2), 2 cells (M–M2) and 3 cells clones (N–N2) derived from single RG in response to the stab injury. In these clones, cells are EGFP+/EdU+ newborn cells (white arrowheads). (O and O1) The size distribution of collected 29 clones. 2-cells clones (20/29) are the most abundant clones. White dashed lines represent the tectal ventricle boundary. A, anterior; P, posterior; D, dorsal; V, ventral; L, lateral; Tel, telencephalon; OT, optic tectum; Ce, cerebellum; SC, spinal cord; TAM, tamoxifen; RG, radial glia; TeO, tectum opticum; TPZ, tectal proliferation zone; PGZ, periventricular gray zone; TS, torus semicircularis; Val, valvula cerebelli. Scale bars, 100 μm (A); 50 μm (B-B3, D-G, and J); 20 μm (K–K3); and 10 μm (L–N2).
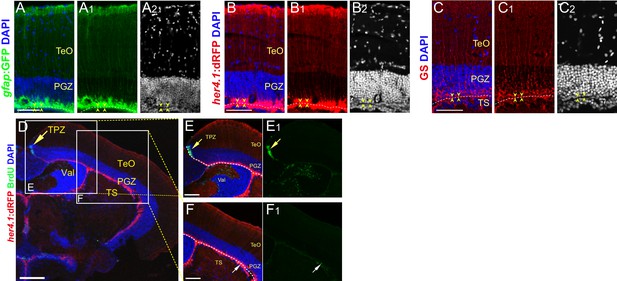
Tectal RG are largely dormant under physiological conditions.
(A–C2) Representative images of Tg(gfap:GFP) (A-A2, green), Tg(her4.1:dRFP) (B-B2, red) and GS (C-C2, red) immunofluorescences showing the RG extend their processes into the superficial neuropils of the optic tectum. The yellow arrowheads indicate the boundary of PGZ and TS. (D–F1) Tg(her4.1:dRFP) (red) and BrdU (green) immunofluorescences show that BrdU signal is restricted to tectal proliferation zone (TPZ) (yellow arrows in (D–E1)) and is absent from the RG layer, very few RG (white arrows in F-F1) is labeled. The fish are administrated with BrdU for one day before analysis. (E–F1) The high-magnification images of the boxed areas shown in (D). White dashed lines represent the boundary of tectal ventricle. RG, radial glia; TeO, tectal opticum; TPZ, tectal proliferation zone; PGZ, periventricular gray zone; Val, valvula cerebelli. Scale bars, 100 μm (D); 50 μm (A–C2, E–F1).
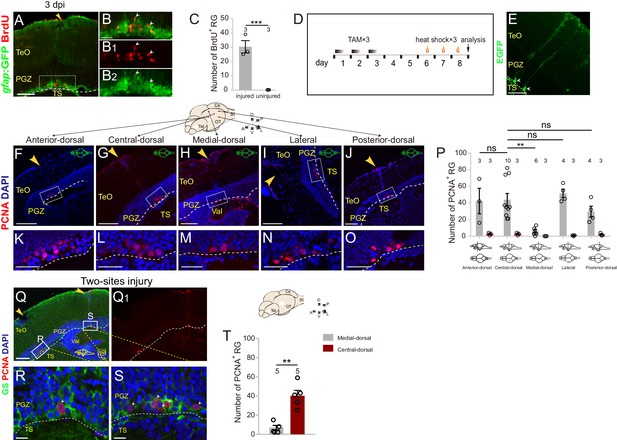
Injury responses of RG in different geographical regions in the optic tectum.
(A–B2) Representative images of Tg(gfap:GFP) (green) and BrdU (red) immunofluorescences showing RG underneath the injury site enter the S phase at 3 dpi. After the injury, the fish are administrated with BrdU for 1 day (2-3dpi) and analyzed at 3 dpi. (B–B2) The high-magnification images of boxed area shown in (A). White arrowheads (in B-B2) indicate the BrdU+/GFP+ RG underneath the injury site. (C) Quantification of BrdU+ RG in (A–B3) showing 30 ± 4 RG enter the S phase. (mean ± SEM, ***p<0.001; Wilcoxon test). (D) Experimental time courses of Cre-loxP-based single RG genetic labeling shown in (E). Double transgenic fish are administrated with tamoxifen (TAM) for three consecutive days and analyzed at day 8, before sacrifice the fish are heat shocked for 3 times (one hour per day) to induce EGFP expression in recombined cells. (E) Representative image of sparsely labeled tectal RG (EGFP+ green cells, white arrows) at day 8. (F–O) Representative images of PCNA (red) immunofluorescence showing injury induces proliferation of RG underneath the injury site at 3 dpi in different regions of optic tecta. (K–O) The high-magnification images of boxed area in (F–J). (P) Quantification of PCNA+ cells in (F–O). Across the five regions, only the RG in the medial-dorsal region shows low proliferation capacity after injury (mean ± SEM; **p<0.01; ns, p>0.05; one-way ANOVA followed by Tukey’s HSD test). (Q–S) Representative images of PCNA (red) and GS (green) immunofluorescences showing that injury induces less proliferation of RG (white arrowhead in (R)) in the medial-dorsal region compared with the RG (white arrowheads in (S)) in the central-dorsal region of a two-sites injured optic tectum. (R and S) The high-magnification images of boxed areas in (Q). (T) Quantification of PCNA+/GS+ RG in (R and S). Injury induces significant more RG proliferation in the central-dorsal region than in the medial-dorsal region (mean ± SEM, **p<0.01; Wilcoxon test). The numbers above the bars indicate the animals used. White dashed lines indicate the boundary of the tectal ventricle. Yellow arrowheads indicate the injury sites. A, anterior; P, posterior; D, dorsal; V, ventral; L, lateral; Tel, telencephalon; OT, optic tectum; Ce, cerebellum; SC, spinal cord; RG, radial glia; TeO, tectal opticum; PGZ, periventricular gray zone; TS, torus semicircularis; Val, valvula cerebelli. Scale bars, 100 μm (F–J and Q); 50 μm (A and E); 30 μm (F–J); 20 μm (K–O); and 10 μm (R and S).
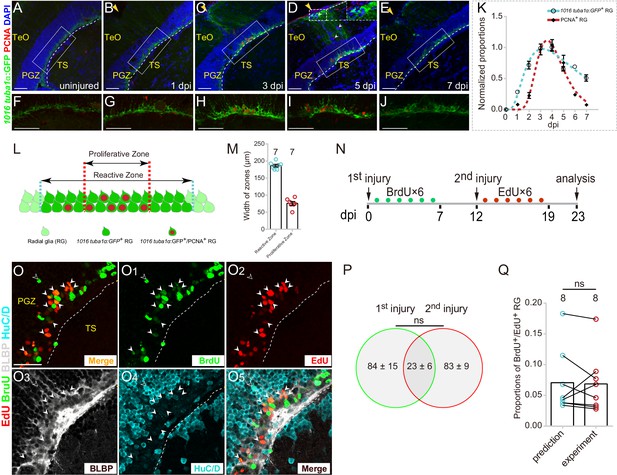
Injury-reactivated RG enter the cell cycle in a stochastic manner.
(A–J) Low- and high-magnification immunofluorescence images showing the dynamics of GFP and PCNA of RG underneath the injury sites (yellow arrows) in the optic tecta of the Tg(1016tuba1α:GFP) fish throughout 0 to 7 dpi. At 3 and 5 dpi, GFP is strongly upregulated in the cell body and radial processes (white dashed box in (D)). Some GFP−/PCNA+ cells (white arrowheads in (D)) are non-RG proliferative cells induced by injury. (F–J) The high-magnification images of boxed areas in (B–E). See also Figure 2—figure supplement 1D-H3. (K) Dot plots showing the change of normalized proportions of GFP+ (open circles, the cyan dashed line representing a fitting curve) and PCNA+ (solid diamonds, the red dashed line representing a fitting curve) RG throughout 0 to 7 dpi. The numbers are fitted by lognormal nonlinear regression. (n = 3, 3, 3, 3, 3, 3, 3, 3 for GFP; n = 3, 4, 4, 10, 9, 3, 3, 2 for PCNA). See also Figure 2—figure supplement 1C. (L and M) Schematic representation and quantification of the physical distribution of Reactive and Proliferative Zones in the injured optic tectum of Tg(1016tuba1α:GFP) fish. GFP is upregulated in RG in Reactive Zone and Proliferative Zone, 88 ± 3% (n = 7, mean ± SEM) proliferative RG (PCNA+/GFP+) are located in Proliferative Zone. (N) Experimental time course of the sequential injury experiment shown in (O–O5). Six consecutive days of BrdU (green dots) are injected after the first injury, and the second injury was applied at 12 dpi followed by EdU injections for six consecutive days (red dots), and finally, fish were analyzed at 23 dpi. (O–O5) BLBP (gray), HuC/D (cyan), BrdU (green) and EdU (red) immunofluorescences show that proliferative RG that respond to the stab injury of either the first time (BrdU+/BLBP+, green cells) or the second time (EdU+/BLBP+, red cells) are distinct but overlapping. RG that respond to both injuries are BrdU+/EdU+/BLBP+ (yellow cells) indicated by white arrowheads. The open white arrowheads indicate a newborn neuron (BrdU+/HuC/D+) generated from the RG respond to the first injury. Venn diagram of the number of RG that enter the cell cycle in a sequential injury experiment. No significant difference is shown between the number of RG induced by the first and the second injury (mean ± SEM; ns, p>0.05; Wilcoxon test). The predicted and experimental proportions of RG entering cell cycle in response to the injury of both times showing no significant difference. The prediction is derived from the multiplication of the reactivation probabilities of either injury, with the assumption of stochastic cell-cycle entry of reactive RG (mean ± SEM; ns, p>0.05; Wilcoxon test). The numbers above the bars indicate the animals used. White dashed lines represent the tectal ventricle boundary. RG, radial glia; TeO, tectum opticum; PGZ, periventricular gray zone; TS, torus semicircularis. Scale bars, 50 μm (A–J); and 30 μm (O–O5). See also Figure 2—figure supplement 1.
-
Figure 2—source data 1
Quantification of the number of RG that enter the cell cycle in the sequential injury experiment.
- https://doi.org/10.7554/eLife.48660.008
-
Figure 2—source data 2
The predicted and experimental proportions of RG entering cell cycle in response to the injury of both times.
- https://doi.org/10.7554/eLife.48660.009
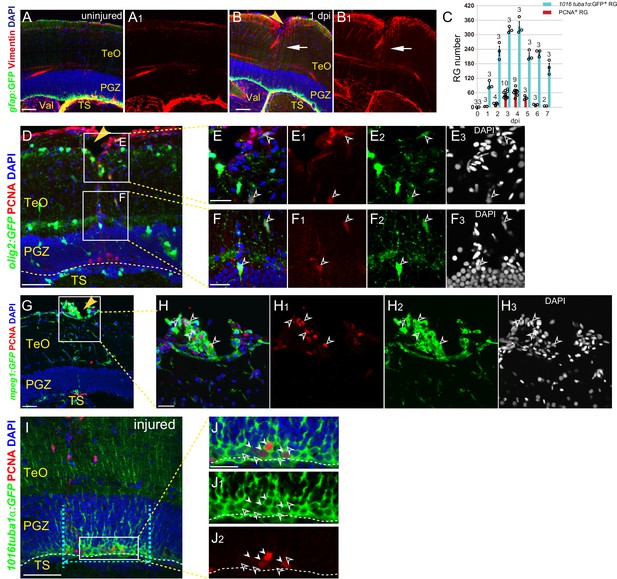
Injury induces proliferation of other cell types.
(A–B1) Representative images of Tg(gfap:GFP) (green) and Vimentin (red) immunofluorescences showing that stab injury results in up-regulation of glial markers Vimentin (red, white arrows in (B and B1)) in RG underneath the injury site compared with the RG in control hemisphere at 1 dpi. (C) Quantification of the RG with robust GFP signal and RG with GFP+/PCNA+ signals underneath the injury site in the optic tecta of the Tg(1016tuba1α:GFP) fish throughout 0 to 7 dpi. (D–F3) Representative images of Tg(olig2:GFP) (green) and PCNA (red) immunofluorescences showing that injury induces proliferation of oligodendrocyte precursor cells in the regions near (E–E3) and underneath (F–F3) the injury site. (E–F3) The high-magnification images of boxed areas shown in (D). Tg(olig2:GFP) is a widely used line that labels oligodendrocytes and their precursor cells in zebrafish. Open white arrowheads in (E–F3) indicate the PCNA+/GFP+ oligodendrocyte precursor cells. (G–H3) Representative images of Tg(mpeg1:GFP) (green) and PCNA immunofluorescences showing injury induces proliferation of microglia/macrophage in the regions near the injury site. (H–H3) The high-magnification images of boxed area shown in (G). Tg(mpeg1:GFP) is a widely used line that labels microglia/macrophage in zebrafish. Open white arrowheads in (H–H3) indicate the PCNA+/GFP+ microglia/macrophage. The GFP-/PCNA+ cells are other proliferative cell types. (I–J2) Injury induces expression of GFP in all of the RG (green, cyan dashed lines) underneath the injury site in the Tg(1016tuba1α:GFP) fish optic tectum, whereas only a subset of the RG express PCNA (white arrowheads in J- J2). (J–J2) The high-magnification images of boxed area shown in (I). Cyan dashed lines indicate the boundary of Reactive Zone; Open white arrowheads indicate PCNA− RG in the Proliferative Zone. The numbers above the bars indicate the animals used. White dashed lines represent the tectal ventricle boundary. RG, radial glia; TeO, tectal opticum; PGZ, periventricular gray zone; TS, torus semicircularis; Val, valvula cerebelli. Scale bars, 50 μm (A-B1, D, G, and I); 20 μm (E1–E3, F1–F3, H–H3 and J–J2).
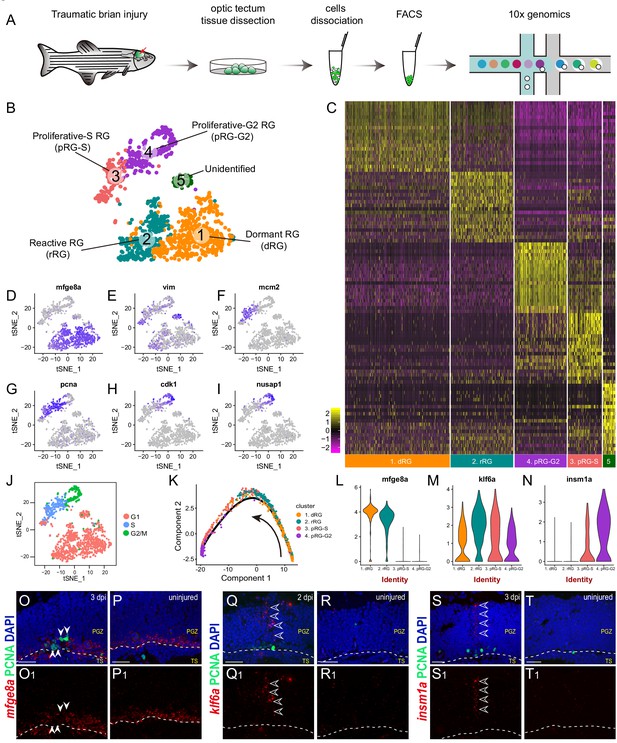
Single-cell RNAseq revealing cellular states underlying the cell-cycle entry of reactive RG.
(A) Workflow for single-cell RNA-seq (scRNA-seq) of tectal RG after stab injury. Optic tecta are dissected from 3 dpi Tg(gfap:GFP) zebrafish brain and dissociated into a single-cell suspension. Single GFP+ RG are sorted by fluorescence-activated cell sorting (FACS) and followed by 10x genomics scRNA-seq. (B) A t-SNE plot of 1174 single tectal RG at 3 dpi revealing 5 cell clusters. Dormant RG (dRG, cluster 1) in orange; Reactive RG (rRG, cluster 2) RG in dark cyan; Proliferative-S RG (pRG-S, cluster 3) in Indian red; Proliferative-G2 RG (pRG-G2, cluster 4) in purple; Unidentified RG (cluster 5) in dark green. (C) Heatmap showing the expression of the top 20 marker genes that characterize each cell clusters. Rows represent genes while columns represent cells. (D–I) t-SNE plots showing expression of state-specific genes of distinct cell states. (J) Cell-cycle characteristics of individual cell states. S phase-related genes are mainly expressed in pRG-S cluster (cluster 3), G2/M-related genes are mainly expressed in pRG-G2 cluster (cluster 4). (K) Pseudo-time developmental trajectory of identified states using Monocle showing that the trajectory is booted from dRG cluster (cluster 1) and end at pRG-G2 cluster (cluster 4). (L–N) Violin plots of expression for genes enriched in dRG cluster (mfge8a, cluster 1), rRG cluster (klf6a, cluster 2) and pRG-S/G2 cluster (insm1a, cluster 3 and 4). (O–T1) In situ hybridization showing the expression of mfge8a (O–P1), klf6a (Q–R1) and insm1a (Q–R1) in the optic tecta after injury. The white arrowheads shown in (O and O1) indicate PCNA+ proliferative RG are mfge8a−, the open white arrowheads indicate klf6a (Q and Q1) or insm1a (S and T1) mRNA signals are located in processes of proliferative RG. White dashed lines represent the tectal ventricle boundary. t-SNE, t-stochastic neighbor embedding; RG, radial glia; PGZ, periventricular gray zone, TS, torus semicircularis. Scale bars, 30 μm. See also Figure 3—figure supplements 1 and 2 and Materials and methods.
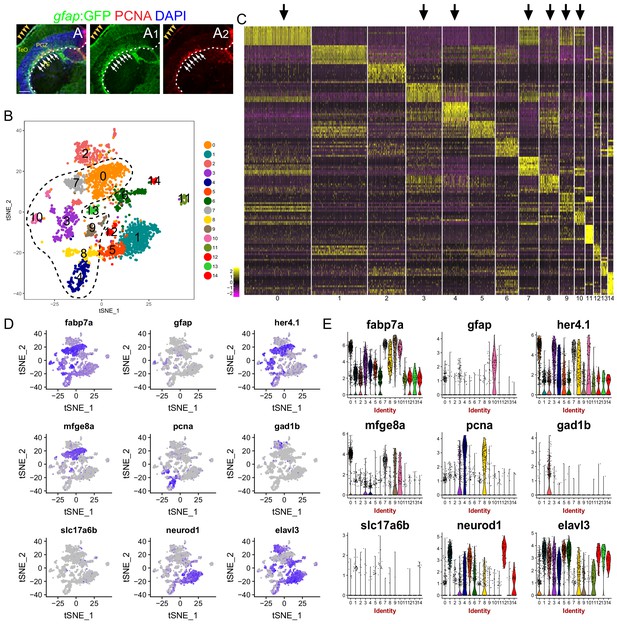
Glial and Non-glial cell clusters identification from the scRNA-seq data.
(A–A2) Tg(gfap:GFP) (green) and PCNA (red) immunofluorescences show large-area injury induces many RG (GFP+/PCNA+ yellow cells, white arrows) to proliferate at 3 dpi. White dashed lines represent the optic tectum boundary. Yellows arrow heads indicate the injury sites. (B) t-SNE plot of 2298 single cells at 3 dpi revealing 15 cell clusters. Black dashed line indicates picked glial cell clusters (cluster 0, 3, 4, 7, 8, 9, 10). (C) Heatmap showing the expression of top 20 marker genes that characterize each cell clusters. Rows represent genes while columns represent cells. Black arrows indicate the picked glial cell clusters (cluster 0, 3, 4, 7, 8, 9, 10). (D) t-SNE plots showing expression of 9 genes utilized to identify neuronal clusters and glial clusters. (E) Violin plots of expression of 9 genes shown in (D). RG, radial glia; TeO, tectal opticum; PGZ, periventricular gray zone; TS, torus semicircularis. Scale bar, 100 μm.
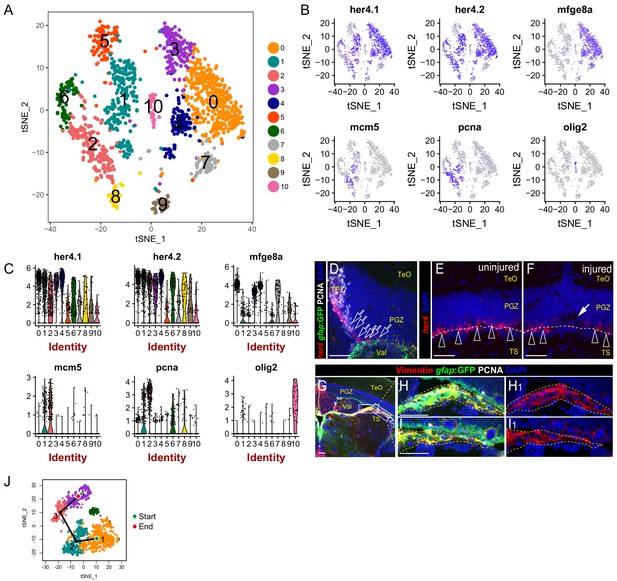
Identification of the clusters representing RG in the TPZ and oligodendrocytes.
(A) t-SNE plot of 1604 glial cells revealing 11 cell clusters. (B) t-SNE plots showing the expression pattern of 6 genes utilized to identify the cluster consists of RG from TPZ (cluster 1) and oligodendrocytes (cluster 10). (C) Violin plots of expression of 6 genes shown in (B). (D–F) In situ hybridization showing her4 mRNA is highly expressed in RG from TPZ (open white arrows in (D)) and dormant RG (open white arrowheads in (E and F)) in central-dorsal region of optic tectum, whereas its expression is down-regulated in RG underneath the injury site ((F), white arrow). White dashed lines represent the tectal ventricle boundary. (G–I1) Representative images of Tg(gfap:GFP) (green), Vimentin (red) and PCNA (white) immunofluorescences showing the RG in the neighboring brain tissues (yellow dashed lines in H-I1) under the PGZ in the midbrain are GFP+/Vimentin+/PCNA−. These cells are likely to cause contamination (cluster 5 in Figure 3B) during the dissection of the optic tecta. (J) Pseudo-time developmental trajectory of identified states using Slingshot showing that the trajectory is booted from dRG cluster (cluster 1) and end at pRG-G2 cluster (cluster 4). RG, radial glia; TeO, tectal opticum; PGZ, periventricular gray zone; TS, torus semicircularis; Val, valvula cerebelli. Scale bars, 50 μm.
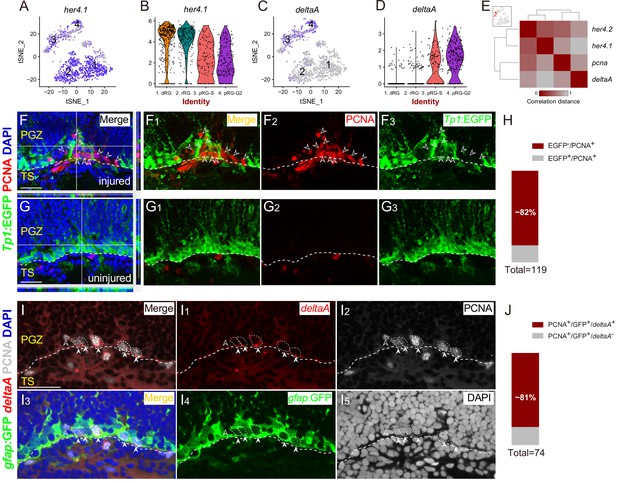
Spatial Distribution of Notch and Delta in reactive RG after the injury.
(A–D) t-SNE plots (A and C) and violin plots (B and D) showing deltaA is mainly expressed in pRG-S (cluster 3) and pRG-G2 (cluster 4) RG, whereas the expression of her4.1, a target gene of Notch signaling, is down-regulated in pRG-S (cluster 3) and pRG-G2 RG (cluster 4). (E) Correlation distance matrix showing deltaA exhibits the high correlation with pcna but the low correlation with her4.1 and her4.2 in pRG-S RG (cluster 3, red dots in the t-SNE plot at the top left, each dot represents a single cell). (F–G3) Tg(Tp1:EGFP) (green) and PCNA (red) immunofluorescences show that PCNA+ proliferative RG (open white arrowheads, red cells) and EGFP+ RG (green cells) are exclusive in the injured optic tectum (F–F3). (G–G3) The representative images of the uninjured optic tectum. (H) Quantification of EGFP−/PCNA+ and EGFP+/PCNA+ RG in (F) showing ~82% PCNA+ RG are EGFP−. Most of the PCNA+ proliferative RG have low Notch activity (97/119 cells in 6 sections from 4 fish). (I–I5) Specific expression of deltaA in PCNA+ tectal RG (white arrowheads, white dashed circles) at 3 dpi is confirmed by in situ hybridization. The open white arrowheads indicate a GFP−/PCNA+/deltaA− non-RG proliferative cells. (J) Quantification of PCNA+/deltaA+ and PCNA+/deltaA− RG in (I) showing ~81% PCNA+ RG express deltaA. (60/74 cells in 10 sections from 5 fish). White dashed lines represent the tectal ventricle boundary. RG, radial glia; PGZ, periventricular gray zone; TS, torus semicircularis. Scale bars, 30 μm.
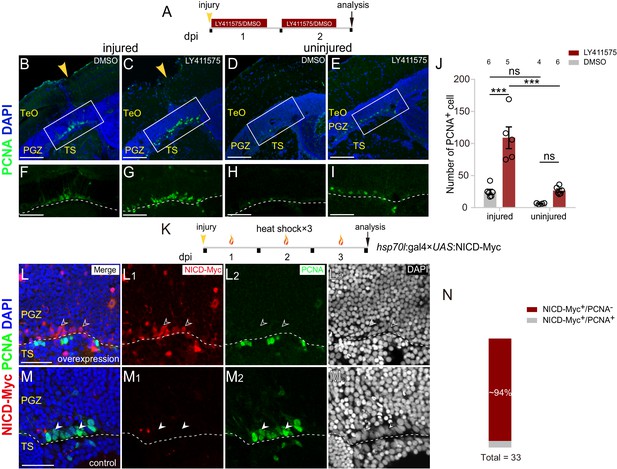
Notch inhibition mediates the proliferation of reactive tectal RG.
(A) Experimental time course of Notch inhibition experiments shown in (B–I). Fish are administrated with LY411575, a Notch inhibitor, or DMSO for two consecutive and are analyzed at 2 dpi. (B–I) LY411575 administration increases the number of proliferative RG (PCNA+, green cells) underneath the injury site in the optic tectum with (B and C) or without injury (D and E). (F–I) The high-magnification images of boxed areas in (B–E). (J) Quantification of PCNA+ cell in (B–E). LY411575 administration significantly increases the number of proliferative RG (PCNA+ green cells) in the optic tectum with or without the injury. Very few RG is proliferative in the uninjured DMSO-treated control optic tectum (mean ± SEM; ***p<0.001, ns, p>0.05; two-way ANOVA followed by Tukey’s HSD test). See also Figure 5—source data 1 for quantification. (K) Experimental time course of heat shock-induced Notch over-activation experiments shown in (L–M3). Tg(hsp70l:gal4 ×UAS:NICD-Myc) fish are injured in the optic tecta and followed by heat shocks for three consecutive days (1 hr per day) and are analyzed at 3 dpi. (L–M3) NICD-overexpressed RG (open white arrowheads, red cells) underneath the injury site are not proliferative after the stab injury whereas RG (white arrowheads, green cells) become proliferative in the control optic tectum with the injury. The expression of NICD-Myc is controlled by the gal4-UAS system. It is a mosaic labeling genetic system so that only a subset of cells could be induced to express NICD-Myc. To avoid obscure the signal, only two representative cells were indicated by arrowheads in (L–M3). See also Figure 5—figure supplement 1C-C3. (N) Quantification of NICD-Myc+/PCNA− and NICD-Myc+/PCNA+ RG in (L–M3) showing ~94% NICD-Myc-overexpressed RG are PCNA−. (31/33 cells in 15 sections of 6 fish). The numbers above the bars indicate the animals used. White dashed lines represent the tectal ventricle boundary. RG, radial glia; TeO, tectum opticum; PGZ, periventricular gray zone; TS, torus semicircularis. Scale bars, 100 μm (B–E); 50 μm (F–I); and 30 μm (L–M3).
-
Figure 5—source data 1
Quantification of PCNA+ cells.
- https://doi.org/10.7554/eLife.48660.016
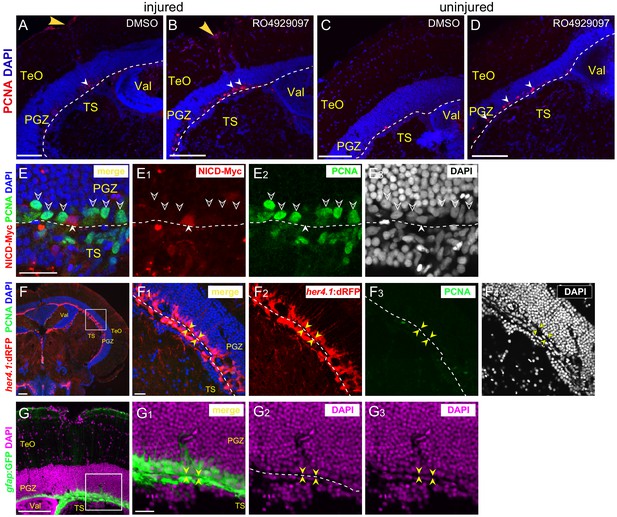
Notch signaling regulates the proliferation of RG.
(A–D) Representative images of PCNA (red) immunofluorescence showing 2 days’ treatment of RO4929097, a putative Notch inhibitor, results in more RG (PCNA+, white arrowheads) proliferation in injured (B) and uninjured (D) optic tecta. (A and C) The representative images of injured and uninjured optic tecta treated with DMSO. (E–E3) Representative images of Myc (red) and PCNA (green) immunofluorescences showing a single RG with NICD-Myc overexpression (red cell, white arrowheads) is lack of PCNA expression (green signals) while many neighboring cells (green cells, open white arrowheads) without NICD-Myc expression are proliferative. This is the most common NICD-Myc and PCNA expression pattern in the heat shock-induced Notch over-activation experiments. (F–F4) Representative images of her4.1:dRFP (red), PCNA (green) and DAPI (gray) immunofluorescences showing that the boundary (white dashed lines and yellow arrowheads) of PGZ and TS could be unambiguously defined by the DAPI signal. The cells under the boundary are her4.1:dRFP+, indicating their glial identity. The lack of PCNA expression in them indicates they are largely dormant. (G–G3) Representative images of gfap:GFP (green) and DAPI (magenta) immunofluorescences showing that the boundary (white dashed lines and yellow arrowheads) of PGZ and TS could be unambiguously defined by the DAPI signal. White dashed lines represent the tectal ventricle boundary. RG, radial glia; TeO, tectal opticum; PGZ, periventricular gray zone; TS, torus semicircularis, Val, valvula cerebelli. Scale bars, 100 μm (A–D, F and G); and 20 μm (E–E3, F1–F4 and G1–G3).
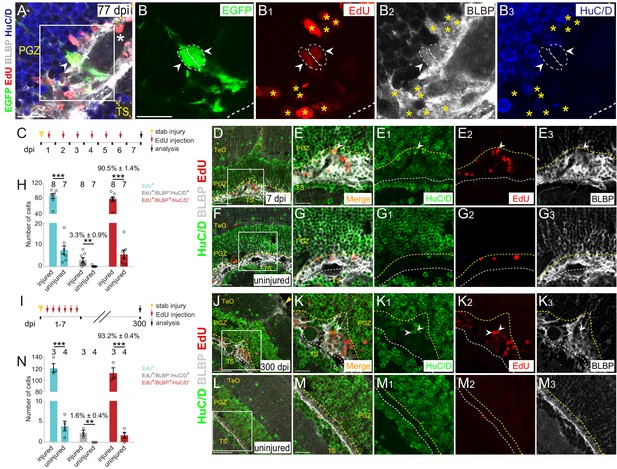
Injury-induced RG are largely undergoing gliogenesis.
(A–B3) Images of a representative 2 cells clone at 77 dpi. Both cells are EdU+/BLBP+/HuC/D− RG (white arrowheads, white dashed circles). The white asterisk in (A) indicates the blood vessel. Yellow asterisks in (B1–B3) indicate other EdU+/BLBP+/HuC/D− RG underneath the injury site at 77 dpi. (B–B3) The high-magnification images of the boxed area in (A) (C) A schematic showing the procedures used for 7 days EdU pulse-and-stain assay. Fish are injected with EdU for six consecutive days after the injury, the injured and uninjured optic tecta are analyzed at 7 dpi. (D–G3) Representative images of EdU (red), BLBP (gray) and HuC/D (green) immunofluorescences showing that most of the newborn cells are EdU+/BLBP+ RG, while a few newborn cells are EdU+/HuC/D+ neurons (white arrowheads). The newborn RG forms a bulge underneath the injury site. (E-E3) The high-magnification images of the boxed area in (D). (F) The representative image of uninjured optic tectum. (G–G3) The high-magnification images of the boxed area in (F). (H) Quantification of EdU+ newborn cells, EdU+/BLBP+/HuC/D− newborn RG and EdU+/BLBP−/HuC/D+ newborn neurons at 7 dpi. The number of EdU+/BLBP−/HuC/D+ newborn neurons on the injured side is significantly increased compared with that on the uninjured side (mean ± SEM; ***p<0.001, **p<0.01, Wilcoxon test). See also Figure 6—source data 1 for quantification. (I) A schematic showing the procedure of EdU pulse-and-staining assay for 300 days long-term tracing. Fish are injected with EdU for six consecutive days after the injury, the injured and uninjured optic tecta are analyzed at 300 dpi. (J–M3) Representative images of EdU (red), BLBP (gray) and HuC/D (green) immunofluorescences showing that EdU+ newborn cells that survive up to 300 dpi are largely EdU+/BLBP+ newborn RG, but a few cells are EdU+/HuC/D+ newborn neurons (white arrowheads). (K–K3) The high-magnification representative images of the boxed area in (J). (L) The representative image of uninjured optic tectum. (M–M3) The high-magnification representative images of the boxed area in (L). (N) Quantification of EdU+ newborn cells, EdU+/BLBP+/HuC/D− newborn RG and EdU+/BLBP−/HuC/D+ newborn neurons at 300 dpi (mean ± SEM, ***p<0.001, *p<0.05, Wilcoxon test). See also Figure 6—source data 2 for quantification. The numbers above the bars indicate the animals used. White dashed lines represent the tectal ventricle boundary. Yellow dashed lines indicate the boundary of bulges. RG, radial glia; TeO, tectum opticum; PGZ, periventricular gray zone; TS, torus semicircularis. Scale bars, 20 μm (A-B3, E-E3, G-G3, K-K3, and M-M3); 50 μm (D, F, J and L).
-
Figure 6—source data 1
Quantification of EdU+, EdU+/BLBP-/HuC/D+ and EdU+/BLBP+/HuC/D- cells at 7 dpi.
- https://doi.org/10.7554/eLife.48660.019
-
Figure 6—source data 2
Quantification of EdU+, EdU+/BLBP-/HuC/D+ and EdU+/BLBP+/HuC/D- cells at 300 dpi.
- https://doi.org/10.7554/eLife.48660.020
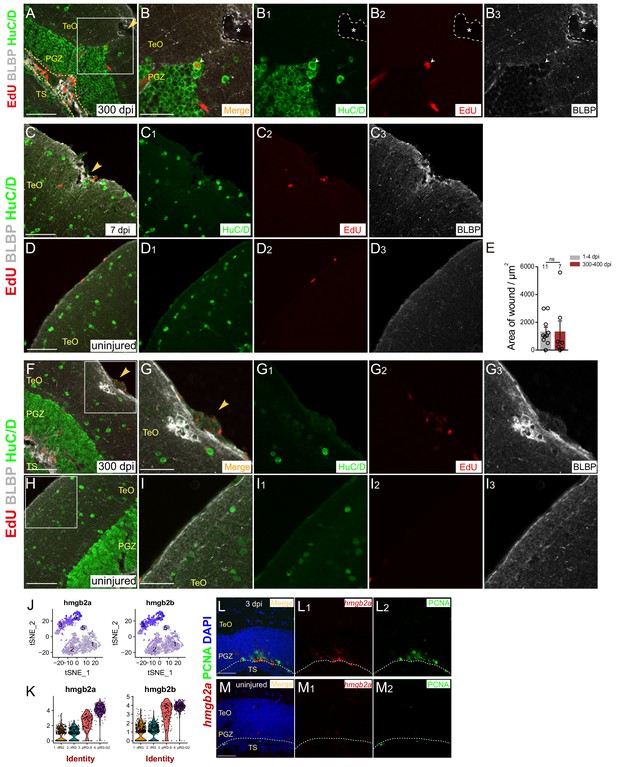
The injury wounds are failed to be restored.
(A–B3) Representative images of EdU (red), HuC/D (green) and BLBP (gray) immunofluorescences showing a EdU+/HuC/D+ neuron migrates to the upper region of injured optic tectum at 300 dpi. (B–B3) The high-magnification images of boxed area in (A). White arrowheads indicate the EdU+/HuC/D+ neuron. Yellow dashed lines indicate the RG layer. White dashed lines and white asterisks indicate the stab wound that is not restored at 300 dpi. (C–D3) Representative images of EdU (red), HuC/D (green) and BLBP (gray) immunofluorescences showing the processes terminal of RG near the injury site are hypertrophic with high level of BLBP expression at 7 dpi. (D–D3) The representative images of uninjured optic tectum. (E) Quantification of the area of stab wounds at 1–4 (1343 ± 315.7 μm2) and 300–400 dpi (1339 ± 768.6 μm2) showing that the stab wounds are not completely restored (mean ± SEM, ns, p>0.05; Wilcoxon test). (F–I3) Representative images of EdU (red), HuC/D (green) and BLBP (gray) immunofluorescences showing stab wound that is surrounded by hypertrophic RG processes terminal (high level of BLBP expression, gray signals) is remained at 300 dpi. (H–H3) are the representative images of uninjured optic tectum. (G–G3) and (I–I3) The high-magnification images of boxed areas in (F) and (H), respectively. (J and K) t-SNE and violin plots showing that hmgb2a and hmgb2b are enriched in proliferative-S and -G2 RG. (L–M2) In situ hybridization showing PCNA+ RG (green cells) express hmgb2a (red signals) at 3 dpi. (M–M2) are the representative images of uninjured optic tectum. Yellow arrow heads indicate the injury site. RG, radial glia; TeO, tectal opticum; PGZ, periventricular gray zone; TS, torus semicircularis; Val, valvula cerebelli. Scale bars, 50 μm (A, F and H); 30 μm (B–B3, C–C3, D–D3, G–G3, I–I3 and L–M2).
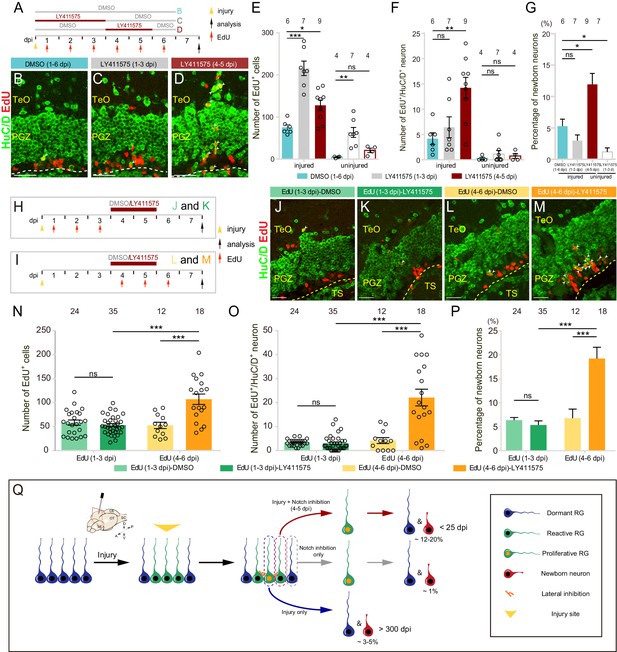
Notch inhibition promotes the neurogenesis of reactive tectal RG.
(A) A schematic of Notch inhibition experiments shown in (B–D). In the control group, fish are administrated with DMSO from 1 to 6 dpi. In experimental groups, fish are administrated with LY411575 during either 1 to 3 dpi or 4 to 5 dpi. EdU is injected for six consecutive days after the injury. All the fish are sacrificed and analyzed at 7 dpi. (B–D) Representative images of HuC/D (green) and EdU (red) immunofluorescences of 7-dpi optic tecta treated with DMSO for 1–6 dpi, LY411575 for 1–3 dpi, or LY411575 for 4–5 dpi showing that significant more EdU+/HuC/D+ newborn neurons are only generated after the treatment of LY411575 during 4–5 dpi. White arrowheads indicate EdU+/HuC/D+ newborn neurons. (E–F) Quantification of EdU+ newborn cells (E) and EdU+/HuC/D+ newborn neurons (F) in (B–D). While Notch inhibition of 1–3 dpi or 4–5 dpi significantly increases the number of EdU+ newborn cells in the injured optic tectum, Notch inhibition during 4–5 dpi but not 1–3 dpi significantly increases the number of EdU+/HuC/D+ newborn neurons in the injured optic tectum. In the uninjured optic tecta, Notch inhibition during both 1–3 dpi and 4–5 dpi increases the number of EdU+ newborn cells, but not EdU+/HuC/D+ newborn neurons (mean ± SEM, ***p<0.001, **p<0.01, *p<0.05, ns, p>0.05; one-way ANOVA followed by Tukey’s HSD test). See also Figure 7—source datas 1 and 2 for quantification. (G) Proportion of EdU+/HuC/D+ newborn neurons to EdU+ newborn cells in (B–D). Notch inhibition during 4–5 dpi increases the proportion of the neuron production, whereas Notch inhibition during 1–3 dpi decreases the proportion (mean ± SEM, **p<0.01; ns, p>0.05; one-way ANOVA followed by Tukey’s HSD test). See also Figure 7—source data 3 for quantification. (H and I) Schematics of the experimental procedure for Notch inhibition experiments shown in (J-M). After the injury, fish are treated with either DMSO or LY411575 during 4–5 dpi and are injected with EdU for three consecutive days during 1–3 dpi (H) or 4–6 dpi (I). All the fish are sacrificed and analyzed at 7 dpi. (J–M) Representative images of HuC/D (green) and EdU (red) immunofluorescences of the 7-dpi optic tecta after the treatment in (H and I). With the treatment of LY411575 during 4–5 dpi, EdU pluses during 4–6 dpi (L and M) but not 1–3 dpi (J and K) label significant more newborn neurons. White arrowheads indicate EdU+/HuC/D+ newborn neurons. (N and O) Quantification of EdU+ newborn cells (N) and EdU+/HuC/D+ newborn neurons (O) in (J–M) (≥3 replicates for each group; mean ± SEM, ***p<0.001, ns, p>0.05; two-way ANOVA followed by Tukey’s HSD test). See also Figure 7—source datas 4 and 5 for quantification. (P) Proportion of EdU+/HuC/D+ newborn neurons to EdU+ newborn cells in (J–M). EdU pulses during 4–6 dpi significantly increase the proportion of neuron production (≥3 replicates for each group; mean ± SEM, ***p<0.001; ns, p>0.05; two-way ANOVA followed by Tukey’s HSD test). See also Figure 7—source data 6 for quantification. (Q) Schematic summary of the working model. Injury induces all RG underneath the injury site to become reactive. Only ~25% of reactive RG enter the cell cycle and become proliferative. The cell-cycle entry of reactive RG is regulated by Notch/Delta lateral inhibition. In the injury condition, proliferative RG largely undergo gliogenesis (~3–5% newborn neurons). The resulting newborn cells could survive up to 300 dpi. In the Notch inhibition condition, dormant RG can become proliferative but only generate ~1% of newborn neurons. However, Notch inhibition during 4–5 dpi drives reactive RG into the cell cycle, giving rise to significant more neurons (~12–20%). Interestingly, these over-produced neurons are largely diminished by 25 dpi. The numbers above the bars indicate the animals used. Yellow dashed lines represent the tectal ventricle boundary. RG, radial glia; TeO, tectum opticum; PGZ, periventricular gray zone; TS, torus semicircularis. Scale bars, 30 μm (B–D); 20 μm (J–M).
-
Figure 7—source data 1
Quantification of EdU+ newborn cells.
- https://doi.org/10.7554/eLife.48660.023
-
Figure 7—source data 2
Quantification of EdU+/HuC/D+ newborn neurons.
- https://doi.org/10.7554/eLife.48660.024
-
Figure 7—source data 3
Percentage of EdU+/HuC/D+ newborn neurons.
- https://doi.org/10.7554/eLife.48660.025
-
Figure 7—source data 4
Quantification of EdU+ newborn cells.
- https://doi.org/10.7554/eLife.48660.026
-
Figure 7—source data 5
Quantification of EdU+/HuC/D+ newborn neurons.
- https://doi.org/10.7554/eLife.48660.027
-
Figure 7—source data 6
Percentage of EdU+/HuC/D+ newborn neurons.
- https://doi.org/10.7554/eLife.48660.028
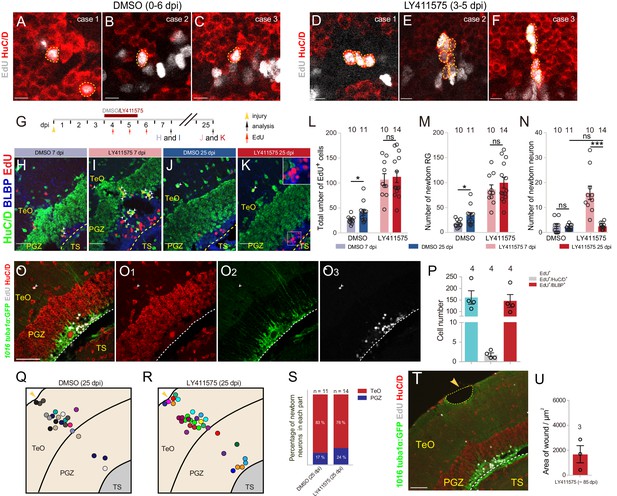
Late Notch inhibition-induced over-produced neurons are short-lived.
(A–F) Representative images of EdU (gray) and HuC/D (red) immunofluorescences showing the over-produced neurons in the optic tecta of the fish treated with LY411575 during 4–5 dpi (D–F) always shown as a cell cluster. (A–C) are the representative images of DMSO-treated optic tecta. Yellow dashed circles indicate the newborn neurons. (G) A schematic of the experimental procedure for Notch inhibition experiments shown in (H–K). Fish are injected with EdU for three consecutive days (4–6 dpi) and administrated with LY411575 or DMSO for two consecutive days (4–5 dpi). Fish are analyzed at 7 dpi or 25 dpi. (H–K) Representative images of HuC/D (green), BLBP (blue) and EdU (red) immunofluorescences showing that many newborn neurons (white arrowheads in (I)) are existed in Notch-inhibited (4–5 dpi) optic tectum at 7 dpi, but only a few newborn neurons (white arrowheads in (K)) are still remained at 25 dpi. (H and J) The representative images of DMSO-treated control optic tecta at 7 and 25 dpi, respectively. (L–N) Quantification of EdU+ newborn cells (L), EdU+/BLBP+ newborn RG (M), and EdU+/HuC/D+ newborn neuron (N) in (H–K) (≥3 replicates for each group; mean ± SEM,***p<0.001; *p<0.05; ns, p>0.05; Wilcoxon test in (L and M), two-way ANOVA followed by Tukey’s HSD test in (N)). (O–O3) Representative images of Tg(1016tuba1α:GFP) (green), EdU (gray) and HuC/D (red) immunofluorescences showing the remaining neuron (white arrowheads) in the optic tectum of the fish treated with LY411575 during 4–5 dpi can survive up to 86 dpi if it is not eliminated by 25 dpi. (P) Quantification of the EdU+ newborn cells, EdU+/HuC/D+ newborn neurons and EdU+/BLBP+ RG in (O–O3) (mean ± SEM, n = 4). (Q and R) Schematic diagrams of the distribution of newborn neurons in the injured 25-dpi optic tecta of the fish treated with DMSO or LY411575 during 4–5 dpi. Circles in different colors represent the newborn neurons from different individuals. (S) Proportion of the newborn neurons in TeO and PGZ of the optic tectum shown in (Q and R). Most of the neurons are existed in the TeO. (T) Representative images of EdU (gray) and HuC/D (red) immunofluorescences showing Notch inhibition during 4–5 dpi does not help to complete restoration of stab wound of the injured optic tectum. Yellow dashed circle indicate the stab wound. (U) Quantification of the area of stab wounds from the ~85 dpi fish treated with LY411575 during 4–5 dpi (1670 ± 704 μm2; mean ± SEM, n = 3). The numbers above the bars indicate the animals used. Yellow arrow heads indicate the injury sites. White dashed lines represent the tectal ventricle boundary. RG, radial glia; PGZ, periventricular gray zone; TeO, tectal opticum; TS, torus semicircularis. Scale bars, 50 μm (O–O3 and T); 20 μm (H–K); 5 μm (A–F).
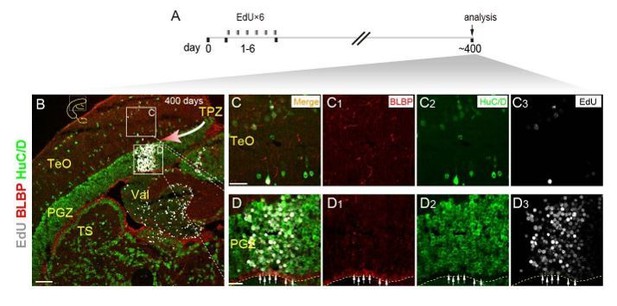
Neurogenic potential of progenitor cells in TPZ of adult zebrafish.
(A) Experimental time courses of long-term tracing of RG in TPZ (panel B-D3). The fish are injected with EdU for 6 consecutive days and analyzed at day 400. (B-D3) Representative images of EdU (red), BLBP (gray) and HuC/D (green) immunofluorescences showing newborn cells in a column migrate toward (red arrow) the central region of optic tectum. Most of the newborn cells are neurons, only the deepest layer of newborn cells become new RG (white arrows in D-D3). Yellow dashed lines indicate the boundary of tectal ventricle. Yellow dashed lines indicate the tectal ventricle boundary. RG, radial glia; TeO, tectal opticum; PGZ, periventricular gray zone; TS, torus semicircularis; Val, valvula cerebelli. Scale bars, 100 μm (B); 20 μm (F-D3).
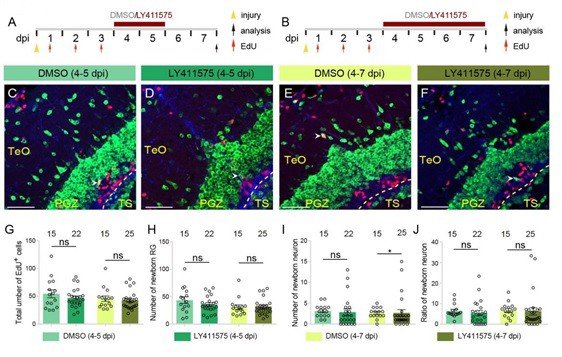
4-days’ Notch inhibition results in a significant decrease in the number of newborn neurons after injury.
(A and B) Schematics of the experimental procedure for Notch inhibition experiments shown in (C-F). After the injury, fish are treated with LY411575 during either 4-5 dpi (A) or 4-7 dpi (B), and are injected with EdU for 3 consecutive days during 1-3 dpi. Control fish are treated with DMSO. All the fish are sacrificed and analyzed at 7 dpi. (C-I) Representative images of HuC/D (green), BLBP (blue) and EdU (red) immunofluorescences of the 7-dpi optic tecta after the treatments in (A and B). In both LY411575-treated and DMSO-treated optic tecta, only few newborn neuron (white arrowheads) is observed. (G-I) Quantifications of EdU+ newborn cells, EdU+/BLBP+ newborn RG and EdU+/HuC/D+ newborn neurons in (C-F). Neither 2-days’ nor 4-days’ Notch inhibition changes the number of newborn cells or newborn RG. 2-days’ Notch inhibition results in a decrease tendency of neuron production, whereas 4-days’ Notch inhibition significantly reduced neuron production (4 replicates for each group; mean ± SEM, ns, not significant, *p < 0.05; Wilcoxon test). (M) Ratios of EdU+/HuC/D+ newborn neurons to EdU+ newborn cells in (C-F). Neither 4-days’ nor 2-days’ Notch inhibition significantly changes the proportion of newborn neurons. (4 replicates for each group; mean ± SEM; ns, not significant; Wilcoxon test). The numbers above the bars indicate the animals used. Yellow dashed lines represent the tectal ventricle boundary. RG, radial glia; TeO, tectum opticum; PGZ, periventricular gray zone; TS, torus semicircularis. Scale bars, 30 μm. See also Figure 7 in main text.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Danio rerio) | gfap:EGFP | Bernardos and Raymond, 2006 | ZDB-FISH-150901–29307 | Tg(gfap:EGFP)mi2001 |
| Strain, strain background (Danio rerio) | her4.1:dRFP | Yeo et al., 2007 | ZDB-TGCONSTRCT-070612–2 | Tg(her4.1:dRFP) |
| Strain, strain background (Danio rerio) | 1016tuba1α:GFP | PMID: 16763038 | ZDB-GENO-070321–4 | Tg(1016tuba1α:GFP) |
| Strain, strain background (Danio rerio) | olig2:GFP | Shin et al., 2003 | ZDB-ALT-041129–8 | Tg(olig2:GFP) |
| Strain, strain background (Danio rerio) | mpeg1:GFP | Ellett et al., 2011 | ZDB-TGCONSTRCT-170801–5 | Tg(mpeg1:GFP) |
| Strain, strain background (Danio rerio) | hsp70l:gal4 | Scheer et al., 2001 | ZDB-TGCONSTRCT-070117–42 | Tg(hsp70l:gal4) |
| Strain, strain background (Danio rerio) | UAS:NICD-Myc | Scheer et al., 2001 | ZDB-TGCONSTRCT-070117–24 | Tg(UAS:NICD-Myc) |
| Strain, strain background (Danio rerio) | Tp1bglob:EGFP | This paper | Tg(Tp1bglob:EGFP) is generated using the plasmid from Dr. Nathan Lawson | |
| Strain, strain background (Danio rerio) | her4.1:mCherryT2ACreERT2 | This paper | Tg(her4.1:mCherryT2ACreERT2) is generated using the plasmid from Dr. Micheal Brand | |
| Strain, strain background (Danio rerio) | hsp70l:DsRed2(floxed)EGFP | This paper | Tg(her4.1:mCherryT2ACreERT2) is generated using the plasmid from Dr. Micheal Brand | |
| Antibody | Mouse monoclonal anti-PCNA | Abcam | Cat#Ab29 RRID:AB_303394 | 1:1000 |
| Antibody | Rabbit polyclonal anti-GFPtag | Abcam | Cat# ab13970 RRID:AB_300798 | 1:500 |
| Antibody | Chicken monoclonal anti-GFP | Proteintech Group | Cat#50430–2-AP RRID:AB_11042881 | 1:2000 |
| Antibody | Rabbit polyclonal anti-DsRed2 | Takara Bio | Cat#632496 RRID:AB_10013483 | 1:1000 |
| Antibody | Mouse monoclonal anti-HuC/D | Invitrogen | Cat#A21271 RRID:AB_221448 | 1:1000 |
| Antibody | Rabbit polyclonal anti-BLBP | Abcam | Cat#ab32423 RRID:AB_880078 | 1:1000 |
| Antibody | Rat monoclonal anti-BrdU | Abcam | Cat#Ab6326 AB_305426 | 1:1000 |
| Recombinant DNA reagent | pTol2-Tp1bglob:EGFP | Quillien et al., 2014 | Addgene plasmid #73586 RRID:Addgene_73586 | Dr. Nathan Lawson (UMass Medical School, Worcester, USA) |
| Recombinant DNA reagent | pTol2-her4.1:mCherryT2ACreERT2 | Kroehne et al., 2011 | Dr. Michael Brand (Technische Universität Dresden, Dresden, Germany) | |
| Recombinant DNA reagent | pTol2-hsp70l:DsRed2(floxed)EGFP | Kroehne et al., 2011 | Dr. Michael Brand (Technische Universität Dresden, Dresden, ermany) | |
| Recombinant DNA reagent | pBS:dlA clone 4 | PMID: 9425133 | Dr. Judith S. Eisen (University of Oregon, Eugene, USA) | |
| Commercial assay or kit | Single Cell 3' Library and Gel Bead kit v2 Chip kit | 10x Genomics | 120237 | |
| Commercial assay or kit | dsDNA High Sensitivity Assay Kit | AATI | DNF-474–0500 | |
| Commercial assay or kit | High Sensitivity Large Fragment −50 kb Analysis Kit | AATI | DNF-464 | |
| Commercial assay or kit | MEGAscriptTM T7 High Yield Transcription Kit | Invitrogen | AM1334 | |
| Commercial assay or kit | Click-iT EdU imaging kit | Invitrogen | C10340 | |
| Commercial assay or kit | DIG RNA labeling kit | Roche | 11277073910 | |
| Chemical compound, drug | Papain | Worthington Biochemical Corporation | LS003126 | 100 µl in 5 ml DEME/F12 |
| Chemical compound, drug | Tamoxifen | Sigma | T5648 | 2.5–5 µM |
| Chemical compound, drug | LY411575 | Selleck Chemical | S2714 | 10 µM |
| Software, algorithm | Cell Ranger Single Cell Software Suite (v2.1.0) | 10x genomics | https://support.10xgenomics.com | |
| Software, algorithm | Seurat R package | Satjia lab | http://satijalab.org/seurat/ | |
| Software, algorithm | R 3.5.1 | R-project | https://www.r-project.org/ | |
| Software, algorithm | GraphPad Prism | GraphPad Software | www.graphpad.com | |
| Software, algorithm | FIJI | PMID: 22743772 | http://fiji.sc/ | |
| Software, algorithm | FV10-ASW 4.0 Viewer | Olympus | www.olympus-global.com |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.48660.029





