Adaptation of hydroxymethylbutenyl diphosphate reductase enables volatile isoprenoid production
Figures
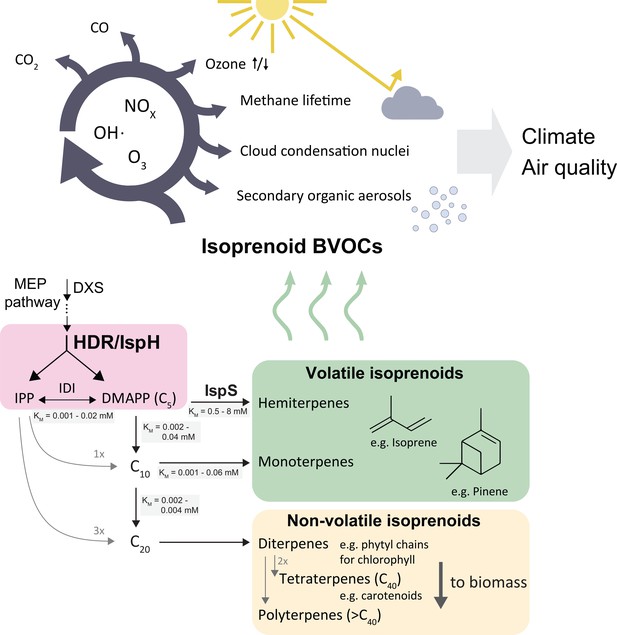
Simplified scheme of the plastidic MEP pathway, important volatile isoprenoids, and their atmospheric reactions.
The MEP pathway makes IPP and DMAPP simultaneously through the action of HDR (pink box), and produces the bulk of volatile isoprenoids, contributing >80 % of total BVOCs (Sindelarova et al., 2014) . Non-volatile isoprenoids are essential and synthesised by all organisms, while volatile isoprenoid production is non-essential and highly species-dependent. The cytosolic MVA pathway contributes most sesquiterpenes (<3 % of BVOCs), but is omitted here for clarity. Emitted volatile isoprenoids are rapidly oxidised, resulting in complex atmospheric photochemistry impacting aerosol and cloud condensation nuclei formation, extension of methane residence time, ozonolysis as well as surface-level ozone formation in the presence of mono-nitrogen oxide (NOx) pollutants (Wennberg et al., 2018). BVOCs, biogenic organic volatile compounds; DMAPP, dimethylallyl pyrophosphate; DXS, deoxyxylulose synthase; IDI, isopentenyl diphosphate isomerase; IPP, isopentenyl pyrophosphate; IspS, isoprene synthase; HDR, hydroxymethylbutenyl diphosphate reductase.
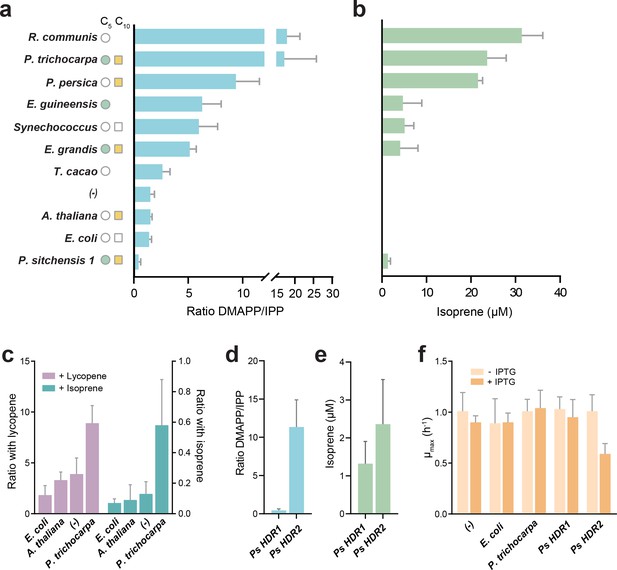
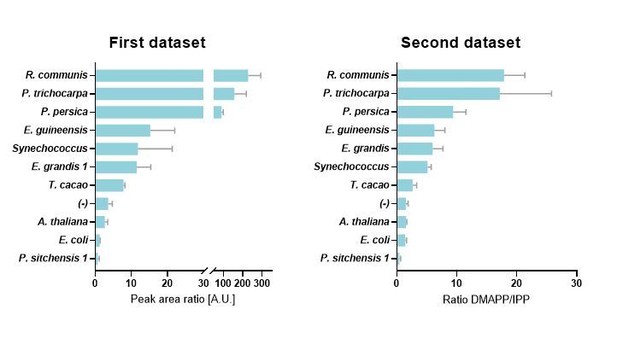
DMAPP:IPP ratio and isoprene production with different HDR enzymes.
(a) In vivo ratio of DMAPP:IPP measured via LC-MS/MS in E. coli overexpressing HDR genes from different species, in the genetic context of dxs and lycopene biosynthetic pathway overexpression. Filled circles and squares indicate that the HDR source species natively emits C5 or C10 isoprenoids. Open symbols indicate no emission, and no symbol indicates no data or conflicting data. (b) Isoprene production in E. coli when the HDR enzymes shown in panel (a) are overexpressed with dxs and an isoprene synthase. (c) Comparison of DMAPP:IPP ratios between selected HDRs co-expressed with dxs and with expression of either lycopene or isoprene as the metabolic sink. (d) Comparison of DMAPP:IPP ratios in E. coli overexpressing Picea sitchensis (Ps) HDR1 or HDR2 in the context of dxs and lycopene biosynthetic pathway overexpression. (e) Isoprene production in E. coli overexpressing P. sitchensis HDR1 or HDR2 along with dxs and an isoprene synthase. (f) The maximum specific growth rate (µmax) of E. coli expressing selected HDRs in the context of dxs and lycopene biosynthetic pathway overexpression, with or without induction of HDR expression by addition of IPTG. All data shown as mean ± SD from > 3 biological replicates; (-) indicates the control strain without HDR overexpression.
-
Figure 2—source data 1
Raw data for metabolomics, proteomics and isoprene measurements shown in Figure 2 and supplements.
- https://cdn.elifesciences.org/articles/48685/elife-48685-fig2-data1-v1.xlsx
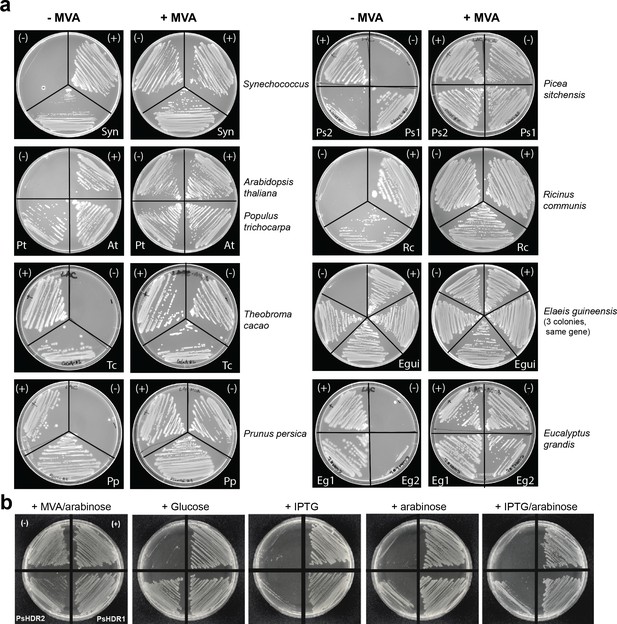
Complementation of lethal knockout of ispH in E. coli using different HDRs, and associated DMAPP toxicity.
(a) An E. coli strain with a ∆ispH knockout and a heterologously expressed lower mevalonate (MVA) pathway depends on mevalonate for survival. When a functional ispH/HDR gene is expressed from a plasmid, growth can be restored in the absence of mevalonate. (-), empty vector negative control; (+), plasmid expressing E. coli ispH as a positive control. E. grandis HDR2 and P. trichocarpa HDR2 did not complement the ∆ispH knockout. MVA, mevalonate. (b) Toxicity of P. sitchensis HDR2 in E. coli ∆ispH. Plasmid-encoded HDR genes are expressed from the trc promoter which can be induced with IPTG or partially repressed with glucose. P. sitchensis HDR2-associated toxicity can be alleviated by adding glucose to repress HDR expression, and is strongest under full IPTG induction. Arabinose, which induces the genomically encoded lower MVA pathway, including a heterologous idi gene, partly alleviates IPTG-induced toxicity. (-), empty vector negative control; (+), plasmid expressing E. coli HDR as a positive control. IPTG, Isopropyl β-D-1-thiogalactopyranoside; MVA, mevalonate.
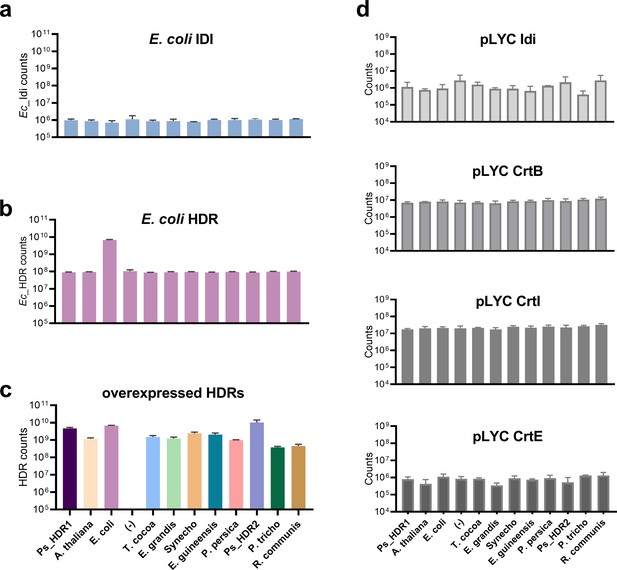
Protein quantification of IDI, HDR and the lycopene biosynthetic pathway.
Relative protein abundance in E. coli overexpressing HDR genes from different species, in the genetic context of dxs and lycopene biosynthetic pathway overexpression. Proteins were quantified using untargeted proteomics via LC-MS/MS, and data represent the three most abundant peptide counts from each protein, normalized across all samples. (a) E. coli native Idi shows no significant difference between strains (one-way ANOVA, p = 0.536). (b) E. coli native HDR quantification. Only the EcHDR overexpression strain is significantly different to the ‘no HDR overexpression’ (-) control (Welch’s one-way ANOVA, p = 0.0048), no significant difference were observed between the other strains (p ≥ 0.743). (c) Overexpressed, heterologous HDR proteins. Comparison of protein abundance between different HDR proteins is not possible due to a lack of shared tryptic peptides across all HDRs. Unique peptides for all overexpressed HDR proteins were detected with high abundance in the respective strains. (d) Plasmid-encoded lycopene biosynthetic pathway proteins. No significant differences were detected across strains for CrtI and CrtB (ordinary or Welch’s ANOVA, respectively, p ≥ 0.05). For Idi and CrtE, differences with p < 0.05 were detected between selected strains (e.g. E. grandis vs. R. communis CrtE p = 0.034); however, no significant differences were observed when comparing any of the strains to the negative control (p ≥ 0.78 for Idi, Welch’s one-way ANOVA; p ≥ 0.46 for CrtE, ordinary one-way ANOVA). For Idi and CrtE, a total of only 3 peptides each were detected in our proteomics analysis, indicating low protein abundance and potentially explaining the higher variability between strains. Data represent means +/- SD from n ≥ 3. All p-values were corrected for multiple hypothesis testing using Dunnett’s method.
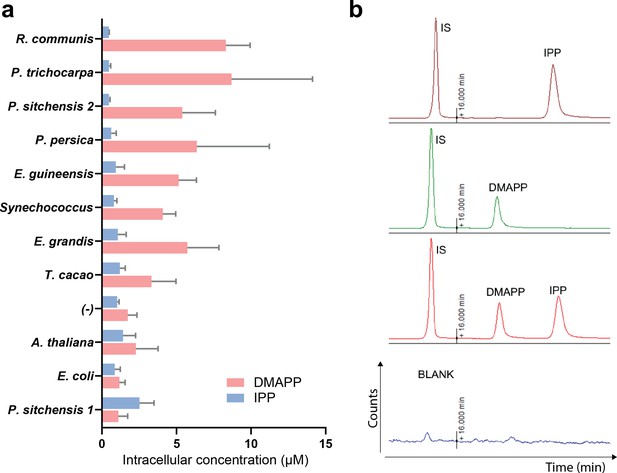
Quantification of DMAPP and IPP using LC-MS/MS.
(a) Absolute quantification of intracellular DMAPP and IPP in E. coli overexpressing HDR genes from different species, in the genetic context of dxs and lycopene biosynthetic pathway overexpression. The difference in product ratio between HDRs at opposite ends of the graph is driven by both an increase in DMAPP and a decrease in IPP concentration. Data represent means +/- SD from n ≥ 3. (b) Representative chromatograms of matrix-matched calibration standards for DMAPP and IPP, including mevalonate-5-phosphate as internal standard (IS). Due to a slow, consistent drift in retention time over the HPLC column lifetime, no fixed retention times are given for DMAPP and IPP; however, with the presented method the analytes remain baseline-separated as shown here.
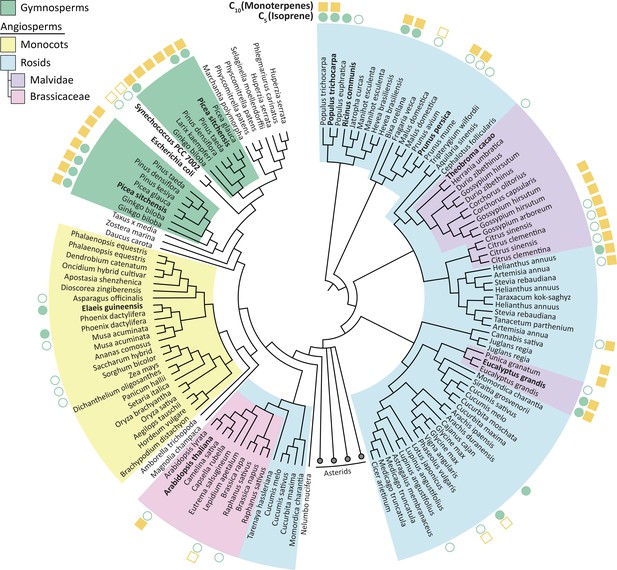
Phylogenetic tree of HDR proteins from land plants, the cyanobacterium Synechococcus and Escherichia coli.
Where known, each species’ C5 (isoprene) and C10 (monoterpenes) emission spectra are shown (Wiedinmyer et al., 2020). High DMAPP-producing HDR proteins (from P. trichocarpa, R. communis and P. persica) cluster together based on high sequence similarity. Homologues within species, such as P. trichocarpa, tend to be highly similar; except for in gymnosperms where two separate groups of likely paralogous HDRs exist. Proteins analysed in this study are highlighted in bold. The Asterids clade is collapsed for clarity. Tree generated from BLAST sequence alignment with A. thaliana HDR against all land plants, using maximum likelihood phylogeny. Empty symbol, no volatile emission; filled symbol, volatile emission; no symbol, no or conflicting data available.
-
Figure 3—source data 1
List of HDR sequences used for phylogenetic analysis in Figure 3.
- https://cdn.elifesciences.org/articles/48685/elife-48685-fig3-data1-v1.txt
-
Figure 3—source data 2
Phylogenetic tree of HDRs in Newick format.
- https://cdn.elifesciences.org/articles/48685/elife-48685-fig3-data2-v1.nwk.txt
Tables
Genetic information and volatile isoprenoid emission profiles for species studied in this work.
Key: blank cell indicates species has not been tested, or genome sequence (or other information) not available; Y indicates significant emissions of isoprene or isoprenoids have been detected, or gene/transcript has been identified; N indicates significant emissions of isoprene or isoprenoids have NOT been detected, or gene/transcript has NOT been identified; MTs, monoterpenes; IspS, isoprene synthase; TPS, terpene synthase.
| Emissions | Gene/transcript* | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kingdom | Phylum/Clade | Clade | Genus, species | Common Name | HDR protein accession number | E. coli construct Genbank ID | Complements?† | Isoprene (C5) | MTs (C10) | IspS | Short chain TPS | Reference | |
| Plantae | Angiosperms | Eudicots | Ricinus communis | castor bean plant | XP_002519102.1 | MH605331 | yes | N | Y | N | Y | Wiedinmyer et al., 2020; Kadri et al., 2011; Xie et al., 2012) | |
| Plantae | Angiosperms | Eudicots | Populus trichocarpa‡ | black cottonwood | 1 | ACD70402 | MH605329 | yes | Y | Y | Y | Y | Wiedinmyer et al., 2020; Tuskan, 2006) |
| 2 | PNT41333.1 | MH605330 | no | ||||||||||
| Plantae | Angiosperms | Eudicots | Prunus persica | peach | XP_007199828.1 | MH605326 | yes | N | Y | N | Y | Wiedinmyer et al., 2020; Verde et al., 2013) | |
| Plantae | Angiosperms | Eudicots | Eucalyptus grandis | flooded gum | 1 | XP_010028563.1 | MH605323 | yes | Y | Y | Y | Y | Wiedinmyer et al., 2020; Myburg et al., 2014 |
| 2 | XP_010047332.1 | MH605324 | no | ||||||||||
| Plantae | Angiosperms | Eudicots | Theobroma cacao | cacao tree | XP_007042717.1 | MH605333 | yes | N | Y | N | Y | Wiedinmyer et al., 2020; Argout et al., 2008 | |
| Plantae | Angiosperms | Eudicots | Arabidopsis thaliana | thale cress | AEE86362.1 | MH605322 | yes | N | Y | N | Y | Sharkey et al., 2005; Chen et al., 2004; Bohlmann et al., 2000 | |
| Plantae | Angiosperms | Monocots | Elaeis guineensis | oil palm | XP_010909277.1 | MH605325 | yes | Y | Y | Wiedinmyer et al., 2020; Wilkinson et al., 2006 | |||
| Plantae | Gymnosperms | Pinophyta | Picea sitchensis | Sitka spruce | 1 | ACN40284.1 | MH605327 | yes | Y | Y | Y | Wiedinmyer et al., 2020; Hayward et al., 2004 | |
| 2 | ACN39959.1 | MH605328 | yes – toxic | ||||||||||
| Bacteria | Cyanobacteria | Synechococcus sp. PCC 7002 | Synechococcus | ACA98524.1 | MH605332 | yes | N | N | N | ||||
-
* Identified from data/genomes available on NCBI (https://www.ncbi.nlm.nih.gov/) and literature search (references noted).
† Whether protein expression was able to functionally complement an E. coli ΔispH knockout in this study.
-
‡ Also known as Populus balsamifera ssp. trichocarpa.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Escherichia coli) | ispH/HDR | NCBI ‘Gene’ | Gene_ID:944777; EcoGene:EG11081; ECK0030; lytB | hydroxymethylbutenyl diphosphate reductase |
| Strain, strain background (Escherichia coli) | Escherichia coli W | ATCC | ATCC:9637 | obtained from L. Nielsen lab, Australia |
| Genetic reagent (Escherichia coli) | E. coli W∆cscR, lacZ::PtDXS, arsB::PaISPS | This paper and PMID: 21782859 (Arifin et al., 2011) | knockout of cscR, knock-in of PtDXS and PaISPS | |
| Genetic reagent (Escherichia coli) | E. coli WΔcscR, lacZ::MVA, ∆ispH | This paper and PMID: 11115399 (Campos et al., 2001) | knock-in of MVA pathway, knockout of ispH | |
| Genetic reagent (Populus trichocarpa) | DXS | NCBI ‘Reference Sequence’ | XP_006378082.1 | Deoxyxylulose phosphate synthase, gene was truncated for expression in E. coli |
| Genetic reagent (Populus alba) | ISPS(del2-52,A3T,L70R,S288C) | Patent WO2012058494 (Beck et al., 2011) | Isoprene synthase (Genbank:EF638224) variant, truncated and mutated | |
| Recombinant DNA reagent | pLacZ-KIKO(cm) plasmid | PMID: 23799955 (Sabri et al., 2013) | Addgene:46764 | used to integrate PtDXS into the genome |
| Recombinant DNA reagent | pArsBKIKO(cm) plasmid | PMID: 23799955 (Sabri et al., 2013) | Addgene:46763 | used to integrate PaISPS into the genome |
| Recombinant DNA reagent | pT-HDR plasmids | This paper | derived from pTrc99a | all HDR genes were cloned into this expression vector |
| Recombinant DNA reagent | pAC-LYC04 | PMID: 7919981 (Cunningham et al., 1994) | ||
| Recombinant DNA reagent | Ricinus communis HDR expression plasmid | Genbank | MH605331 | HDR protein XP_002519102.1 |
| Recombinant DNA reagent | Populus trichocarpa HDR 1 expression plasmid | Genbank | MH605329 | HDR protein ACD70402 |
| Recombinant DNA reagent | Populus trichocarpa HDR 2 expression plasmid | Genbank | MH605330 | HDR protein PNT41333.1 |
| Recombinant DNA reagent | Prunus persica HDR expression plasmid | Genbank | MH605326 | HDR protein XP_007199828.1 |
| Recombinant DNA reagent | Eucalyptus grandis HDR 1 expression plasmid | Genbank | MH605323 | HDR protein XP_010028563.1 |
| Recombinant DNA reagent | Eucalyptus grandis HDR 2 expression plasmid | Genbank | MH605324 | HDR protein XP_010047332.1 |
| Recombinant DNA reagent | Theobroma cacao HDR expression plasmid | Genbank | MH605333 | HDR protein XP_007042717.1 |
| Recombinant DNA reagent | Arabidopsis thaliana HDR expression plasmid | Genbank | MH605322 | HDR protein AEE86362.1 |
| Recombinant DNA reagent | Elaeis guineensis HDR expression plasmid | Genbank | MH605325 | HDR protein XP_010909277.1 |
| Recombinant DNA reagent | Picea sitchensis HDR 1 expression plasmid | Genbank | MH605327 | HDR protein ACN40284.1 |
| Recombinant DNA reagent | Picea sitchensis HDR 2 expression plasmid | Genbank | MH605328 | HDR protein ACN39959.1 |
| Recombinant DNA reagent | Synechococcus sp. PCC 7002 HDR expression plasmid | Genbank | MH605332 | HDR protein ACA98524.1 |
| Commercial assay or kit | Astec Cyclobond I2000 chiral HPLC column | Sigma Aldrich | 20024AST | HPLC column used for IPP/DMAPP separation |
| Chemical compound, drug | Isoprene | Sigma Aldrich | Cat. # I19551 | |
| Chemical compound, drug | Isopentenyl pyrophosphate | Sigma Aldrich | Cat. # I0503 | |
| Chemical compound, drug | Dimethylallyl pyrophosphate | Sigma Aldrich | Cat. # D4287 | |
| Chemical compound, drug | (±)-Mevalonic acid 5-phosphate | Sigma Aldrich | Cat. # 79849 | |
| Chemical compound, drug | Mevalonolactone | Sigma Aldrich | Cat. # M4667 | |
| Software, algorithm | CLC Main Workbench | Qiagen | RRID:SCR_000354 | |
| Software, algorithm | iTOL | PMID: 27095192 (Letunic and Bork, 2016) | https://itol.embl.de/ | Interactive Tree of Life |