Tyr1 phosphorylation promotes phosphorylation of Ser2 on the C-terminal domain of eukaryotic RNA polymerase II by P-TEFb
Figures
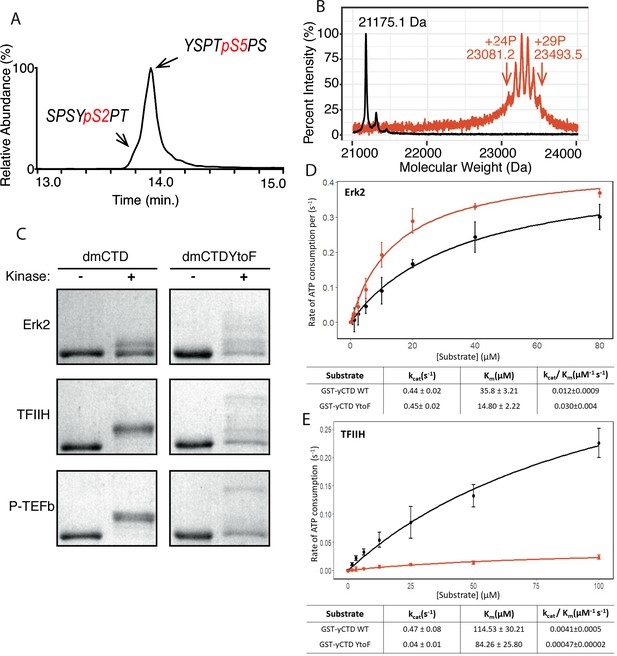
P-TEFb in vitro activity and the effect of phenylalanine replacement of Tyr1 in CTD phosphorylation.
(A) LC-UVPD-MS/MS analysis of yCTD treated with P-TEFb alone showing extracted ion chromatogram for two CTD heptads. (B) Portions of MALDI mass spectra of 3C-protease digested yCTD variant treated with P-TEFb alone (red) and no kinase control reaction (black). Mass labels indicate m/z at the various peak maxima. Arrows indicate the range of m/z peaks for kinase treated sample. ‘+#P’ notation indicates an approximate number of phosphates added based on mass shifts relative to no kinase control. (C) SDS-PAGE EMSA of dmCTD and dmCTDYtoF (as indicated) treated with Erk2 (top, right bands), TFIIH (middle, right bands), or P-TEFb (bottom, right bands) and paired no kinase control reactions (left bands). (D–E) Kinase activity assay of wild-type yCTD (shown in black) and yCTDYtoF (shown in red) variant by Erk2 (D) and TFIIH (E) fitted to the Michaelis-Menten kinetic equation. The Michaelis-Menten kinetic parameters kcat(s−1), Km(µM), and kcat/Km(µM−1 s−1) are given below the graphs for each respective fit. Each measurement was conducted in triplicate with standard deviations shown as error bars.

Amino acid sequences of GST-CTD constructs.
(A) Sequence of CTD portion of GST-CTD constructs used in the manuscript. Sequences presented N to C-terminal as individual heptads. Residues mutated are indicated above in blue font.
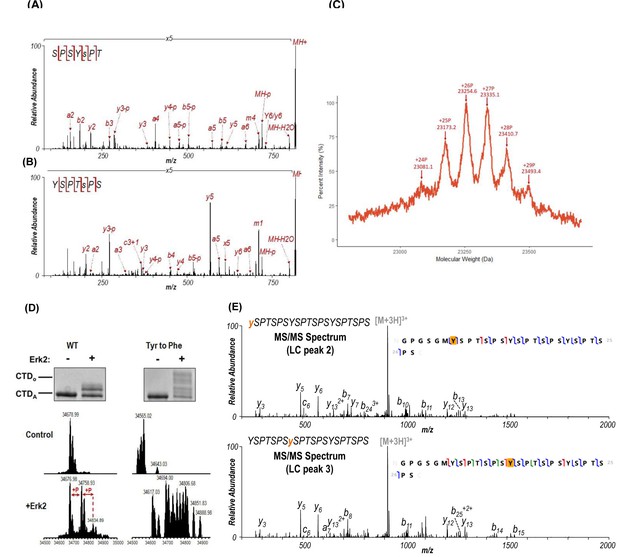
Mass spectrometry analysis of yCTD treated by kinases.
(A, B) LC-UVPD-MS/MS MS2 spectra of yCTD treated with P-TEFb: (A) SPSYsPT and (B) YSPTsPS. These spectra correspond to the LC traces in Figure 1A. The locations of the confirmed phosphorylation sites are indicated as lowercase y or s in the sequences. (C) MALDI-TOF of yCTD treated with P-TEFb, zoomed view to show detail. A portion of MALDI-MS spectra of 3C-protease digested yCTD constructs treated with P-TEFb. Mass labels indicate m/z at the various peak maxima. Arrows indicate the maximum and minimum m/z peaks for kinase treatment sample. ‘+#P’ notation indicates an approximate number of phosphates added based on mass shifts relative to no kinase control. (D) EMSA to show the effect of YtoF mutation on CTD gel shift upon kinase phosphorylation. EMSA and intact mass analysis result for Drosophila CTD 9mer before and after treatment with Erk2. (E) LC-UVPD-MS/MS spectra of 3CTD treated with c-Abl. Diagnostic fragmentation of mono-phosphorylated peptides corresponding to LC peaks 2 and 3 from Figure 2B are shown in the top and bottom spectra, respectively.
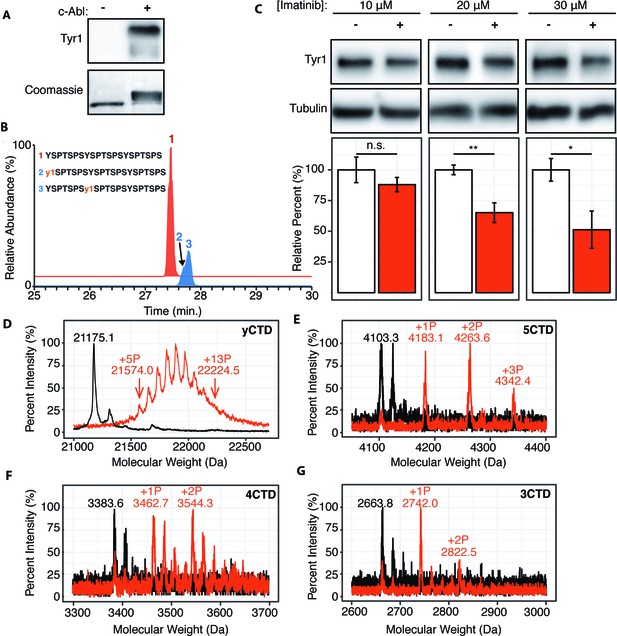
c-Abl kinase phosphorylates Tyr1 of RNA polymerase II CTD in cells and in vitro.
(A) Representative image of western blot against phosphorylated Tyr1 (top) of yCTD (containing 26 heptad repeats) treated in vitro with c-Abl (right) and paired no kinase control (left). Coomassie-stained blot included indicating loading (bottom). Data representative of three experimental replicates. (B) LC-UVPD-MS/MS analysis of 3CTD treated with c-Abl showing extracted ion chromatograms for 3CTD (m/z 888.76, 3+ charge state, red trace) and mono-phosphorylated 3CTD (m/z 915.36, 3+ charge state, blue trace) peptides. (C) Representative images (top) and quantification (bottom) of a western blot of imatinib dosage series (10–30 μM, as indicated, red) and paired DMSO vehicle controls (left band, white) of 20 μg total protein from HEK293T cells. Phospho-specific Tyr1 antibody (clone 3D12) was used. Imatinib decreases pTyr1 epitope abundance to 88.0% (10μM imatinib, not significant), 65.2% (20μM imatinib), and 51.3% (30μM imatinib) relative to paired vehicle controls. Epitope signals normalized against tubulin loading control. Significance determined by Welch’s t-test (*=p value<0.05, **=p value<0.01, n.s. = not significant (p-value>0.05)), n = 6, error bars indicate SEM. (D–G) Portions of MALDI mass spectra of 3C-protease digested yeast CTD (D), 5CTD (E), 4CTD (F), and 3CTD (G) construct treated with c-Abl (red) and no kinase control reaction (black). Mass labels indicate m/z at various peak maxima. Arrows indicate the range of m/z peak for kinase treated sample. ‘+#P’ notation indicates an approximate number of phosphates added based on mass shifts. Satellite peaks, prevalent in (F), correlate well with sodium adducts (M+23 Da).
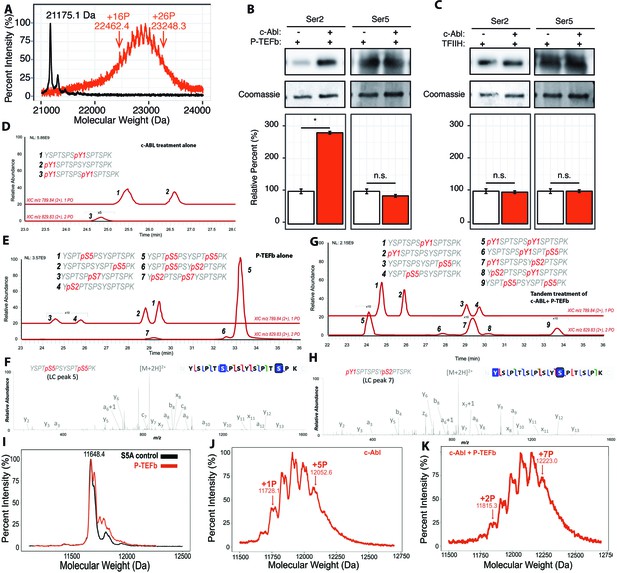
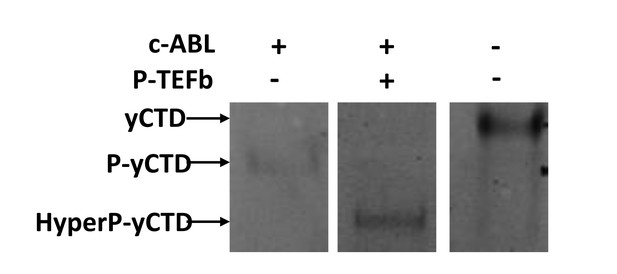
Effect of Tyr1 phosphorylation by c-Abl on the function of P-TEFb.
(A) Portions of MALDI mass spectra of 3C-protease digested yCTD construct treated tandemly with c-Abl followed by P-TEFb (red) and no kinase control reaction (black). Mass labels indicate m/z peaks for kinase treated sample. ‘+#P’ notation indicates an approximate number of phosphates added based on mass shifts. (B) Representative images (top) and quantification (bottom) of western blot analysis of yCTD treated with P-TEFb alone (left, white) and tandemly with c-Abl followed by P-TEFb (right, red). Tandem treatment of c-Abl followed by P-TEFb increases phosphorylated Ser2 epitope abundance to 279% of P-TEFb only treatment control. Ser5 phosphorylation levels are not significantly altered. Significance determined by Welch’s t-test (*=p value<0.05, n.s. = not significant (p-value>0.05)), n = 3, error bars indicate SEM. (C) Representative images (top) and quantification (bottom) of western blot analysis of yCTD treated with TFIIH alone (left, white) and tandemly with c-Abl followed by TFIIH (right, red). Tandem treatment of c-Abl followed by TFIIH does not significantly alter the epitope abundance of phosphorylated Ser2 or Ser5. Significance determined by Welch’s t-test (n.s. = not significant (p-value>0.05)), n = 3, error bars indicate SEM. (D–H) LC-UVPD-MS/MS analysis of yeast CTD with inserted Lys in every other heptad repeat (yCTD-Lys) treated with kinases or combination of kinases. Biological triplet samples were independently measured with exemplary spectra shown (n = 3). (D) yCTD-Lys treated with c-Abl showing extracted ion chromatograms for mono-phosphorylated (m/z 789.84, 2+ charge state) and doubly phosphorylated (m/z 829.83, 2+ charge state) peptides of sequence (YSPTSPSYSPTSPK). The LC traces are shown in red, and the phosphorylation sites determined by UVPD-MS/MS are highlighted in red font with ‘p’ to indicate phosphorylation. (E) LC-UVPD-MS/MS analysis of yCTD-Lys treated with P-TEFb showing extracted ion chromatograms for mono-phosphorylated (m/z 789.84, 2+ charge state) and doubly phosphorylated (m/z 829.83, 2+ charge state) peptides. (F) Representative UVPD spectra that demonstrate the diagnostic fragmentation pattern of the peptides shown in the inset from (E). The one shown is peak 5, which is the predominant product from yCTD-Lys treated by P-TEFb alone. (G) LC-UVPD-MS/MS analysis of yCTD-Lys with c-Abl followed by P-TEFb showing extracted ion chromatograms for mono-phosphorylated (m/z 789.84, 2+ charge state) and doubly phosphorylated (m/z 829.83, 2+ charge state) peptides. (H) Representative UVPD spectra that demonstrate the diagnostic fragmentation pattern of the peptides shown in the inset of (G). The one shown is peak 7, which is the predominant product from yCTD-Lys treated by c-Abl followed by P-TEFb. (I–K) Portions of MALDI-MS spectra of 3C-digested S5A. Panels included no kinase control (I), P-TEFb only treated (J), c-Abl only treated (I), and tandemly treated with c-Abl followed by P-TEFb (K). Mass labels indicate m/z at the various peak maxima. Arrows indicate the maximum and minimum m/z peak for kinase treated sample. ‘+#P’ notation indicates an approximate number of phosphates added based on mass shifts.
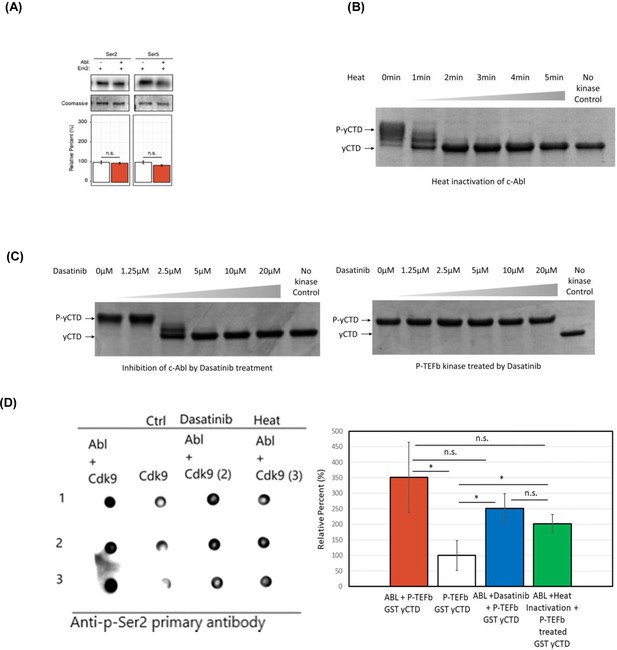
Phosphorylation of different CTD constructs by CTD kinases.
(A) Western blot analysis of yCTD treated in tandem with c-Abl and Erk2. Representative images (top) and quantification (bottom) of western blot analysis of yeast CTD treated with Erk2 alone (left, white) and tandemly with c-Abl followed by Erk2 (right, red). Tandem treatment of c-Abl followed by Erk2 does not significantly alter the epitope abundance of phosphorylated Ser2 or Ser5. Significance determined by Welch’s t-test (n.s. = not significant (p-value>0.05)), n = 3, error bars indicate SEM. (B) c-Abl is inactivated by heat treatment, as shown in the time-dependent course. (C) Dasatinib inhibition of c-Abl towards CTD substrate. Dasatinib inhibits c-Abl in a dose-dependent manner (left) but has no effect on the kinase activity of P-TEFb (right). (D) Dot blot analysis of the tandem treatment of c-Abl and P-TEFb. c-Abl treated GST yCTD followed by Dasatinib inhibition before P-TEFb treatment (blue), c-Abl treated GST yCTD followed by heat inactivation of c-Abl before treatment with P-TEFb (green) with corresponding P-TEFb only treated GST yCTD (white) and c-Abl followed by P-TEFb treated GST yCTD (red). Significance determined by the t-test (*=p value<0.05) n = 3, n.s. = not significant (p-value>0.05), n = 3, error bars indicate standard deviation from three technical replicates.
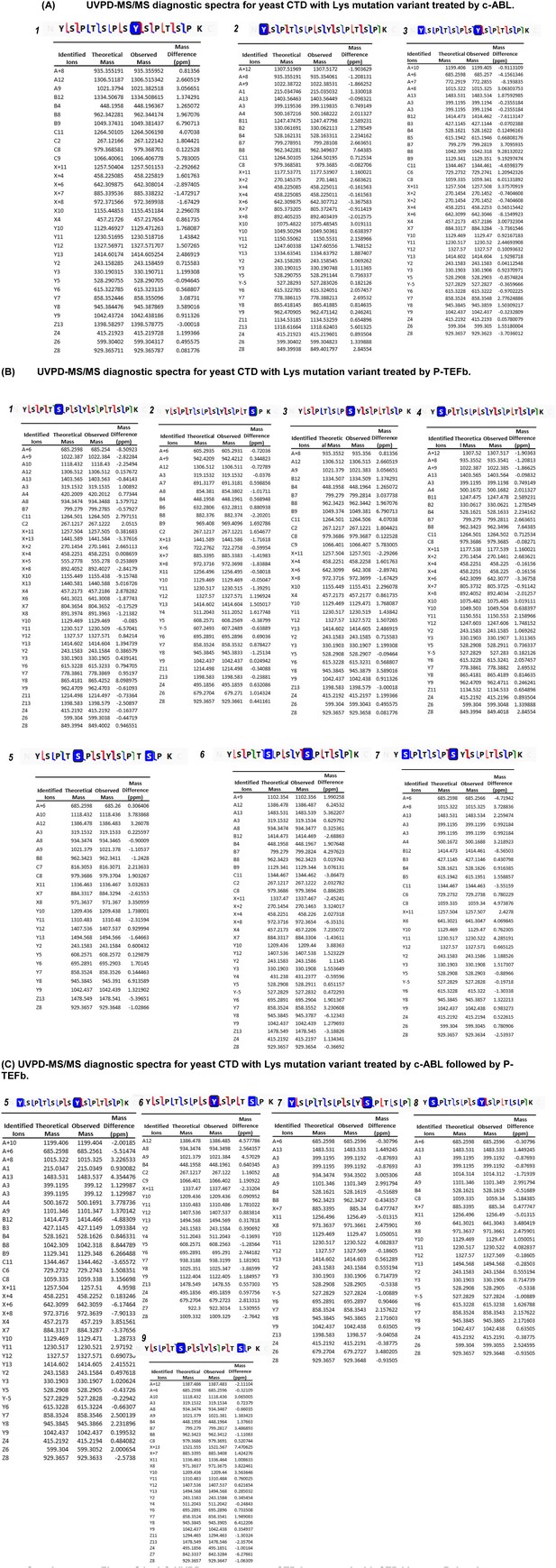
LC-UVPD-mass spectra yCTD-Lys treated with CTD kinases.
The tables show the fragments matched from UVPD spectra. The confirmed phosphorylated residues are highlighted with blue boxes in the sequence, and backbone cleavages that produce diagnostic fragment ions are designated by color-coded slash marks (a/x green, b/y blue, c/z red) in the sequence.
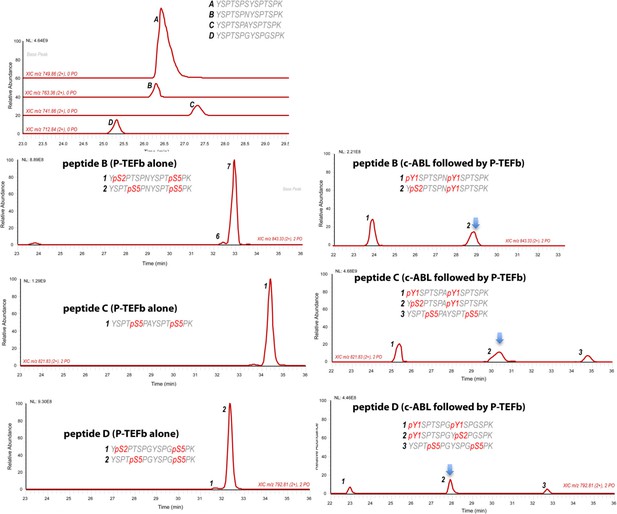
LC-UVPD-mass spectra yCTD-Lys treated with CTD kinases.
LC-UVPD-MS/MS analysis of yeast CTD with inserted Lys in every other repeat treated with c-Abl showing extracted ion chromatograms. For clarity, only doubly phosphorylated species (m/z 792.81, 2+ charge state) peptides are shown. Peptide A is most abundant, and its spectra are shown in Figure 3G–H. The less abundant peptides (peptide B, C, D) are shown here when different kinase treatment was applied. For clarity, only doubly phosphorylation species were shown. The phosphoryl-species containing both pTyr1 and pSer2 are highlighted with arrows.
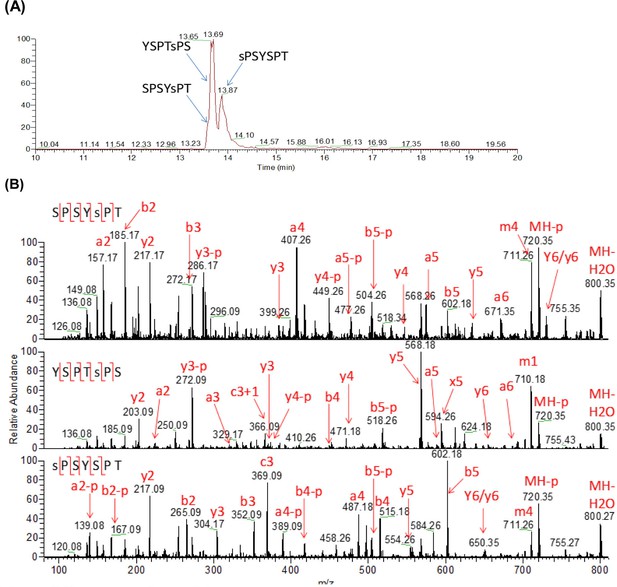
Analysis of yCTD treated in tandem with TFIIH and P-TEFb.
(A) Extracted ion chromatogram of mono-phosphorylated heptad (m/z 818, 2+ charge state) peptides. (B) UVPD mass spectra of three mono-phosphorylated heptads. The locations of the confirmed phosphorylation sites are indicated as lowercase y or s in the sequences.
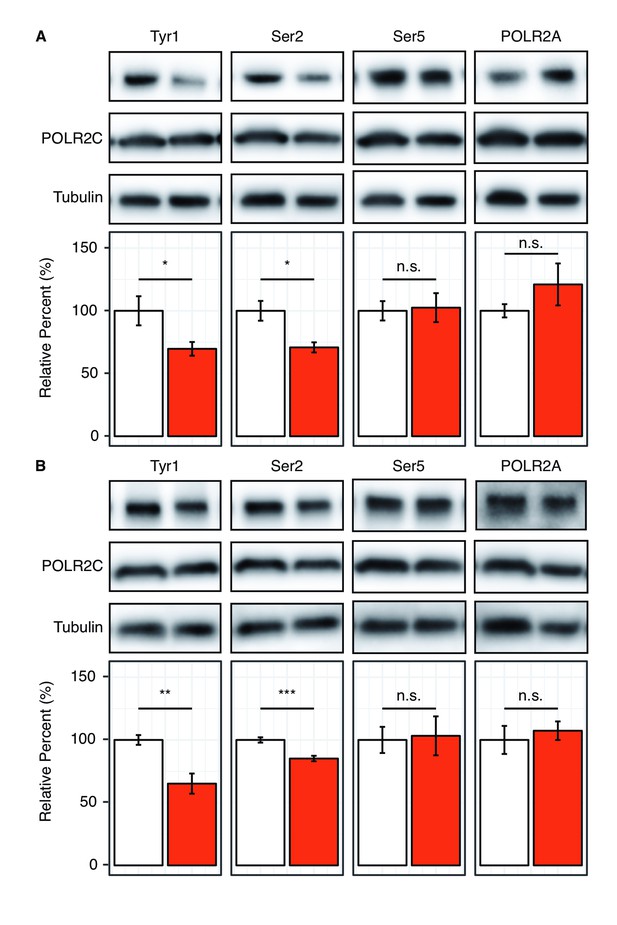
Reduction of Tyr1 levels specifically reduces Ser2 levels in cells.
(A) Representative images (top) and quantification (bottom) of western blot against 20 μg total protein from HEK293T cells treated with paired DMSO vehicle control (left, white) or 10 μM dasatinib (right, red). Immuno-blotting against epitopes left to right: phosphorylated Tyr1 reduced by treatment to 69.7% control (n = 6), phosphorylated Ser2 reduced by treatment to 70.8% control (n = 4), phosphorylated Ser5 unaltered (n = 6), POLR2A unaltered (n = 6) (POLR2C quantification provided in Figure 4—figure supplement 1A). (B) Representative images (top) and quantification (bottom) of western blot against 20 μg total protein from HEK293T cells treated with paired DMSO vehicle control (left, white) or 20 μM imatinib (right, red). Immuno-blotting against epitopes, left to right: Phosphorylated Tyr1 reduced by treatment to 65.2% control (n = 6), phosphorylated Ser2 reduced by treatment to 85.2% control (n = 6), phosphorylated Ser5 unaltered (n = 6), POLR2A unaltered (n = 6) (POLR2C quantification supplied in Figure 4—figure supplement 1A). Epitope signals normalized against tubulin loading control. Significance determined by Welch’s t-test (*=p value<0.05, **=p value<0.01, ***=p value<0.001, n.s. = not significant (p-value>0.05)), error bars indicate SEM.
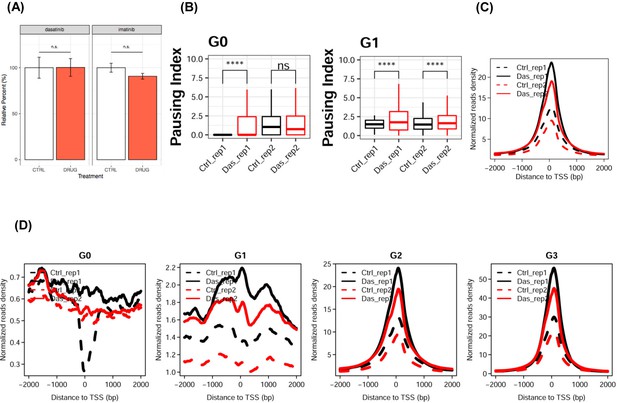
Cell-based analysis of phosphorylation of RNA polymerase II.
(A) Quantification of POLR2C western blot from Figure 4. POLR2C bands from dasatinib and imatinib blots (indicated) from Figure 4 were quantified. Dasatinib treatment POLR2C quantification was performed on the same blots providing the Ser2 and Ser5 data. Imatinib treatment POLR2C quantification was performed on the same blots providing POLR2A and Ser5 data. No significant change in POLR2C levels was observed for any blot analyzed. Epitope signals normalized against tubulin loading control. Significance determined by Welch’s t-test (n.s. = not significant (p-value>0.05)), error bars indicate SEM, n = 6). (B) Boxplot of pausing index changes of HEK293T cells on the genes from G0 and G1 clusters upon Tyr1 phosphorylation. ‘ns’ indicates a p-value higher than 0.05. ‘****” indicates p-value less than 0.0001. (C) Line plot of pausing indexes on all genes. Solid and dotted lines are representing two biological replicates. (D) Line plot of pausing indexes on genes for each cluster.
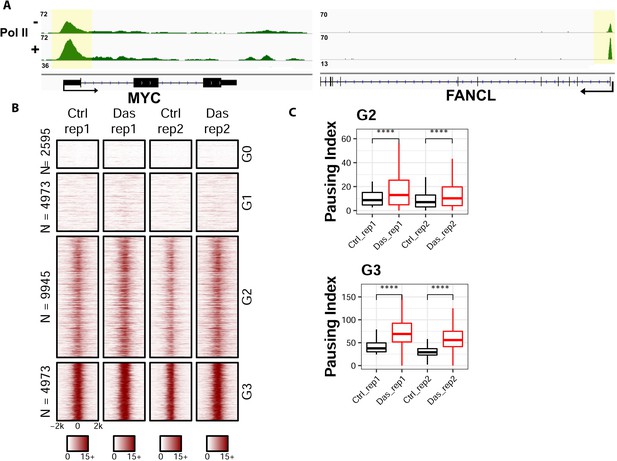
ChIP-seq analyses on the distribution of RNA polymerase II upon the inhibition of Tyr1 phosphorylation.
(A) ChIP-seq example illustrating the association of RNA polymerase II along with the active transcribing genes. Antibody 8WG16 was used to detect RNA polymerase II regardless of its phosphorylation state. The promoter regions of the genes are shaded in yellow for highlighting. (B) Heatmaps of ChIP-seq signal intensity of RNA polymerase II (8WG16) [±2 kb windows around the center of transcription start site (TSS)] for genes in each group. (C) Boxplots on the pausing index changes on the genes from G2 (9945 genes) and G3 (4973 genes) clusters upon Tyr1 phosphorylation. ‘****” indicates p-value≤0.0001.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line | HEK293T | ATCC | ||
| Antibody | Anti-Tyr1 (Clone 3D12)(rat monoclonal) | Millipore | Cat# MABE350 | (1:1000) |
| Antibody | Anti-beta Tubulin (rabbit polyclonal) | Abcam | Cat# ab6046, RRID:AB_2210370 | (1:10000) |
| Antibody | Anti-Ser2 (Clone 3E10) (rat monoclonal) | Millipore | Cat# 04–1571, RRID:AB_11212363 | (1:10000) for in vittro phosphorylated samples, (1:5000) for cell lysates and dot blot |
| Antibody | Anti-Ser5 (Clone 3E8)(rat monoclonal) | Millipore | Cat# 04–1572, RRID:AB_10615822 | (1:10000) for in vitro phosphorylated samples, (1:5000) for cell lysates |
| Antibody | Anti-POLR2A (Clone 4F8) (rat monoclonal) | Millipore | Cat# 04–1569, RRID:AB_11213378 | (1:5000) |
| Antibody | Anti-POLR2C (rabbit monoclonal) | Abcam | Cat# ab182150 | (1:5000) |
| Antibody | Goat Anti-Rat IgG Antibody HRP conjugate | Millipore | Cat# AP136P, RRID:AB_91300 | (1:20000) |
| Antibody | Goat Anti-Rabbit IgG H and L | Abcam | Cat# ab6721, RRID:AB_955447 | (1:20000) |
| Antibody | Anti-RNA polymerase II CTD repeat YSPTSPS antibody [8WG16] (mouse monoclonal) | Abcam | Cat# ab817, RRID:AB_306327 | 5 µl/Chip |
| Antibody | RNA Pol II Ser2-P antibody [3E10](rat monoclonal) | Chromotek | RRID: AB_2631403 | 500 µl/Chip |
| Recombinant DNA reagent | 3CTD, 4CTD, 5CTD and Y to F gene fragments | IDT | Cloned into pET28a derived vector with His GST tag | |
| Recombinant DNA reagent | pET-His6-ERK2 MEK1_R4F_coexpression vector | Gift from Melanie Cobb | Addgene plasmid Cat#39212 | |
| Recombinant DNA reagent | S5A 13 repeat CTD | Biomatik | Cloned into pET28a derived vector with His GST tag | |
| Recombinant DNA reagent | S7K spaced CTD | Biomatik | Cloned into pET28a derived vector with His GST tag | |
| Recombinant DNA reagent | human ABL1 kinase domain (residues 229–511) | Kind gift from Kuriyan Lab | ||
| Peptide, recombinant protein | TFIIH(Cdk7/Cyclin H/MAT1 (CAK complex)) | Millipore | Cat# 14–476 | Used at a concentration of 0.025 µg/µl |
| Peptide, recombinant protein | P-TEFb (Cdk9/Cyclin T1) | Millipore | Cat# 14–685 | Used at a concentration of 0.0075 µg/µl |
| Peptide, recombinant protein | c-Abl kinase | ProQinase | Cat# 0992-0000-1 | Used at a concentration of 0.0035 µg/µl |
| Commercial assay or kit | Pierce BCA Protein Assay Kit | Thermo Fischer Scientific | 23252 | |
| Commercial assay or kit | NEBNext Ultra II DNA Library Prep Kit for Illumina | NEB | E7645S | |
| Commercial assay or kit | NEBNext Multiplex Oligos for Illumina(Index Primers Set 1) | NEB | E7335S | |
| Chemical compound, drug | Imatinib | Selleck Chemicals | S1026 | 20 µM concentration |
| Chemical compound, drug | Dasatinib | Sigma Aldrich | CDS023389 | 10 µM concentration |
| Chemical compound, drug | 10 nCi/µl radiolabeled ATP | Perkin Elmer | NEG002A 100UC | |
| Chemical compound, drug | 0.45 µm nitrocellulose filters | Sigma Aldrich | WHA10401114 | |
| Chemical compound, drug | Econo-Safe Economical Biodegradable Counting Cocktail | Research Products International | SKU: 111175 | |
| Chemical compound, drug | HALT protease and phosphatase inhibitor cocktail | Thermo Fischer Scientific | Cat# 78440 | |
| Chemical compound, drug | Ribonuclease A | VWR lifesciences | CAS# 9001-99-4 | |
| Chemical compound, drug | Proteinase K | Ambion | Cat# 2542 | |
| Chemical compound, drug | Glycogen | Thermofischer scientific | Cat# R0561 | |
| Chemical compound, drug | 16% Formaldehyde solution (w/v), Methanol-free | Thermo scientific | Ref# 28908 | |
| Chemical compound, drug | SuperSignal West Pico Chemiluminescent Substrate | Pierce | 34079 | |
| Software, algorithm | ggplot2, R smoothing package | R-Studio | https://www.rstudio.com/ | |
| Software, algorithm | DataExplorer (AB) | Matrix Science | http://www.matrixscience.com/help/instruments_data_explorer.html | |
| Software, algorithm | Image J | NIH | https://imagej.nih.gov/ij/download.html | |
| Software, algorithm | XCalibur Qual Browser | Thermo Fischer Scientific | XCALI-97617 | |
| Software, algorithm | ProSight Lite | Proteomics Center of Excellence Northwestern University | http://prosightlite.northwestern.edu/ | |
| Other | Ni-NTA | Qiagen | 30210 | |
| Other | Dynabeads Protein G | ThermoFischer Scientific | Cat# 10004D | |
| Other | AMPure XP beads | Beckman Coulter | Ref# A63881 | |
| Other | Vivaspin | Sartorius | VS2002 | |
| Other | Picofrit 75 µm id analytical columns | New Objective | ||
| Other | Picofrit 75 µm id analytical columns | New Objective | ||
| Other | Waters Xbridge BEH C18 | Milford | ||
| Other | Orbitrap Fusion Lumos Tribrid mass spectrometer | Thermo Fischer Scientific | ||
| Other | Velos Pro dual linear ion trap mass spectrometer | Thermo Fischer | ||
| Other | G:BOX imaging systems | Syngene |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.48725.015