A single H/ACA small nucleolar RNA mediates tumor suppression downstream of oncogenic RAS
Figures
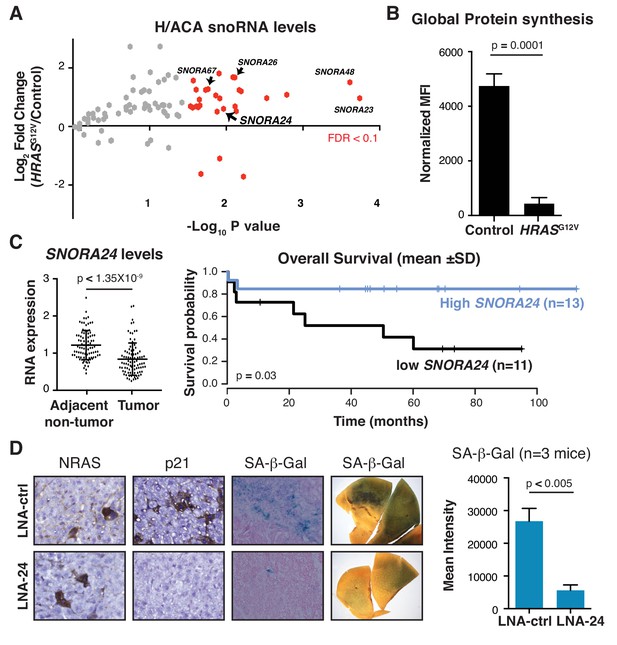
RAS-induced H/ACA snoRNAs are required for oncogene-induced senescence in vivo.
(A) Volcano plot displays Log2 fold change in H/ACA snoRNA levels 5 days following HRASG12V expression in primary human skin fibroblasts measured by snoRNA qPCR array from three independent experiments. SnoRNAs in red exhibit statistically significant fold change in expression in HRASG12V expressing cells compared to controls (p<0.05, unpaired Student’s t-test or FDR < 0.1). SnoRNAs highlighted with labels were independently validated as shown in Figure 1—figure supplement 1B. (B) Graph illustrates mean ± SD mean fluorescent intensity (MFI) of the amount of de novo protein synthesis in primary human fibroblasts 5 days following expression of HRASG12V compared to control treated cells by measuring OPP incorporation into newly synthesized protein from three independent experiments. Statistical analysis was performed using an unpaired Student’s t-test, p=0.0001. (C) Analysis of SNORA24 levels in HCC specimens compared to adjacent non-tumor tissue of 91 HCC patients (GSE25097) (paired Student’s t-test, p<1.35×10−9) (left panel) and Kaplan-Meier curve showing overall survival of HCC patients with high or low SNORA24 levels (mean ± 1 SD of SNORA24 levels) (right panel). Statistical significance was calculated using the log-rank test, with p=0.03. (D) Representative image for NRAS, p21, and SA-β-Gal staining in liver sections or resected liver lobes (SA-β-gal wholemount staining) 6 days following delivery of NRASG12V and treated with control LNA (LNA-ctrl) or LNA targeting Snora24 (LNA-24). Graph shows mean ± SD mean intensity of SA-β-gal staining in liver from mice treated with LNA-ctrl (n = 3 mice) or LNA-24 (n = 3 mice) 6 days following NRASG12V expression. Statistical analysis was performed using an unpaired Student’s t-test, p=0.005.
-
Figure 1—source data 1
H/ACA snoRNA levels upon oncogenic HRAS expression.
- https://doi.org/10.7554/eLife.48847.010
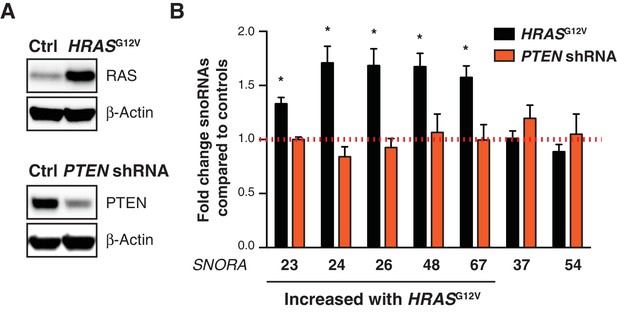
Select RAS-induced snoRNAs are not altered upon PTEN reduction.
(A) Analysis of RAS protein 5 days following HRASG12V or empty vector expression in primary human skin fibroblasts by western blot. β-Actin was used as a loading control (top panel). Analysis of PTEN protein 5 days following shRNA targeting PTEN or control non-targeting shRNA in primary human skin fibroblasts by western blot. β-Actin was used as a loading control (bottom panel). (B) qPCR analysis of the indicated snoRNAs 5 days following HRASG12V expression (black bars) or PTEN reduction (PTEN shRNA, orange bars) in primary human skin fibroblasts measured by qPCR. Graph shows mean fold change expression ± SD relative to control cells and normalized to the levels of RN7SK from three independent experiments. Statistical analysis was performed using an unpaired Student’s t-test, *p < 0.05.
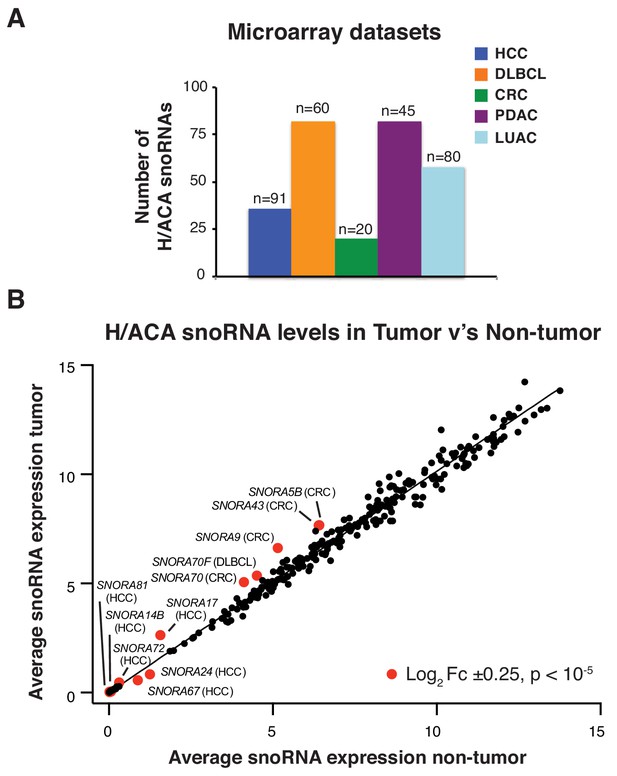
Altered expression of distinct H/ACA snoRNAs in human cancers.
(A) Graph shows the number of distinct H/ACA snoRNAs analyzed in the indicated microarray gene expression datasets; HCC GSE25097, Diffuse large B-cell lymphoma (DLBCL) GSE22898, Colorectal cancer (CRC) GSE20916, Pancreatic ductal adenocarcinoma (PDAC) GSE28735, and Lung adenocarcinoma (LUAC) GSE43458. The number of patient samples analyzed in each dataset is indicated over the corresponding column. (B) Graph shows average H/ACA snoRNA expression in all tumor samples compared to controls from each microarray gene expression dataset indicated in A. Red dots highlight H/ACA snoRNAs that exhibit changes in expression between tumor and controls of greater than Log2 fold change ± 0.25 and p<0.00005.
-
Figure 1—figure supplement 2—source data 1
H/ACA snoRNA levels in human cancer.
- https://doi.org/10.7554/eLife.48847.006
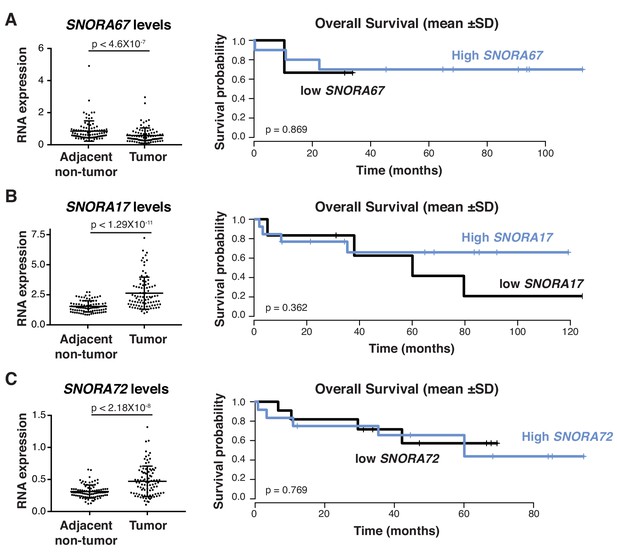
Association between H/ACA snoRNA levels and patient survival in HCC.
(A) Analysis of SNORA67 levels in HCC specimens compared to adjacent non-tumor tissue of 91 HCC patients (GSE25097) (paired Student’s t-test, p<4.6×10−7) (left panel) and Kaplan-Meier curve showing overall survival of HCC patients with high or low SNORA67 levels (mean ± 1 SD of SNORA67 levels) (right panel). (B) Analysis of SNORA17 levels in HCC specimens compared to adjacent non-tumor tissue of 91 HCC patients (GSE25097) (paired Student’s t-test, p<1.29×10−11) (left panel) and Kaplan-Meier curve showing overall survival of HCC patients with high or low SNORA17 levels (mean ± 1 SD of SNORA17 levels) (right panel). (C) Analysis of SNORA72 levels in HCC specimens compared to adjacent non-tumor tissue of 91 HCC patients (GSE25097) (paired Student’s t-test, p<2.18×10−8) (left panel) and Kaplan-Meier curve showing overall survival of HCC patients with high or low SNORA72 levels (mean ± 1 SD of SNORA72 levels) (right panel).
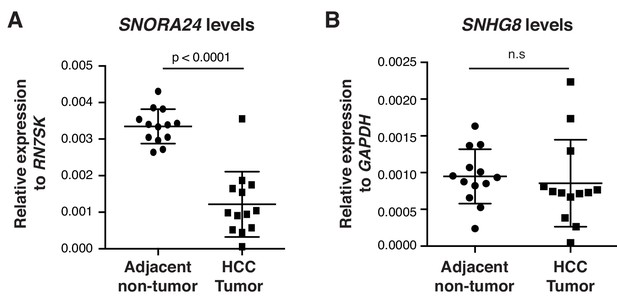
SNORA24 is reduced in primary human HCC specimens.
(A) qPCR analysis of SNORA24 in HCC tissue and matched adjacent non-tumor tissue. Graph shows mean ± SD SNORA24 levels normalized to the expression of RN7SK. Statistical analysis was performed using a paired Student’s t-test, p<0.0001. (B) qPCR analysis of SNHG8 in HCC tissue and matched adjacent non-tumor tissue. Graph shows mean ± SD SNHG8 levels normalized to the expression of GAPDH. Statistical analysis was performed using a paired Student’s t-test, n.s = non significant.
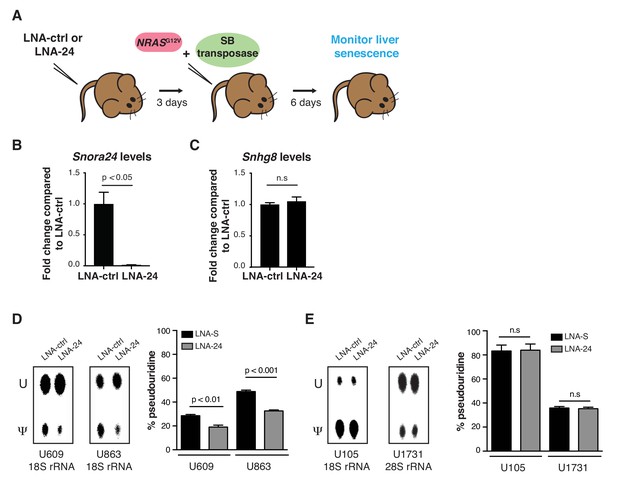
Loss of Snora24 cooperates with RAS to promote the development of HCC resembling human SH-HCC.
(A) Cartoon outlines the experimental approach to target Snora24 in vivo and test the requirement of Snora24 in RASG12V-induced senescence in the mouse liver using a previously established system (SB(+)NRASG12V). (B) qPCR analysis of Snora24 in liver from LNA-ctrl and LNA-24 treated mice. Graph shows mean fold change expression ± SD relative to LNA-ctrl treated mice and normalized to the levels of Rn7sk from n = 4 mice. Statistical analysis was performed using an unpaired Student’s t-test, p<0.05.(C) qPCR analysis of Snhg8 in liver from LNA-ctrl and LNA-24 treated mice. Graph shows mean fold change expression ± SD relative to LNA-ctrl treated mice and normalized to the levels of Gapdh from n = 4 mice. Statistical analysis was performed using an unpaired Student’s t-test, n.s = non significant. (D) Representative thin layer chromatography (TLC) of site-specific amounts of pseudouridine (Ψ) or uridine (U) present at position U609 and U863 on 18S rRNA in primary fibroblasts transfected with LNA-S or LNA-24 for 48 hours (hrs) (left panel). Quantification of TLC showing the percentage pseudouridine at position U609 (left) and U863 (right) on 18S rRNA. Graph shows mean ± SD percentage pseudouridine for the indicated residue from three independent experiments. Statistical analysis was performed using an unpaired Student’s t-test, p<0.01 and p<0.001 (right panel). (E) Representative TLC of site-specific amounts of pseudouridine (Ψ) or uridine (U) present at position U105 on 18S rRNA and U1731 on 28S rRNA in primary fibroblasts transfected with LNA-S or LNA-24 for 48 hrs (left panel). Quantification of TLC showing the percentage pseudouridine at position U105 (left) and U1731 (right) on 18S rRNA and 28S rRNA, respectively. Graph shows mean ± SD percentage pseudouridine for the indicated residue from three independent experiments. Statistical analysis was performed using an unpaired Student’s t-test, n.s = non significant (right panel).
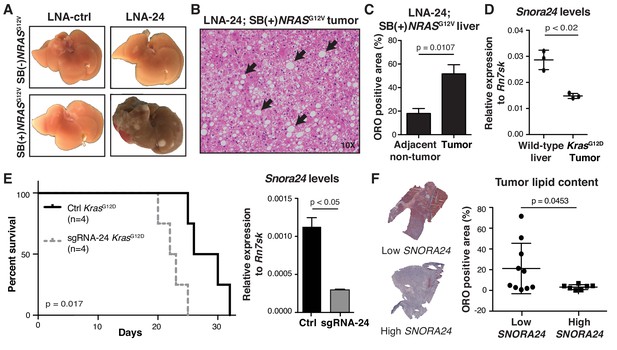
Snora24 plays a role in the initiation and maintenance of RAS-driven hepatocellular carcinoma.
(A) Representative images of explanted livers from control (SB(-)NRASG12V) or SB(+)NRASG12V mice treated with either LNA-ctrl or LNA-24. (B) H and E staining of a liver section from SB(+)NRASG12V mouse treated with LNA-24. Black arrows highlight the presence of fat droplets. (C) Graph shows mean ± SD percentage Oil Red O (ORO) positive area in liver tumor nodules and adjacent non-tumor tissue from n = 3 SB(+)NRASG12V mice treated with LNA-24. For each mouse liver section, the amount of ORO positive stain per total area from at least four distinct tumor and non-tumor regions (as determined by H and E staining) was measured (see Materials and methods and Figure 2—figure supplement 1). Statistical analysis was performed using a paired Student’s t-test, p=0.0107). (D) Quantitative PCR (qPCR) analysis of Snora24 levels in wild-type liver or age- and sex-matched liver tumors from Alb-cre;KrasG12D mice. Graph shows mean ± SD Snora24 expression normalized to the levels of Rn7sk from n = 3 mice per condition. Statistical analysis was performed using an unpaired Student’s t-test, p<0.02. (E) Kaplan-Meier curves showing survival in male C57BL/6 wild-type mice following intrahepatic orthotopic injection of Ctrl KrasG12D HCC cells (black line, n = 4 mice) and sgRNA-24 KrasG12D HCC cells (gray dashed line, n = 4 mice), p=0.017, log-rank test (left panel). qPCR analysis of Snora24 in Ctrl KrasG12D and sgRNA-24 KrasG12D HCC cells (right panel). Graph shows mean relative expression ± SD normalized to the levels of Rn7sk from three independent experiments. Statistical analysis was performed using an unpaired Student’s t-test, *p < 0.05. (F) Representative image of ORO staining in HCC from a patient with high SNORA24 (bottom) or low SNORA24 (top) expression. Quantification of ORO stain in tissue sections from HCC patients dichotomized into high or low by identifying samples with SNORA24 expression greater than ±one SD from the mean (n = 17 HCC specimens). Graph shows mean ± SD percentage Oil Red O (ORO) positive area present in HCC tissue specimens from patients with high or low SNORA24 expression and statistical analysis was performed using an unpaired Student’s t-test, p=0.0453.
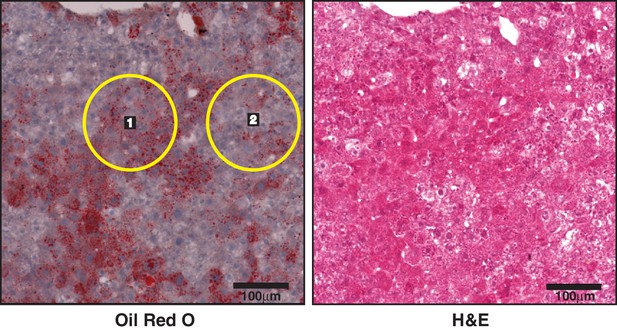
Increased lipid content in tumor regions of LNA-24; SB(+)NRASG12V mice.
Representative image of ORO stain (left) and H and E (right) from liver of a SB(+)NRASG12V mouse treated with LNA-24. Lipid content (ORO stain) in tumor (yellow circle numbered 1) or adjacent non-tumor liver tissue (yellow circle numbered 2), as indicated by H and E staining, was quantified from at least four distinct tumor and non-tumor regions per mouse liver section (from n = 3 mice) and presented in Figure 2C.
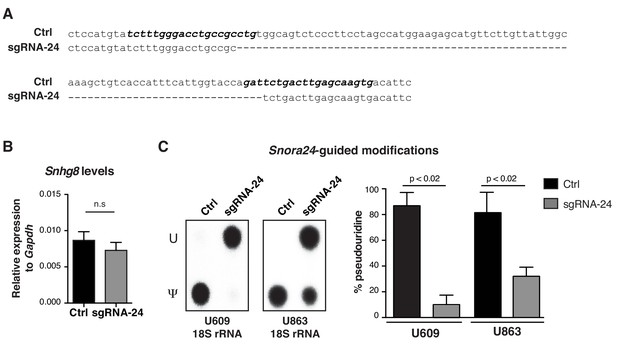
Reduction of Snora24-guided modifications in mouse KrasG12D liver cancer cells using CRISPR-Cas9 gene editing.
(A) The mouse Snora24 gene sequence is shown (Ctrl) aligned to the sequence of the Snora24 gene upon generating an 81 nucleotide deletion following CRISPR-Cas9 gene editing in KrasG12D HCC cells (sgRNA-24). The binding sites of the sgRNAs used to target mouse Snora24 are highlighted in bold and italics. (B) qPCR analysis of Snhg8 in Ctrl KrasG12D and sgRNA-24 KrasG12D HCC cells. Graph shows mean relative expression ± SD normalized to the levels of Gapdh from three independent experiments. Statistical analysis was performed using an unpaired Student’s t test, n.s = non significant. (C) Representative TLC of site-specific amounts of pseudouridine (Ψ) or uridine (U) present at position U609 and U863 on 18S rRNA using SCARLET in Ctrl KrasG12D and sgRNA-24 KrasG12D HCC cells (left panel). Quantification of TLC showing the percentage pseudouridine at position U609 (left) and U863 (right) on 18S rRNA using SCARLET in Ctrl KrasG12D and sgRNA-24 KrasG12D HCC cells. Graph shows mean ± SD percentage pseudouridine for the indicated residue from three independent experiments. Statistical analysis was performed using an unpaired Student’s t-test, p<0.02 (right panel).
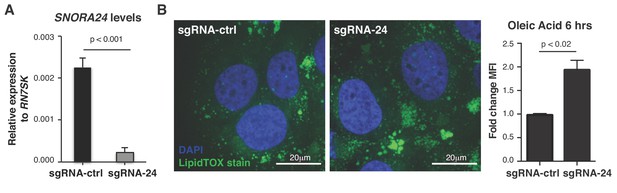
Reduction of SNORA24 in HuH-7 cells using CRISPR-Cas9 gene editing.
(A) qPCR analysis of SNORA24 in HuH-7 sgRNA-24 cells compared to HuH-7 sgRNA-ctrl cells. Graph shows mean SNORA24 expression ± SD normalized to the levels of RN7SK from three independent experiments. Statistical analysis was performed using an unpaired Student’s t-test, p<0.001. (B) Representative image of LipidTOX (neutral lipid stain) and DAPI (nuclei) stain in HuH-7 sgRNA-24 (right image) compared to sgRNA-ctrl (left image) cells 6 hrs following Oleic Acid (OA) addition to the media. Graph shows mean ± SD fold change MFI LipidTOX stain from n = 3 independent experiments. Statistical analysis was performed using an unpaired Student’s t-test, p<0.02.
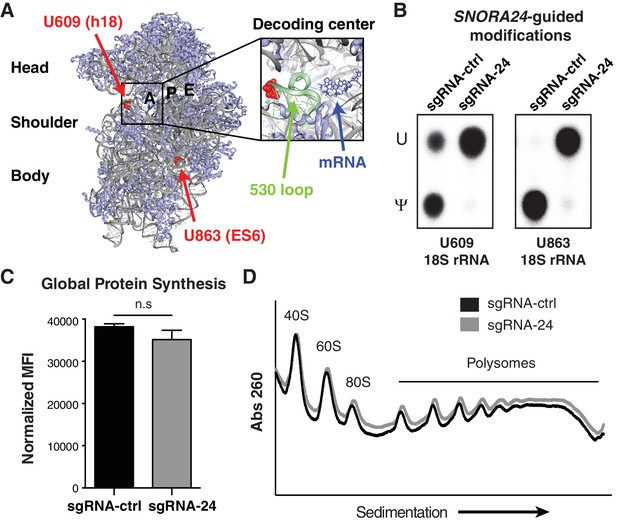
Loss of SNORA24-guided pseudouridine modifications does not impact global protein production in HCC cells.
(A) SNORA24 target residues U609 and U863 are highlighted (in red) on the structure of the mammalian 40S ribosomal subunit (Protein Data Bank (PDB) ID: 5LZS [Shao et al., 2016] and visualized with PyMOL). 18S rRNA is in gray, the mRNA in the decoding center is highlighted in blue, and the ‘530 loop’ is highlighted in pale green. (B) Representative thin layer chromatography (TLC) of site-specific amounts of pseudouridine (Ψ) or uridine (U) present at position U609 and U863 on 18S rRNA using SCARLET in HuH-7 sgRNA-ctrl or sgRNA-24 cells. (C) Graph illustrates mean ± SD mean fluorescent intensity (MFI) of the amount of de novo protein synthesis in HuH-7 sgRNA-24 cells compared to HuH-7 sgRNA-ctrl cells by measuring OPP incorporation into newly synthesized protein from three independent experiments. Statistical analysis was performed using an unpaired Student’s t-test, n.s = non significant. (D) Representative polysome profiles of HuH-7 sgRNA-ctrl (black line) and sgRNA-24 (gray line) cells. Lower molecular weight (MW) complexes (40S and 60S) are on the left side of the x axis and higher MW complexes (polysomes) are on the right side.
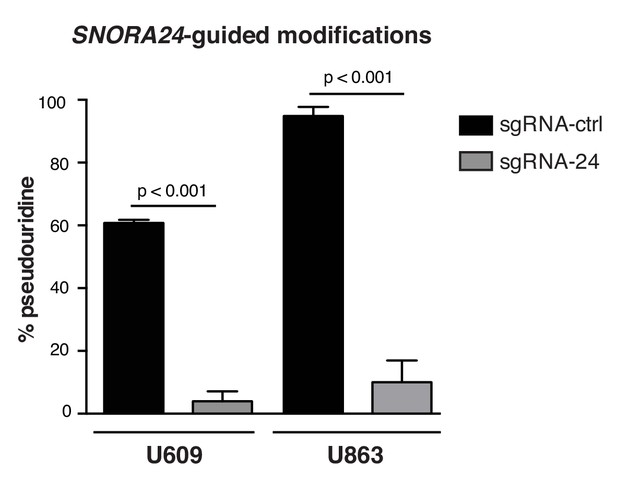
Loss of SNORA24-guided modifications in HuH-7 cells with reduced SNORA24.
Quantification of TLC showing the percentage pseudouridine at position U609 (left) and U863 (right) on 18S rRNA using SCARLET in HuH-7 cells described in Figure 2—figure supplement 3A. Graph shows mean ± SD percentage pseudouridine for the indicated residue from three independent experiments. Statistical analysis was performed using an unpaired Student’s t-test, p<0.001. Relates to Figure 3B.
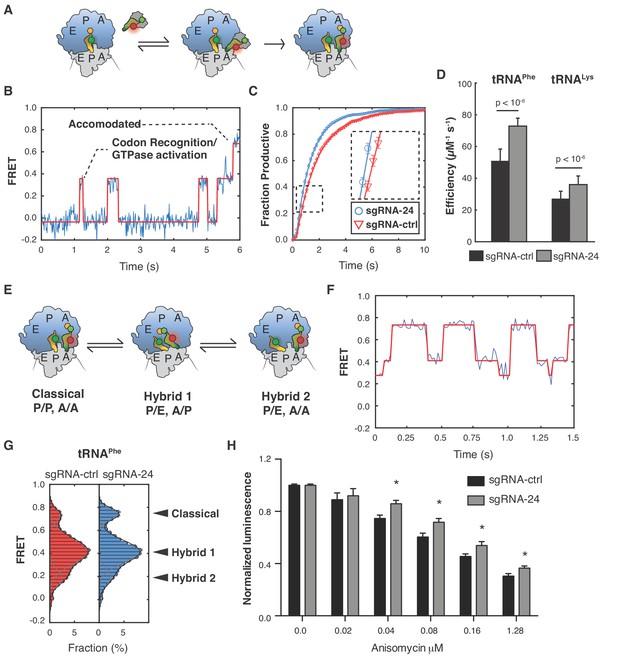
Ribosomes lacking SNORA24-guided modifications display alterations in aa-tRNA selection and pre-translocation complex dynamics.
(A) Schematic representation of the reaction assayed in the smFRET experiments. Ternary complex consisting of eEF1A, GTP, and fluorescently labeled aa-tRNA (either LD655-tRNAPhe or LD650-tRNALys) binds to ribosomes, carrying Cy3 labeled Met-tRNAfMet in the P site and displaying a cognate codon (UUC or AAA) in the A site, bringing the two dyes close enough for FRET. After binding, the ternary complex either dissociates or codon recognition, GTP hydrolysis, and subsequent dissociation of eEF1A takes place followed by accommodation of the CCA end of the tRNA into the peptidyl transferase center and subsequent peptide bond formation. These conformational changes within the decoding ribosome complex lead to a stepwise increase in FRET until stable accommodation of the tRNA occurs. (B) Representative smFRET trace of ribosome purified from HuH-7 sgRNA-ctrl cells displaying a UUC codon reacting with LD655-tRNAPhe containing ternary complex. Non-productive events are characterized by rapid fluctuations in FRET values between 0.2 ± 0.075 and 0.46 ± 0.075, followed by dissociation of the ternary complex and loss of FRET. Productive events are characterized by a stepwise progression from 0.2 ± 0.075, through several intermediate FRET values to a final FRET of 0.72 ± 0.075. The red line represents a hidden Markov-model idealization of the smFRET trace. (C) Cumulative distributions for ribosomes purified from HuH-7 sgRNA-ctrl (red line) and sgRNA-24 (blue line) cells displaying a UUC codon reacting with LD655-tRNAPhe containing ternary complex. Distributions were constructed from all recorded individual smFRET traces by estimating the number of productive events that occurred at each movie frame. The solid lines represent exponential functions fitted to the data. The error bars represent SEM for each data point. For simplicity, data from every tenth movie frame is shown. (D) Graph shows catalytic efficiency (kcat/KM) for LD655-tRNAPhe or LD650-tRNALys containing ternary complexes reacting on ribosomes purified from either HuH-7 sgRNA-ctrl or sgRNA-24 cells and displaying the respective cognate codons, UUC or AAA, in the A site. The error bars represent SEM for the estimated kcat/KM values. Statistical analysis was performed using Welch’s t-test, p<10−6. (E) Schematic representation of the dynamics of ribosome pre-translocation complexes. Each ribosome can occupy three distinct conformational states, the classical state; with both tRNAs in classical binding conformations with their anticodon stems and CCA ends occupying corresponding binding sites on the small and large ribosomal subunits. The first hybrid state; with both tRNAs in hybrid binding conformations with their anticodon stems and CCA ends occupying different binding sites on the small and large subunits, and the second hybrid state where the A-site tRNA is in a classical binding conformation while the P-site tRNA is in a hybrid binding conformation. (F) Representative smFRET trace of a pre-translocation complex containing P-site bound tRNAfMet and A-site bound Met-Phe-tRNAPhe. The highest FRET state corresponds to the classical state, the middle FRET state corresponds to the first hybrid state, and the lowest FRET state corresponds to the second hybrid state. The solid red line represents a hidden Markov-model idealization of the smFRET trace. (G) Histograms of FRET values attained by pre-translocation complexes from HuH-7 sgRNA-ctrl (red bars) and sgRNA-24 (blue bars) cells containing P-site bound tRNAfMet and A-site bound Met-Phe-tRNAPhe. The solid lines represent fits of three gaussian functions (gray lines) to the data and their sum (black line). The error bars represent SEM for the mean count in each histogram bin. Statistical analysis was performed using Welch’s t-test, p<10−6. (H) Graph shows mean ± SD normalized luminescence from CellTiter-Glo Luminescent Cell Viability Assay 48 hrs post-treatment of HuH-7 sgRNA-ctrl (black bars) or sgRNA-24 (gray bars) cells with increasing concentrations of Anisomycin from three independent experiments. Statistical analysis was performed using an unpaired Student’s t-test, *p<0.05.
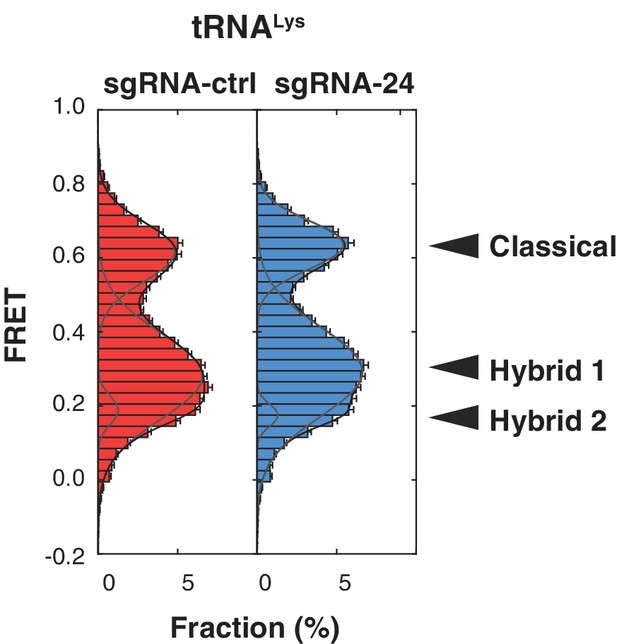
Loss of SNORA24-guided modifications does not impact the dynamics of pre-translocation complexes containing A-site bound Met-Lys-tRNALys.
Histograms of FRET values attained by pre-translocation complexes isolated from HuH-7 sgRNA-ctrl (red bars) and sgRNA-24 (blue bars) cells containing P-site bound tRNAfMet and A-site bound Met-Lys-tRNALys. The solid lines represent fits of three Gaussian functions (gray lines) to the data and their sum (black line). The error bars represent SEM for the mean count in each histogram bin.
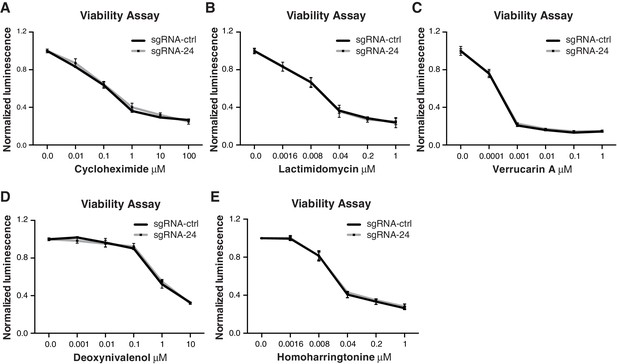
Effects of specific ribosome targeting drugs on the growth of HCC cells lacking SNORA24-guided rRNA modifications.
Graphs show mean ± SD normalized luminescence from CellTiter-Glo Luminescent Cell Viability Assay 48 hrs post-treatment of HuH-7 sgRNA-ctrl (black line) or sgRNA-24 (gray line) cells with increasing concentrations of (A) Cycloheximide (B) Lactimidomycin (C) Verrucarin A (D) Deoxynivalenol, and (E) Homoharringtonine from three independent experiments. Statistical analysis was performed using an unpaired Student’s t-test.
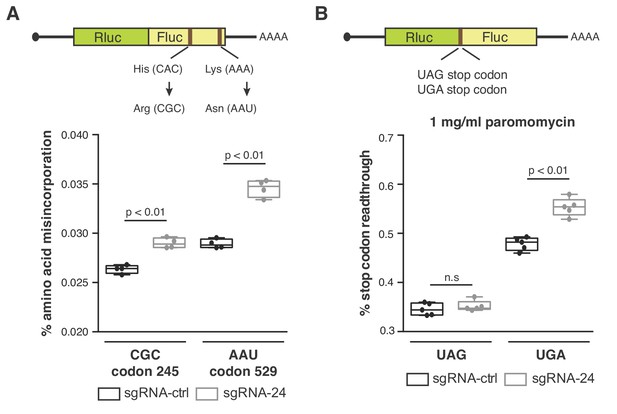
SNORA24-guided modifications impact translation accuracy.
(A) Diagram of luciferase reporters used to monitor amino acid misincorporation. Point mutations in Fluc at either His 245 or Lys 529 to near-cognate codons are highlighted (top panel). Amino acid misincorporation (%) of the indicated luciferase reporters (CGC codon or AAU codon) in HuH-7 sgRNA-ctrl cells (black) and HuH-7 sgRNA-24 cells (gray) are shown (bottom panel). On the box and whisker plots, the center line is the medium amino acid misincorporation (%), box limits are minimum and maximum values, and whiskers are S.D. from four independent experiments. Statistical analysis was performed using an unpaired Student’s t-test, p<0.01. (B) Diagram of luciferase reporters used to monitor stop codon readthrough (top panel). Stop codon readthrough (%) of the indicated luciferase reporters (UAG stop codon or UGA stop codon) in the presence of 1 mg/ml paromomycin in HuH-7 sgRNA-ctrl cells (black) and HuH-7 sgRNA-24 cells (gray) are shown (bottom panel). On the box and whisker plots, the center line is the medium stop codon readthrough (%), box limits are minimum and maximum values, and whiskers are S.D. from five independent experiments. Statistical analysis was performed using an unpaired Student’s t-test, p<0.01 and n.s = non significant.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Mus musculus) | Wild-type C57BL/6J | The Jackson Laboratory | #000664; RRID:IMSR_JAX:000664 | |
| Genetic reagent (Mus musculus) | KrasG12D | PMID:15093544 | MGI:2429948 | |
| Genetic reagent (Mus musculus) | Alb-cre | PMID:9867845 | MGI:2176228 | |
| Cell line (Homo sapiens) | Primary human skin fibroblasts | Coriell Cell Repositories | Cat. # GM00730; RRID:CVCL_L944 | |
| Cell line (Homo sapiens) | HuH-7 | Japanese Collection of Research Bioresources Cell Bank | Cat. # JCRB0403; RRID:CVCL_0336 | |
| Cell line (Homo sapiens) | 293T | American Type Culture Collection | Cat. # CRL-3216; RRID:CVCL_0063 | |
| Cell line (Mus musculus) | KrasG12DHCC cell line | PMID:30643286 | Laboratory of Davide Ruggero (UCSF) | |
| Antibody | RAS (rabbit polyclonal) | Cell Signaling Technology | Cat. #3965; RRID:AB_2180216 | (1:1000) |
| Antibody | PTEN (rabbit monoclonal) | Cell Signaling Technology | Cat. #9188; RRID:AB_2253290 | (1:1000) |
| Antibody | β-actin (mouse monoclonal) | Sigma-Aldrich | Cat. # A5316; RRID:AB_476743 | (1:10,000) |
| Antibody | p21 (mouse monoclonal) | BD Biosciences | Cat. # 556431 (clone SXM30); RRID:AB_396415 | (1:50) |
| Antibody | NRAS (mouse monoclonal) | Santa Cruz Biotechnology | Cat. # sc-31 (clone F155); RRID:AB_628041 | (1:50) |
| Recombinant DNA reagent | NRASG12V | Addgene, PMID:19147555 | Plasmid #20205; RRID:Addgene_20205 | (pT/Caggs-NRASV12) |
| Recombinant DNA reagent | SB13 | Addgene, PMID:19147555 | Plasmid #20207; RRID:Addgene_20207 | (PT2/C-Luc//PGK-SB13) |
| Recombinant DNA reagent | HRASG12V | Addgene | Plasmid #9051; RRID:Addgene_9051 | (pBABE puro H-Ras V12) |
| Recombinant DNA reagent | PTEN shRNA | PMID:29720449 | (pLKO.1-PTEN-shRNA) Laboratory of Davide Ruggero (UCSF) | |
| Recombinant DNA reagent | Rluc-Fluc control | PMID:30576652 | (pCMV-WT: CMV promoter, Rluc-Fluc) Laboratory of Maria Barna (Stanford University) | |
| Recombinant DNA reagent | CGC codon 245 | Other | (pCMV-245 CGC: CMV promoter, Rluc-Fluc) Laboratory of Maria Barna (Stanford University) | |
| Recombinant DNA reagent | AAU codon 529 | This paper | Generated by site-directed mutagenesis of plasmid pCMV-WT at codon 529 from AAA to AAT (pCMV-529 AAU) | |
| Recombinant DNA reagent | Readthrough control | PMID:22099312 | (pJD175f (pHDL-SV40-control)) Laboratory of Jonathan Dinman (University of Maryland) | |
| Recombinant DNA reagent | UAG stop codon | Other | (pJD1644 (pHDL-SV40-UAG)) Laboratory of Jonathan Dinman (University of Maryland) | |
| Recombinant DNA reagent | UGA stop codon | Other | (pJD1645 (pHDL-SV40-UGA)) Laboratory of Jonathan Dinman (University of Maryland) | |
| Sequence-based reagent | Oligonucleotides for qPCR analysis | This paper | Supplementary file 2 | |
| Sequence-based reagent | Synthetic mRNA Met-Phe | Dharmacon | CAACCUAAAACUUACACACCCUUAGAGGGACAAUCGAUGUUCAAAGUCUUCAAAGUCAUC | |
| Sequence-based reagent | Synthetic mRNA Met-Lys | Dharmacon | CAACCUAAAACUUACACACCCUUAGAGGGACAAUCGAUGAAAUUCGUCUUCAAAGUCAUC | |
| Commercial assay or kit | Dual-Luciferase Reporter Assay System | Promega | Cat. # E1910 | |
| Commercial assay or kit | Senescence Detection Kit | Calbiochem-Millipore | Cat. # QIA117 | |
| Commercial assay or kit | CellTiter-Glo Luminescent Cell Viability Assay | Promega | Cat. # G7570 | |
| Chemical compound, drug | Anisomycin | Sigma-Aldrich | Cat. # A9789 | |
| Chemical compound, drug | Paromomycin | Sigma-Aldrich | Cat. # P9297 | |
| Chemical compound, drug | Cycloheximide | Sigma-Aldrich | Cat. # C7698 | |
| Chemical compound, drug | O-propargyl-puromycin | Medchem Source LLP | Cat. # JA-1024 | |
| Chemical compound, drug | Cy3-Maleimide | GE Healthcare | Cat. # PA23031 | |
| Chemical compound, drug | LD655-NHS | Lumidyne Technologies | Cat. # 08 | |
| Chemical compound, drug | LD650-NHS | Lumidyne Technologies | Cat. # 99 | |
| Software, algorithm | PyMOL | Schrödinger, NY, USA | https://www.pymol.org/2/ | |
| Software, algorithm | ImageJ | National Institute of Health, USA | https://imagej.nih.gov/ij/ | |
| Software, algorithm | GraphPad Prism six software | GraphPad | https://www.graphpad.com/ | |
| Software, algorithm | Spartan | Other | Available at: https://www.scottcblanchardlab.com/software |
Additional files
-
Supplementary file 1
Summary of HCC patients demographics and staging.
- https://doi.org/10.7554/eLife.48847.021
-
Supplementary file 2
Sequences of primers used.
- https://doi.org/10.7554/eLife.48847.022
-
Transparent reporting form
- https://doi.org/10.7554/eLife.48847.023





