Cytoplasmic retention and degradation of a mitotic inducer enable plant infection by a pathogenic fungus
Figures
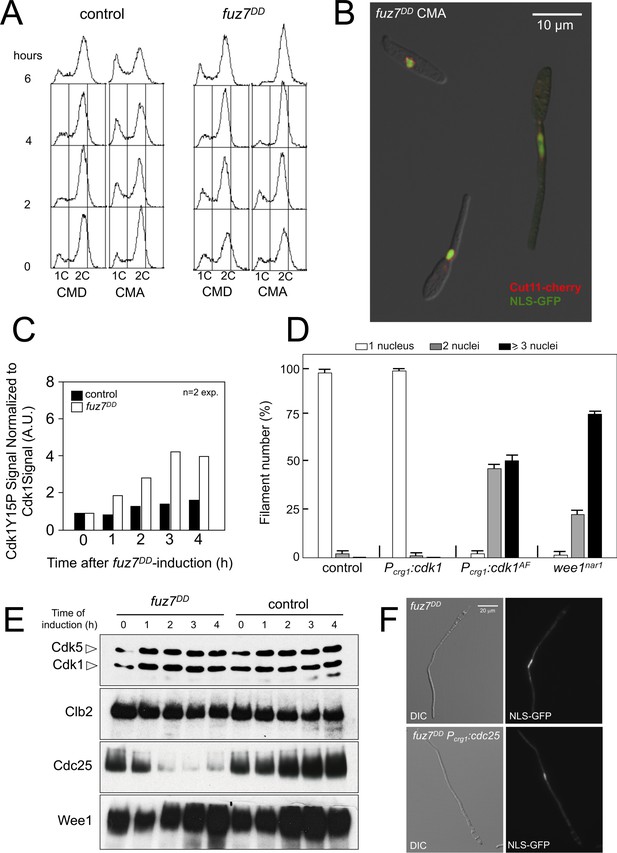
Expression of fuz7DD allele promotes a G2 cell cycle arrest that depends on Cdk1 inhibitory phosphorylation.
(A) Cells expressing the fuz7DD allele accumulated with a 2C DNA content. Fluorescence/Activated Cell Sorter (FACS) analysis of the DNA content of a control strain and a strain carrying an ectopic copy of the fuz7DD allele under the control of the crg1 promoter growing in inducing (Complete Medium Arabinose, CMA) and non-inducing (Complete Medium Glucose, CMD) conditions (Figure 1—figure supplement 1). The period of incubation in testing media is indicated (hours). (B) Cells expressing the fuz7DD allele induce conjugative hyphae that are arrested in G2 phase. Representative image of cells expressing the fuz7DD allele and carrying NLS-GFP and Cut11-Cherry fusions to detect the nucleus and the nuclear envelope, growing in CMA for 6 hr. This image was a composition from various images to show different stages during the production of the conjugation hyphae. Bar: 15 μm. (C) Cells expressing the fuz7DD showed increased levels of Cdk1 inhibitory phosphorylation (Cdk1Y15P). Data acquisition is described in Figure 1—figure supplement 2A and. Means are shown (Figure 1—source data 1). (D) Interfering with the Cdk1 inhibitory phosphorylation resulted in inability to arrest cell cycle upon fuz7DD allele expression. Fuz7DD-derived strains carrying the NLS-GFP reporter as well as the indicated mutations were incubated in inducing conditions (CMA) for 6 hr. Filaments were sorted as carrying 1, 2 or 3 and more nuclei. The graph shows the result from three independent experiments, counting more than 100 filaments each. Means and SDs are shown (Figure 1—figure supplement 2 and Figure 1—source data 2). (E) Protein levels of G2/M regulators upon fuz7DD allele expression. Strains carrying HA-tagged versions of Clb2, Cdc25 and Wee1 and carrying the fuz7DD allele or not (control) were incubated for the indicated time in induction conditions (CMA). Similar amount of protein extracts was separated by SDS-PAGE. Immunoblots were incubated with an antibody against HA. As loading control, we used the Cdk1 protein, which can be detected using anti-PSTAIRE (which recognizes both Cdk1 and Cdk5). (F) Overexpression of cdc25 does not abrogate the fuz7DD-dependent cell cycle arrest. Representative images of cultures growing in inducing conditions (CMA) for 6 hr, from a control strain expressing the fuz7DD allele, and a strain co-expressing both the fuz7DD allele and an ectopic copy of cdc25 (Figure 1—figure supplement 4).
-
Figure 1—source data 1
Data for Figure 1C.
- https://doi.org/10.7554/eLife.48943.008
-
Figure 1—source data 2
Data for Figure 1D.
- https://doi.org/10.7554/eLife.48943.009
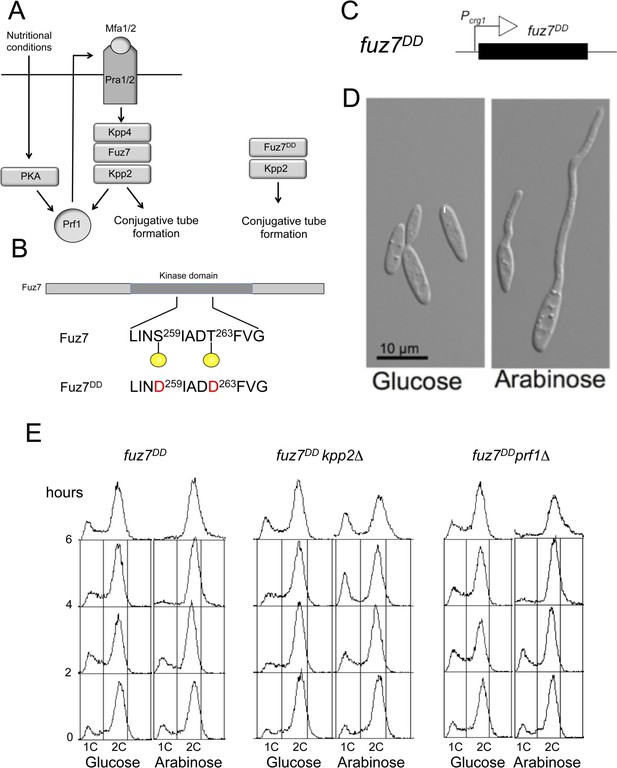
Activation of the pheromone response cascade upon expression of the fuz7DD allele.
(A) Scheme of the pheromone response cascade stressing the requirement of Prf1 phosphorylation via PKA and MAPK to induce the genes encoding the receptors and pheromone (left) and a simplified activation using the fuz7DD allele (right). Mating requires poor nutritional conditions, which through the cAMP/PKA cascade together with the MAPK cascade activate the transcriptional regulator Prf1, allowing the transcription of the mating type genes encoding the pheromone and pheromone receptor and thereby promoting a positive feedback loop that enables the pheromone response (Hartmann et al., 1999; Kaffarnik et al., 2003). (B) Amino acid changes responsible of fuz7DD allele. (C) Scheme of the transgene driving fuz7DD allele under the control of Pcrg1. (D) Representative images of cells carrying the fuz7DD allele in conditions of repression (glucose) or activation (arabinose) of crg1 promoter. Cells were grown by 6 hr in complete medium (CM) amended with the indicated carbon source. (E) DNA content analyzed by flow cytometry of strains expressing the fuz7DD allele and carrying deletions of kpp2, encoding the pheromone MAPK, or prf1, encoding the transcriptional activator responsible of pheromone and receptors gene activation. Cells were incubated by the indicated time in CMD (glucose) or CMA (arabinose).
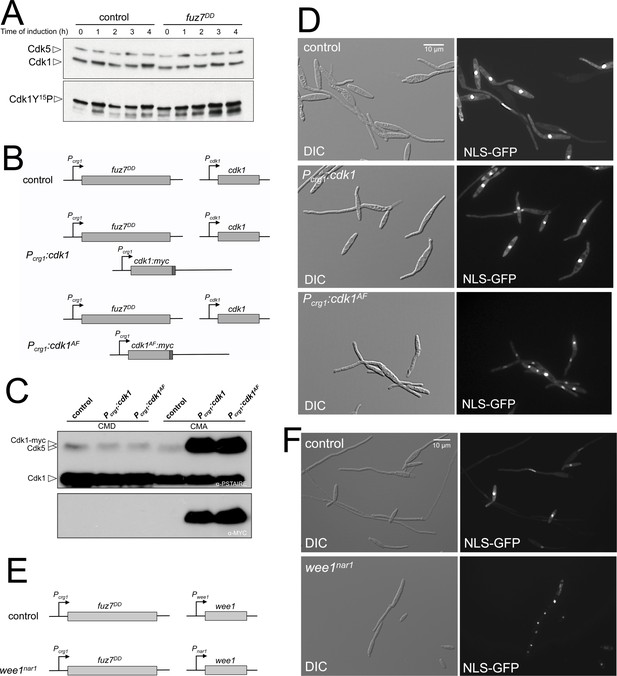
Fuz7DD-dependent cell cycle arrest requires inhibitory phosphorylation of Cdk1.
(A) Western blot analysis of inhibitory phosphorylation of Cdk1. Cells carrying or not the fuz7DD allele were grown for the indicated time in inducing conditions (CMA) and samples were taken and submitted to Western blot analysis. Immunoblots were incubated successively with an antibody that recognizes the Cdk1 phosphorylated form (anti-Cdc2-Y15P) and anti-PSTAIRE, which recognizes both Cdk1 and Cdk5. Levels of Cdk1 phosphorylation were determined by quantifying the level of antibody signal using a ChemiDoc (Bio-Rad). Differences in loading of samples were corrected by dividing each phosphopeptide-specific antibody signal by the Cdk1 (anti-PSTAIRE) antibody signal. This experiment was carried out two independent times and the values shown in Figure 1C are the average from both repeats. (B and E) Schemes of the strains used for the expression of a cdk1 allele refractory to inhibitory phosphorylation as well as the conditional strain for wee1. The ectopic copy of cdk1-myc alleles, was inserted at the intergenic region (IG) between the putative ORF UMAG_10893 and UMAG_04177 at chromosome 12, which is a permissive region for integration. The conditional wee1nar1 allele was expressed in minimal medium amended with nitrate and repressed in complete medium (CM). (C) Western blot analysis to show the level of expression of the Cdk1 ectopic transgenes (tagged with myc epitope) growing in non-inducing conditions (CMD) and inducing conditions (CMA) for 6 hr. (D and F) Representative images of cultures of strains used to acquisition of data showed in the graph of Figure 1D. Cultures were incubated for 6 hr in inducing conditions for fuz7DD and repression conditions for nar1 promoter (CMA).
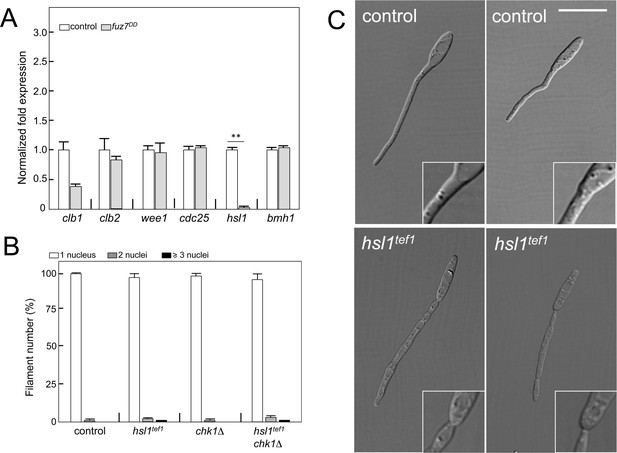
The mechanism of fuz7DD-dependent cell cycle arrest is unrelated to b-dependent cell cycle arrest.
(A) Quantitative real time-PCR for the indicated genes in a control as well as fuz7DD-expressing strains. RNA was isolated after 6 hr of induction of crg1 promoter (arabinose complete medium, CMA). As internal control, the expression of tub1 (encoding Tubulin α) was used. Values are referred to the expression of each gene in control strain. Each column represents the mean value of three independent biological replicates. Error bars represent the SD; **p<0.01 based on a two-tailed Student´s t-test to control sample. (B) Ability to arrest the cell cycle of strains carrying the fuz7DD allele as well as mutations that disable the b-dependent cell cycle arrest: loss of function of Chk1 (chk1Δ) and constitutive expression of hsl1 by exchange of its native promoter with tef1 promoter, which is not repressed (hsl1tef1). The indicated strains, which also carried the NLS-GFP transgene, were incubated in inducing conditions (CMA) for 6 hr. Filaments were sorted as carrying 1, 2 or 3 and more nuclei. The graph shows the result from three independent experiments, counting more than 100 filaments each. Means and SDs are shown. (C) Effects of the absence of repression of hsl1 in the filament morphology. Representative images of strains expressing fuz7DD and carrying (hsl1tef1) or not (control) the constitutive expression of hsl1 growing for 6 hr in CMA. Note that keeping the transcription of hsl1 active resulted in affection of the morphology of the filament as well as in the apparition of constrictions in the filament neck (inset). These morphological defects were described previously in the b-dependent filament and attributed to some interaction of Hsl1 kinase with septins (Castanheira and Pérez-Martín, 2015). Bar: 15 μm.
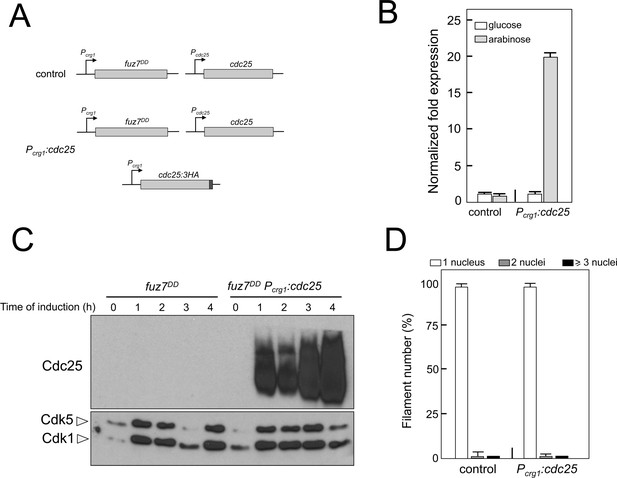
Overexpression of cdc25 does not abrogate the fuz7DD-dependent cell cycle arrest.
(A) Schemes of the strains used for the expression of an ectopic copy of cdc25-3HA allele. (B) Quantitative real time-PCR for cdc25 in the indicated strains. RNA was isolated after 4 hr of induction of crg1 promoter (arabinose complete medium, CMA). As internal control, the expression of tub1 (encoding Tubulin α) was used. Values are referred to the expression of cdc25 in control strain growing in glucose. Each column represents the mean value of three independent biological replicates. Error bars represent the SD. (C) Representative western blot analysis to show the level of expression of the cdc25 ectopic transgene (tagged with 3 HA epitope) growing in inducing conditions (CMA) for the indicated time. (D) Ability to arrest the cell cycle upon expression of fuz7DD of a strain overexpressing an ectopic copy of cdc25. The indicated strains, which also carried the NLS-GFP transgene, were incubated in inducing conditions (CMA) for 6 hr. Filaments were sorted as carrying 1, 2 or 3 and more nuclei. The graph shows the result from three independent experiments, counting more than 100 filaments each. Means and SDs are shown.
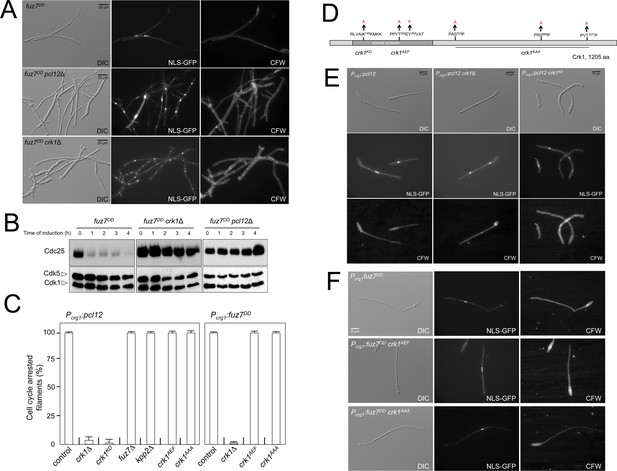
Fuz7DD-induced cell cycle arrest depends on an alternative cyclin interacting with a Cdk-like kinase.
(A) The cyclin Pcl12 and the Cdk-like kinase Crk1 were required for Fuz7DD-dependent cell cycle arrest. Representative images of cultures of strains carrying the fuz7DD allele and the indicated mutations. Cultures were incubated for 12 hr in inducing conditions for fuz7DD (CMA). Cells carried a constitutively expressed NLS-GFP reporter to detect nuclei and were stained with Calcofluor White (CFW) to detect septa. Note that filaments in the mutants were composed of cell compartments carrying one nucleus each and separated by septa. Bar: 20 μm. (B) The cyclin Pcl12 and the Cdk-like kinase Crk1 were required for Fuz7DD-dependent decrease of Cdc25 levels. Western blot analysis to show the level of Cdc25 (upper blot) upon expression of fuz7DD allele in cells growing in inducing conditions (CMA) for the indicated time. Levels of Cdk1 were used as loading control (bottom blot). (C) Ability to arrest the cell cycle upon expression of fuz7DD or pcl12 in distinct mutant strains. The indicated strains, which also carried the NLS-GFP transgene, were incubated in inducing conditions (CMA) for 6 hr. Filaments from each culture were counted and sorted as carrying 1 (cell cycle arrested) or more than one nucleus (not arrested). The graph shows the result from three independent experiments, counting more than 100 filaments each. Means and SDs are shown (Figure 2—source data 2). Representative images corresponding to the respective cultures could be found at Figure 2E and F as well as at Figure 2—figure supplement 3. (D) Scheme of Crk1, showing the mutant alleles used in this work. These mutants were already described: crk1KD, is a kinase-dead loss of function mutant; crk1AEF is refractory to T-loop activation by Fuz7; crk1AAA is refractory to phosphorylation by the MAPK Kpp2 (Garrido et al., 2004). (E) Crk1 is required for cell cycle arrest promoted upon expression of pcl12. Representative images of cultures of strains carrying an ectopic copy of pcl12 under crg1 promoter and the indicated mutations. Crk1KD carried the K145A mutation that inactivates its kinase catalytic activity (Garrido et al., 2004). Cultures were incubated for 6 hr in inducing conditions for pcl12 (CMA). Cells carried a constitutively expressed NLS-GFP reporter to detect nuclei and were stained with Calcofluor White (CFW) to detect septa. Bar: 15 μm. (F) Representative images of cultures of strains carrying the fuz7DD allele as well as the indicated mutations. Cultures were incubated for 6 hr in inducing conditions for fuz7DD (CMA). Cells carried a constitutively expressed NLS-GFP reporter to detect nuclei and were stained with Calcofluor White (CFW). Bar: 15 μm.
-
Figure 2—source data 1
Data from LC/MS.
- https://doi.org/10.7554/eLife.48943.015
-
Figure 2—source data 2
Data for Figure 2C.
- https://doi.org/10.7554/eLife.48943.016
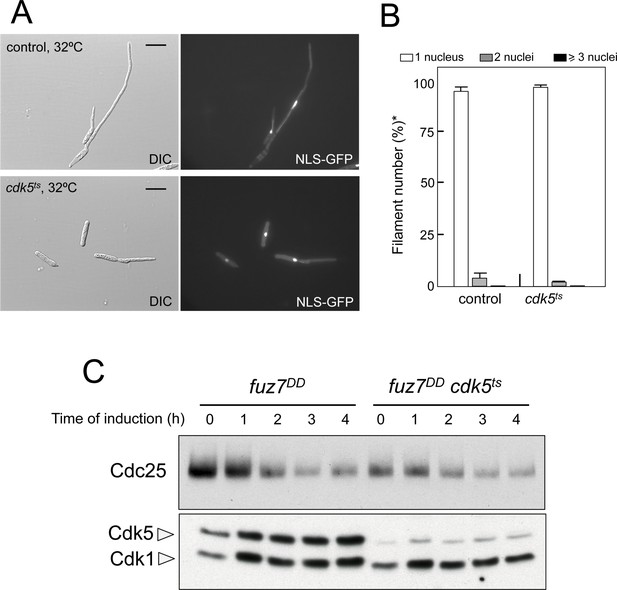
Cdk5 is not required for cell cycle arrest upon expression of the fuz7DD allele.
(A) Representative images of cultures of strains carrying the fuz7DD allele as well as a thermosensitive cdk5 allele. Cultures were incubated for 6 hr in inducing conditions for fuz7DD (CMA) and restrictive conditions for cdk5ts (32°C). Cdk5 was required for the maintenance of strong polar growth required during the formation of conjugation tubes, and in its absence the cells undergo isotropic growth resulting in the formation of a bulbous structure at the tip of the cell (Castillo-Lluva et al., 2007). Bar: 15 μm. (B) Ability to arrest the cell cycle upon expression of fuz7DD of a strain defective for Cdk5. The indicated strains, which also carried the NLS-GFP transgene, were incubated in inducing conditions (CMA) for 6 hr at 32°C. Filaments were sorted as carrying 1, 2 or 3 and more nuclei. The graph shows the result from three independent experiments, counting more than 100 filaments each. Means and SDs are shown. *In the case of cdk5ts mutant, the cells carrying bulbous structure at their tips were considered as filaments for counting purposes. (C) Western blot analysis to show the level of Cdc25 protein (upper blot) as well as the levels of Cdk5 (bottom blot). Cdk1 levels were used as loading control. Extracts were obtained from cells growing in inducing conditions (CMA) for the indicated time at 32°C.
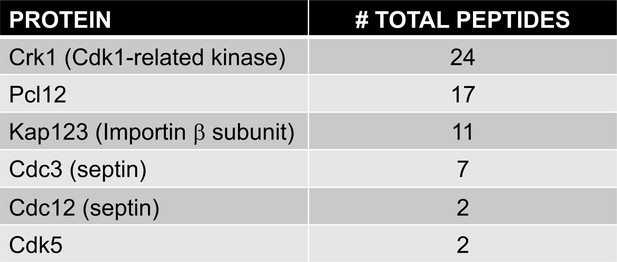
Protein interacting with Pcl12-GFP.
List of specifically bound peptides to Pcl12-GFP and found in LC/MS analysis is shown. Comprehensive data from the proteomic analysis of Pcl12-GFP-interacting proteins is included as a single Excel spreadsheet (Figure 2—source data 1).
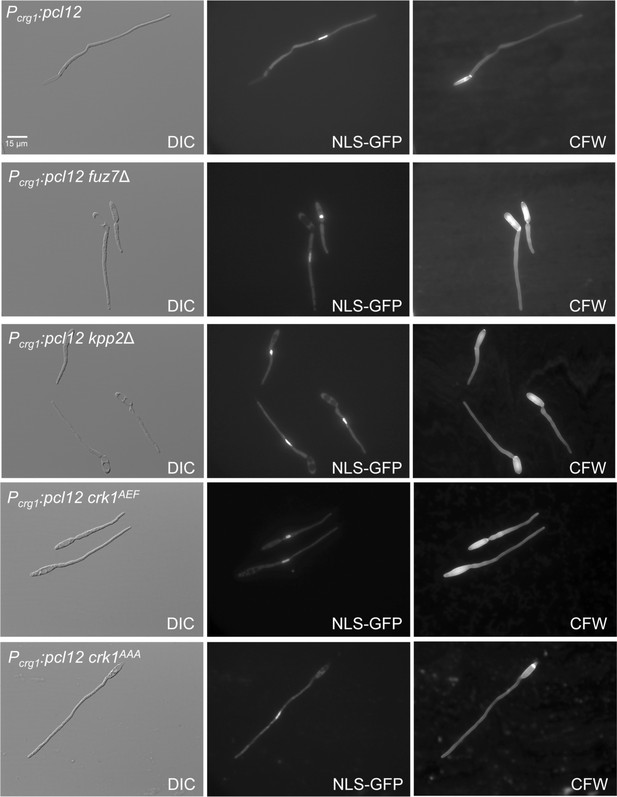
Cell cycle induced by the ectopic expression of pcl12 is independent on MAPK-mediated phosphorylation of Crk1.
Representative images of cultures of strains expressing an ectopic copy of pcl12 (Pcrg1:pcl12) as well as the indicated mutations, used for the graph showed in Figure 2C. Cultures were incubated for 6 hr in inducing conditions for Pcrg1:pcl12 (CMA). Cells carried a constitutively expressed NLS-GFP reporter to detect nuclei and were stained with Calcofluor White (CFW). Note that mutants in MAPK signaling showed an aberrant deposition of CFW staining. Bar: 15 μm.
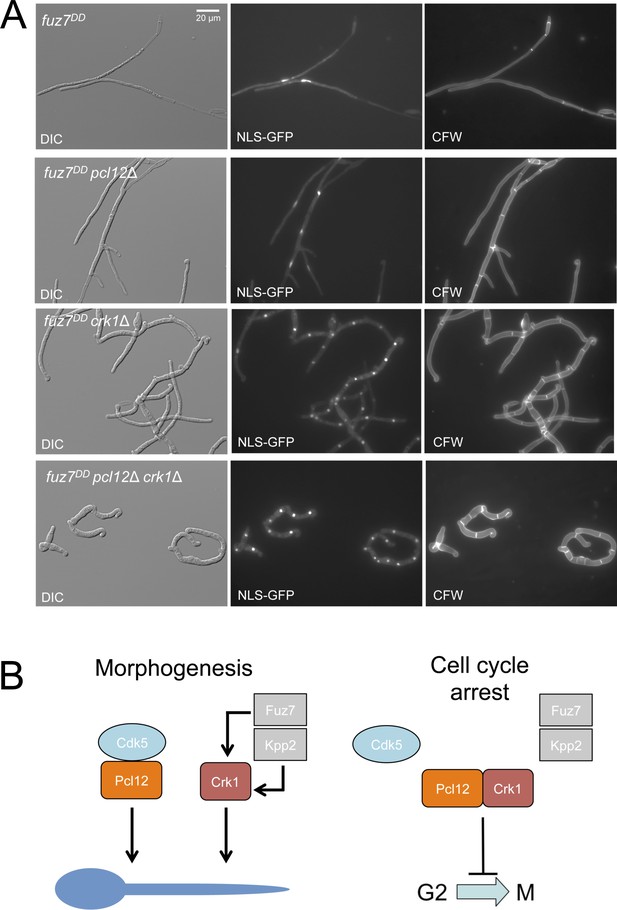
Loss-of-function mutations in crk1 and pcl12 showed additive defects in morphology.
(A) Representative images of cultures of strains expressing the fuz7DD allele, and carrying single and double mutations in crk1 and pcl12. Cultures were incubated for 12 hr in inducing conditions for fuz7DD (CMA). Cells carried a constitutively expressed NLS-GFP reporter to detect nuclei and were stained with Calcofluor White (CFW) to detect septa. Note the different morphology of filaments in single mutants as well as the enhancement of the morphological defects in double mutants. Bar: 20 μm. Control image is the same as showed in Figure 2A. (B) Scheme proposing the formation of distinct complexes of Pcl12 and Crk1 depending on their roles in morphogenesis or cell cycle arrest.
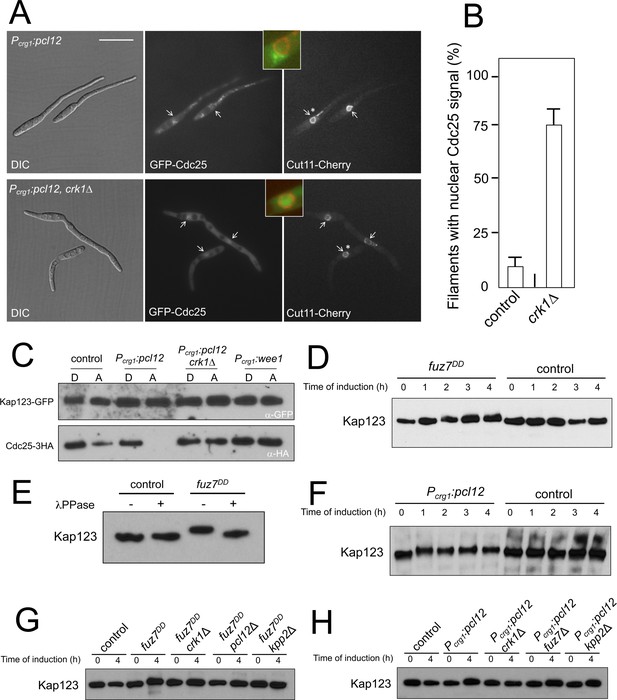
Kap123 seems to be phosphorylated upon activation of the pheromone cascade.
(A) Expression of an ectopic copy of pcl12 resulted in exclusion of Cdc25 from nucleus. Representative images of cultures of strains expressing an ectopic copy of pcl12 and the indicated mutations. The cells carried an endogenous GFP-Cdc25 fusion as well as a Cut11-cherry fusion to detect nuclear membrane. Arrows pointed to nuclei in the filaments. Insets show merged images of selected nuclei (asterisk) from the respective filaments. Cultures were incubated for 6 hr in inducing conditions (CMA). Bar: 20 μm. (B) Quantification of number of filaments showing GFP fluorescence associated with the nucleus in control (Pcrg1:pcl12) and crk1 loss of function mutant (crk1Δ). The graph shows the result from three independent experiments, counting 50 filaments each (Figure 3—source data 1). Means and SDs are shown. (C) The presence of Crk1-Pcl12 complex inhibits the interaction between Cdc25 and its importin, Kap123. Soluble extracts from strains carrying Cdc25-3HA and Kap123-GFP tagged in their corresponding endogenous loci, and carrying ectopic copies of pcl12 or wee1 under the control of crg1 promoter, were incubated with GFP-trap beads and the immunoprecipitates submitted to Western blot with anti-HA (Cdc25) and anti-GFP (Kap123) antibodies in succession. Cells were grown in inducing conditions (CMA, (A) or repressive conditions (CMD, (D) for crg1 promoter during 6 hr. (D and F) Decrease in the electrophoretic mobility of Kap123 in response to pheromone-cascade activation. Extracts from cells carrying a Kap123-3HA allele and expressing fuz7DD or pcl12 grown in inducing conditions (CMA) for the indicated time were submitted to Western blot with anti-HA. (E) The observed decrease in the electrophoretic mobility is sensitive to the treatment with phosphatase. Anti-HA immunoprecipitates of cell extracts from cultures expressing or not fuz7DD for 6 hr (CMA) were incubated at 30°C for 20 min in the absence (-) or presence (+) of lambda protein phosphatase (λ PPase) and were then subjected to immunoblot analysis with anti-HA. (G) Fuz7DD-dependent phosphorylation of Kap123 requires Pcl12, Crk1 and Kpp2. Western blot analysis of extracts from strains carrying kap123-3HA allele and the indicated mutations, incubated in inducing conditions for fuz7DD expression (CMA) for the indicated time. (H) Ectopic expression of pcl12 induces a decreased electrophoretic mobility of Kap123 that is dependent on Crk1 but not on Kpp2 and Fuz7. Western blot analysis of extracts from strains carrying kap123-3HA allele and the indicated mutations, incubated in inducing conditions for pcl12 expression (CMA) for the indicated time.
-
Figure 3—source data 1
Data for Figure 3B.
- https://doi.org/10.7554/eLife.48943.020
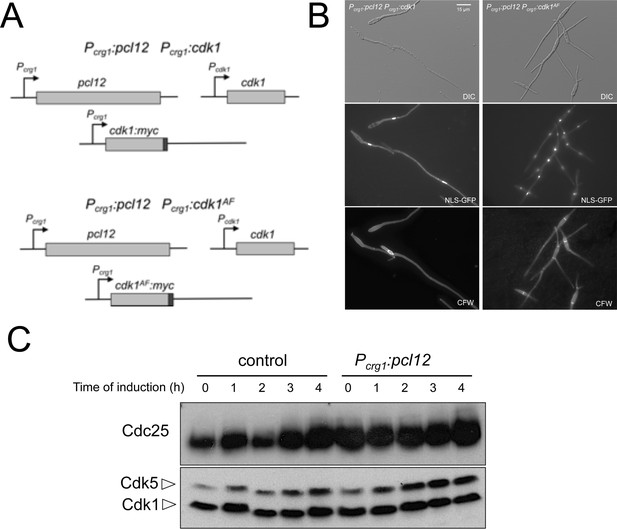
Cell cycle arrest upon ectopic expression of pcl12 relies on Cdk1 inhibitory phosphorylation, although does not down-regulate Cdc25 levels.
(A) Schemes of the strains used for the co-expression of an ectopic copy of pcl12 as well as a cdk1 allele refractory to inhibitory phosphorylation. (B) Representative images of cultures of indicated strains. Cultures were incubated for 6 hr in inducing conditions for crg1 promoter (CMA). Cells carried a constitutively expressed NLS-GFP reporter to detect nuclei and were stained with Calcofluor White (CFW) to detect the septa. Bar: 15 μm. (C) Western blot analysis showing the levels of Cdc25 (upper blot) upon ectopic expression of pcl12 in cells growing in inducing conditions (CMA) for the indicated time. Levels of Cdk1 were used as loading control (bottom blot).
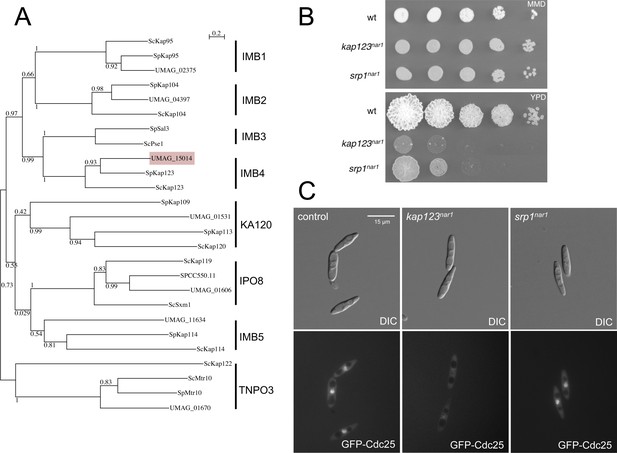
Kap123 is the importin of Cdc25.
(A) Dendrogram of β-importins from fission (Sp) and budding (Sc) yeast including putative β-importins from U. maydis (UMAG). The graph was created by the distance-based minimum-evolution method, based on 500 replicates. Bootstrap values are given (1 = 100), and branching points and the scale bar denote substitutions per site. Note that U. maydis showed β-importins from each family but for family IMB3. However, β-importins from IMB3 and IMB4 families show significant functional redundancy (Chook and Süel, 2011), opening the possibility that in U. maydis Kap123 could fulfill the roles of both families. (B) α-importin Srp1 (UMAG_10699) and β-importin Kap123 are essential in U. maydis. Conditional strains in which the importin proteins could be depleted were constructed by exchanging the endogenous promoter of srp1 and kap123 with the nar1 promoter, which is induced by growing the cells in nitrate as nitrogen source and repressed by adding ammonium or amino acids as nitrogen source. The figure shows representative plates of serial tenfold dilutions of cultures from wild-type strain and the conditional strains, spotted in solid minimal medium amended with nitrate (MMD, permissive conditions) and rich medium (YPD, restrictive conditions). Plates were incubated for 3 days at 28°C. (C) Kap123 is required for nuclear localization of Cdc25. Strains carrying an endogenous GFP-Cdc25 fusion and the conditional alleles for srp1 and kap123, were incubated in restrictive conditions (YPD) for 6 hr. Note that while Kap123 seems to be required for nuclear localization of the GFP-Cdc25 fusion, Srp1 seems to be dispensable. β-importins often are able to import cargos independently of α-importin (Xu et al., 2010).
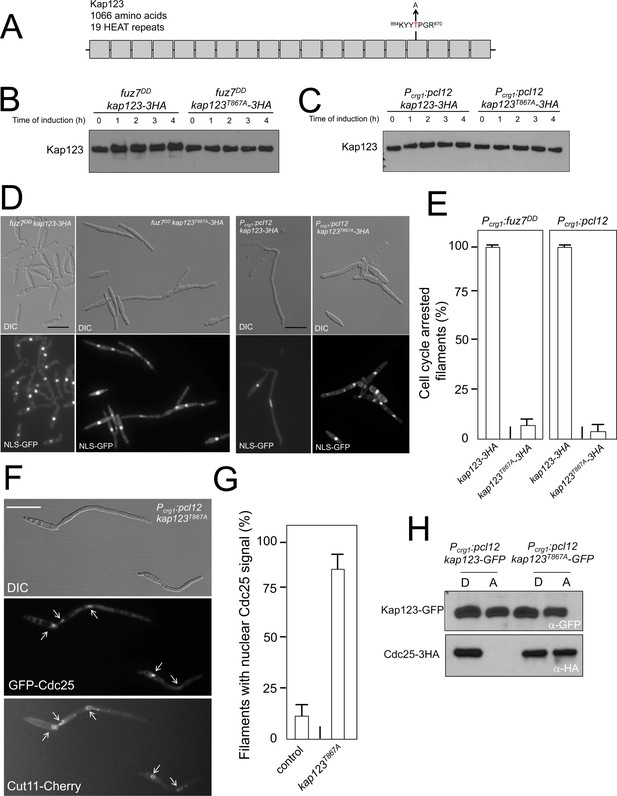
Pheromone cascade-induced cell cycle arrest is mediated by the phosphorylation of Cdc25 importin, Kap123.
(A) Scheme of Kap123 showing the change to Alanine in Threonine 867, which is located at the 16th HEAT repeat. (B and C) Absence of electrophoretic mobility decrease in the Kap123T867A mutant upon expression of fuz7DD or pcl12. Extracts from cells carrying the indicated kap123 allele and expressing fuz7DD or pcl12 grown in inducing conditions (CMA) for the indicated time were submitted to Western blot with anti-HA. (D and E) Absence of cell cycle arrest in the Kap123T867A mutant. Control cells as well as cells carrying the mutant kap123T867A allele and expressing either fuz7DD or pcl12 were incubated for 6 hr in CMA. Bar: 20 μm. Filaments from each culture were counted and sorted as carrying 1 (cell cycle arrested) or more than one nucleus (not arrested). The graph shows the result from three independent experiments, counting more than 100 filaments each (Figure 4—source data 1). Means and SDs are shown. (F) GFP-Cdc25 is not excluded from nucleus in the kap123T867A mutant strain. Representative image of strain expressing an ectopic copy of pcl12 and carrying the kap123T867A allele. The cells also carried an endogenous GFP-Cdc25 fusion as well as a Cut11-cherry fusion to detect nuclear membrane. Arrows pointed to nuclei in the filaments. Cultures were incubated for 6 hr in inducing conditions (CMA). Bar: 20 μm. (G) Quantification of number of filaments showing GFP fluorescence associated with the nucleus in control and kap123T867A mutant cells expressing pcl12 and incubated for 6 hr in inducing conditions (CMA). The graph shows the result from three independent experiments, counting 50 filaments each (Figure 4—source data 2). Means and SDs are shown. (H) Kap123T867A binds Cdc25 in the presence of a Crk1-Pcl12 complex. Soluble extracts from strains carrying Cdc25-3HA and Kap123-GFP or Kap123T867A-GFP alleles, and carrying ectopic copies of pcl12 under the control of crg1 promoter, were incubated with GFP-trap beads and the immunoprecipitates submitted to Western blot with anti-HA (Cdc25) and anti-GFP (Kap123) antibodies in succession. Cells were grown in inducing conditions (CMA, (A) or repressive conditions (CMD, (D) for crg1 promoter during 6 hr.
-
Figure 4—source data 1
Data for Figure 4E.
- https://doi.org/10.7554/eLife.48943.023
-
Figure 4—source data 2
Data for Figure 4G.
- https://doi.org/10.7554/eLife.48943.024
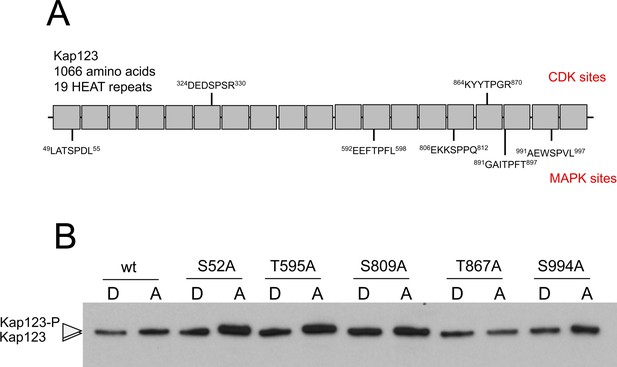
Analysis of the phosphorylation of Kap123.
(A) Scheme of Kap123 showing putative CDK as well as MAPK phosphorylation sites based on sequence. Since Crk1 has been defined both as a CDK-like and as a MAPK, and no consensus sequence for its phosphorylation sites was known, we considered the established consensus sequences for both CDK and MAPK. (B) Analysis of the electrophoretic mobility reduction of Kap123 alleles carrying distinct threonine or serine to alanine changes. In all cases the mutation was introduced in the endogenous copy of the gene at the same time the C-terminal 3 HA epitope was inserted. Ser327 and Thr894 changes were not obtained, most likely because compromised the functionality of the mutant.
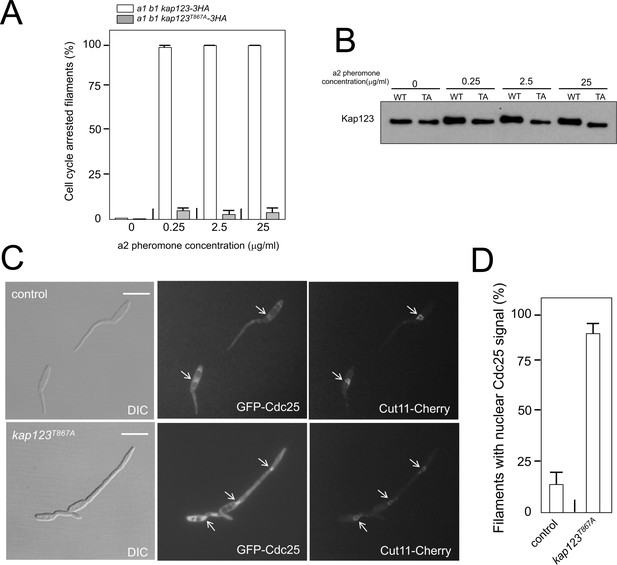
Cell cycle arrest in response to pheromone requires the phosphorylation of Kap123.
(A) kap123T867A mutant is unable to arrest the cell cycle in response to synthetic pheromone. The indicated strains were incubated for 6 hr in the presence of the indicated synthetic a2 pheromone concentrations in CMD medium. Filaments from each culture were counted and sorted as carrying 1 (cell cycle arrested) or more than one nucleus (not arrested). The graph shows the result from three independent experiments, counting more than 100 filaments each (Figure 5—source data 1). Means and SDs are shown. (B) Pheromone treatment results in a decrease in electrophoretic mobility of Kap123. Western blot from extracts obtained from a1 mating-type cells carrying HA-tagged wild-type or kap123T867A alleles that were incubated for 6 hr in the presence of the indicated synthetic a2 pheromone concentrations in CMD medium. (C) kap123T867A mutant is unable to exclude Cdc25 from nucleus in response to synthetic pheromone. Representative images of cultures of wild-type and kap123T867A mutant cells carrying endogenous GFP-Cdc25 and Cut11-Cherry gene fusions, in the presence of 0,25 μg/ml of a2 synthetic pheromone for 6 hr in CMD. Note the presence of more than one nucleus that accumulates GFP-Cdc25 in the kap123T867A mutant strain. Bar: 15 μm. (D) Quantification of number of filaments showing GFP fluorescence associated with the nucleus in control and kap123T867A mutant cells in the presence of 0,25 μg/ml of a2 synthetic pheromone for 6 hr in CMD. The graph shows the result from three independent experiments, counting 50 filaments each (Figure 5—source data 2). Means and SDs are shown.
-
Figure 5—source data 1
Data for Figure 5A.
- https://doi.org/10.7554/eLife.48943.026
-
Figure 5—source data 2
Data for Figure 5D.
- https://doi.org/10.7554/eLife.48943.027
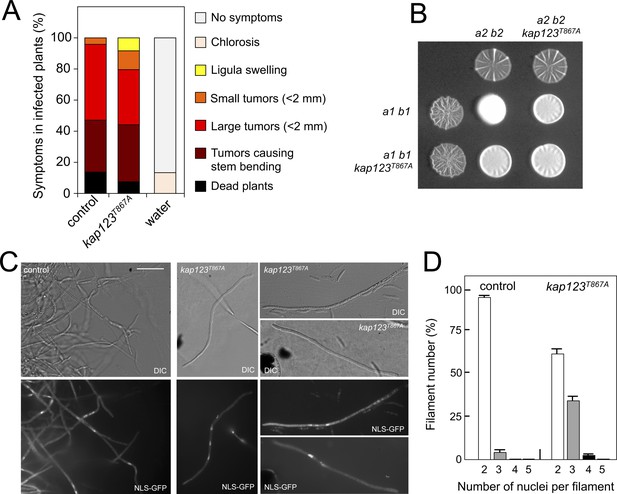
Effects of the absence of cell cycle arrest in the ability to mate and to infect plants.
(A) kap123T867A mutant strains are able to infect plants. Disease symptoms caused by crosses of wild-type and kap123T867A mutant strains. The symptoms were scored 14 days after infection. Two independent experiments were carried out and the average values are expressed as percentage of the total number of infected plants (n: 30 plants in each experiment) (Figure 6—source data 1). (B) kap123T867A mutant is able to mate. Crosses of control as well as kap123T867A mutant strains carrying compatible mating types (a1 b1 and a2 b2) in charcoal-containing agar plates. Positive fuzzy phenotype can be detected as a white-appearance mycelial growth. Note that mutant combinations were slightly affected in the ability to produce fuzzy phenotype. Plates were incubated at 22°C for two days. (C) kap123T867A mutant filaments were affected in nuclear number. Crosses from compatible strains (wild-type or kap123T867A mutant) carrying a NLS-GFP fusion under control of the b-factor-dependent dik6 promoter were scrapped from agar surface, mounted on microscopy slides and epifluorescence was observed. Representative images show DIC and fluorescence in GFP channel. Bar: 20 μm. (D) Quantification of the nuclear content of filaments obtained from charcoal plates. The graph shows the result from two independent experiments, counting more than 50 filaments each (Figure 6—source data 2). Means and SDs are shown.
-
Figure 6—source data 1
Data for Figure 6A.
- https://doi.org/10.7554/eLife.48943.030
-
Figure 6—source data 2
Data for Figure 6D.
- https://doi.org/10.7554/eLife.48943.031
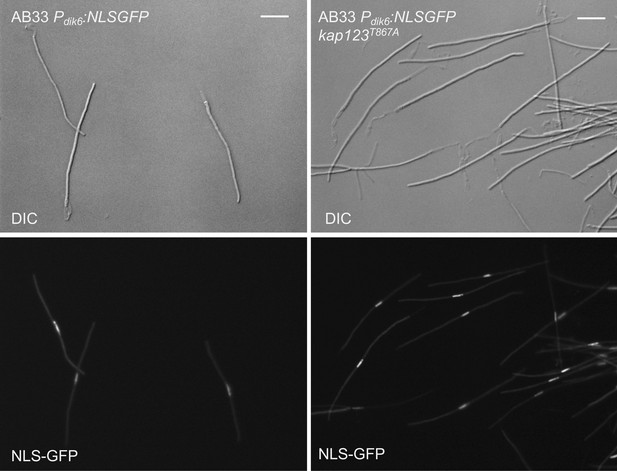
The presence of the kap123T867A allele did not affect the ability of b-factor to arrest the cell cycle.
To prove that, we took advantage of the AB33 strain, a special haploid strain carrying compatible b alleles under the control of a nitrate-induced promoter, which upon growth in minimal medium containing nitrate resulted in the formation of the b-dependent G2-arrested filament (Brachmann et al., 2001). We have introduced the kap123T867A allele in AB33-derived cells carrying a NLS-GFP reporter that is expressed in b-activation conditions. The figure shows representative images of cultures from the indicated strains and growing in inducing conditions for b transcription (minimal medium with nitrate, 8 hr). Bar: 20 μm. Note that the mutant cells were cell cycle arrested at the same extent as the control strain in similar conditions.
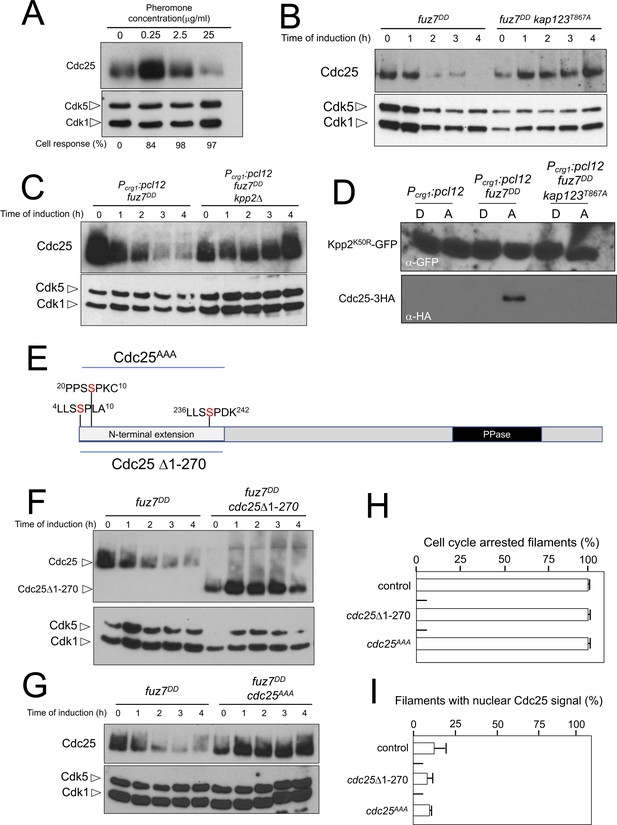
Cdc25 is negatively regulated by Kpp2, the pheromone MAPK.
(A) High levels of pheromone resulted in a decrease in Cdc25 levels. Western blot from extracts obtained from a1 mating-type cells carrying a HA-tagged Cdc25 endogenous allele that were incubated for 6 hr in the presence of the indicated synthetic a2 pheromone concentrations in CMD medium. Similar amount of protein extracts was separated by SDS-PAGE. Immunoblots were incubated with an antibody against HA. As loading control, we used the Cdk1 protein, which can be detected using anti-PSTAIRE (which recognizes both Cdk1 and Cdk5). Cell response refers to percentage of cells showing conjugation tube in each culture. (B) Inability to retain Cdc25 at the cytoplasm resulted in stabilization of Cdc25 levels in response to expression of the fuz7DD allele. Western blot analysis to show the level of Cdc25 (upper blot) in cells growing in inducing conditions (CMA) for the expression of fuz7DD allele during the indicated time. Levels of Cdk1 were used as loading control (bottom blot). (C) The MAPK Kpp2 is required for down-regulation of Cdc25 levels. Strains expressing at the same time fuz7DD and pcl12 and carrying or not a loss of function allele of kpp2, were grown during the indicated time in inducing conditions (CMA). Extracts were analyzed by Western blot to detect Cdc25-3HA protein levels (upper blot) and the levels of Cdk1, which were used as loading control (bottom blot). (D) Kpp2 is able to interact with Cdc25 upon activation of the pheromone cascade. Soluble extracts from strains carrying Cdc25-3HA and a Kpp2KD-GFP tagged in their corresponding endogenous loci, and carrying ectopic copies of the indicated genes under the control of crg1 promoter, as well as additional mutations (also indicated) were incubated with GFP-trap beads and the immunoprecipitates submitted to Western blot with anti-HA (Cdc25) and anti-GFP (Kpp2KD) antibodies in succession. Cells were grown in inducing conditions (CMA, (A) or repressive conditions (CMD, (D) for crg1 promoter during 6 hr. (E) Scheme of U. maydis Cdc25, showing the N-terminal specific extension, and the postulated MAPK phosphorylation sites, with the Serine residue (in red) exchanged to Alanine in the mutant allele cdc25AAA. (F) The N-terminal extension of Cdc25 determines its down-regulation in response to expression of the fuz7DD allele. Western blot analysis to show the level of Cdc25 or Cdc25 Δ1–270 (upper blot) upon expression of fuz7DD allele in cells growing in inducing conditions (CMA) for the indicated time. Levels of Cdk1 were used as loading control (bottom blot). (G) Putative MAPK phosphorylation sites were involved in the down-regulation of Cdc25 in response to expression of the fuz7DD allele. Western blot analysis to show the level of Cdc25 or Cdc25AAA (upper blot) upon expression of fuz7DD allele in cells growing in inducing conditions (CMA) for the indicated time. Levels of Cdk1 were used as loading control (bottom blot). (H) Cell cycle arrest in response to expression of the fuz7DD allele is unaffected by the absence of down-regulation of Cdc25. fuz7DD-expressing strains carrying the indicated cdc25 alleles, which also carried the NLS-GFP transgene, were incubated in inducing conditions (CMA) for 6 hr. Filaments from each culture were counted and sorted as carrying 1 (cell cycle arrested) or more than one nucleus (not arrested). The graph shows the result from three independent experiments, counting more than 100 filaments each (Figure 7—source data 1). Means and SDs are shown. (I) Quantification of number of filaments showing GFP fluorescence associated with the nucleus in control and strains carrying the indicated cdc25 alleles, expressing pcl12 and incubated for 6 hr in inducing conditions (CMA). The graph shows the result from three independent experiments, counting 50 filaments each (Figure 7—source data 2). Means and SDs are shown.
-
Figure 7—source data 1
Data for Figure 7H.
- https://doi.org/10.7554/eLife.48943.033
-
Figure 7—source data 2
Data for Figure 7I.
- https://doi.org/10.7554/eLife.48943.034
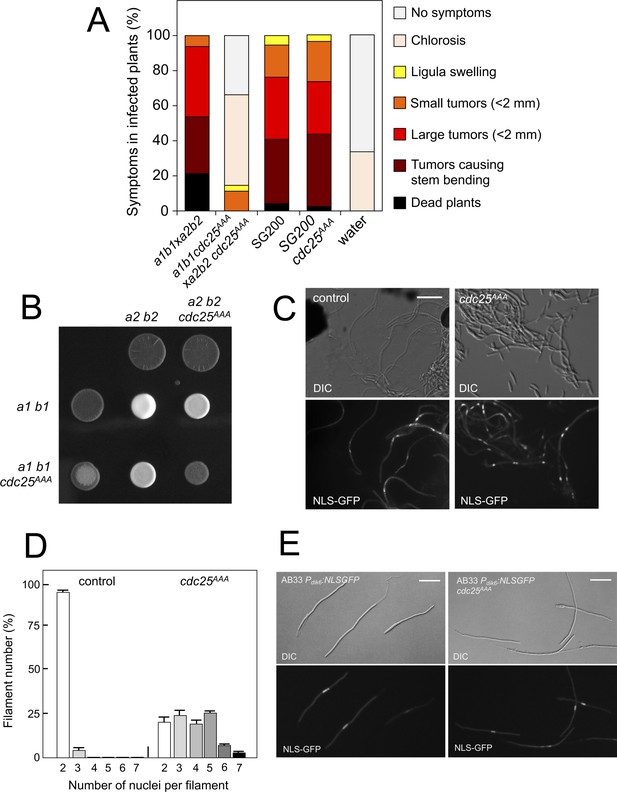
Pheromone-dependent down-regulation of Cdc25 is required for full virulence.
(A) cdc25AAA mutant strains are affected in the ability to infect plants. Disease symptoms caused by crosses of wild-type and cdc25AAA mutant strains. Strikingly, solopathogenic SG200-derived strains were not affected. The symptoms were scored 14 days after infection. Two independent experiments were carried out and the average values are expressed as percentage of the total number of infected plants (n: 30 plants in each experiment) (Figure 8—source data 1). (B) cdc25AAA mutant is affected in the formation of dikaryotic hyphae. Crosses of control as well as cdc25AAA mutant strains carrying compatible mating types (a1 b1 and a2 b2) in charcoal-containing agar plates. Positive fuzzy phenotype can be detected as a white-appearance mycelial growth. Note that mutant combinations were affected in the ability to produce fuzzy phenotype. Plates were incubated at 22°C for two days. (B) cdc25AAA mutant cells are able to fuse. Crosses from compatible strains (wild-type or cdc25AAA mutant) carrying a NLS-GFP fusion under control of the b-factor-dependent dik6 promoter were scrapped from agar surface, mounted on microscopy slides and epifluorescence was observed. Representative images show DIC and fluorescence in GFP channel. Bar: 20 μm. (C) cdc25AAA mutant filaments were affected in nuclear number. Graph shows the quantification of the nuclear content of filaments obtained from charcoal plates. The graph shows the result from two independent experiments, counting more than 50 filaments each (Figure 8—source data 2). Means and SDs are shown. (E) b-induced cell cycle arrest de novo is not affected by the presence of cdc25AAA mutant allele. Representative images of cultures from AB33-derived strains that were incubated in inducing conditions (minimal medium with nitrate) for 8 hr. Bar: 20 μm.
-
Figure 8—source data 1
Data for Figure 8A.
- https://doi.org/10.7554/eLife.48943.036
-
Figure 8—source data 2
Data for Figure 8D.
- https://doi.org/10.7554/eLife.48943.037
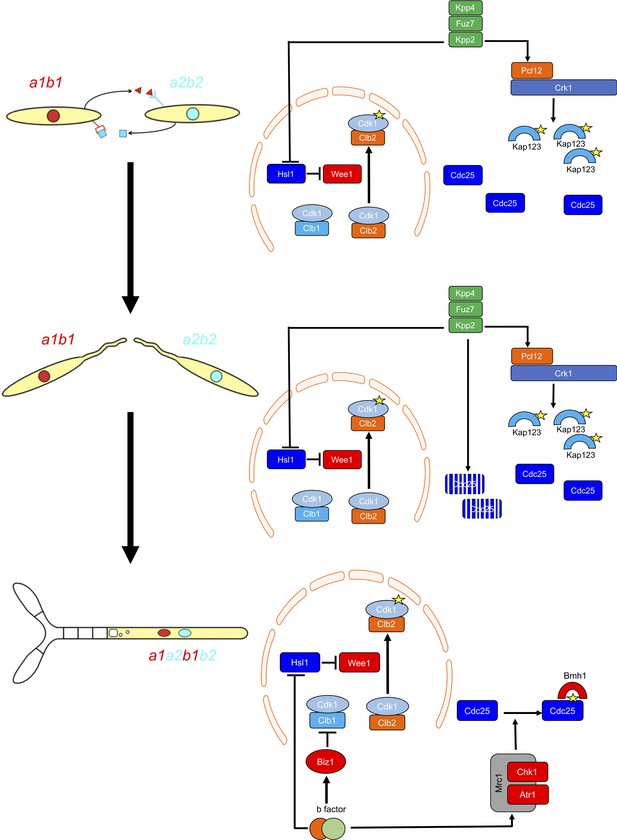
Two regulatory networks recruit both shared and specific elements to induce and sustain a G2 cell cycle arrest during the production of the infective structures in U.
maydis. Scheme showing the distinct steps at cell cycle level occurring during the formation of the infective filament of U. maydis. Arrows and bars denote positive and negative interactions. Yellow stars denote phosphorylation. For details, see discussion section.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Ustilago maydis | Strains used in this study are listed inSupplementary file 1 | Perez-Martin lab | N/A | |
| E. coli strain | DH5α | CGSC | 12384 | |
| Antibody | Anti-PSTAIRE (Rabbit polyclonal) | Santa Cruz Biotechnology, Inc | Sc-53 | RRID:AB_2074908 |
| Antibody | Anti-phospho-Cdc2 (Tyr15) (10A11, Rabbit monoclonal) | Cell Signaling | 4539 | RRID:AB_560953 |
| Antibody | Anti-HA, High affinity (3F10, Rat monoclonal) | Roche | 1 867 423 | RRID:AB_390919 |
| Antibody | Anti-HA-HRP conjugate, High affinity (3F10, Rat monoclonal) | Roche | 12 013 819 001 | |
| Antibody | Anti-c-Myc-HRP conjugate (9E10, mouse monoclonal) | Roche | 1 814 150 | |
| Antibody | Anti-GFP, Living Colors (JL-8, mouse monoclonal) | Clontech | 632380 | RRID:AB_2314359 |
| Antibody | Anti-rabbit IgG-HRP conjugate (Donkey polyclonal) | Amersham Biosciences | NA934 | RRID:AB_772206 |
| Antibody | Anti-rat IgG-HRP conjugate (Mouse, monoclonal) | SIGMA | R7636 | RRID:AB_1840005 |
| Antibody | Anti-mouse IgG-HRP conjugate (Goat polyclonal) | SIGMA | A0168 | RRID:AB_257867 |
| Chemical compound, drug | Hygromycin B | Roche | 834 555 | |
| Chemical compound, drug | clonNAT (nourseothricin) | Werner BioAgents | CAS#96736-11-7 | |
| Chemical compound, drug | G418 | Formedium | G418-1 | |
| Chemical compound, drug | Carboxine | SIGMA | 45371 | |
| Chemical compound, drug | Phleomycin | InvivoGen | ant-ph-1 | |
| Chemical compound, drug | Roche Protease Inhibitor Cocktail | Roche | 11-697-498-001 | |
| Chemical compound, drug | PhosSTOP | Roche | 04-906-837-001 | |
| Chemical compound, drug | Synthetic U. maydis a2 pheromone | Proteomic Services from National Center of Biotechnology, CSIC, Madrid | N/A | |
| Commercial assay or kit | High Pure RNA isolation kit | Roche | 11828665001 | |
| Commercial assay or kit | GFP-Trap_MA | Chromotek | gtma-400 | |
| Commercial assay or kit | IgG coupled to Dynabeads (M-270 Epoxy) | Thermo Fisher | 14304 | |
| Commercial assay or kit | High-Capacity cDNA Reverse Transcription Kit | Applied Biosystems | 4368814 | |
| Commercial assay or kit | SsoAdvanced SYBR Green supermix | BioRad | 172–5260 | |
| Commercial assay or kit | Supersignal West Femto Maximum Sensitivity Substrate | Thermo Scientific | 34095 | |
| Commercial assay or kit | Clarity Western ECL Substrate | Bio-Rad | 170–5061 | |
| Commercial assay or kit | Mini-Protean TGX gels | Bio-Rad | 456–1095 | |
| Software, algorithm | Image J | NIH | https://imagej.nih.gov/ij/ | |
| Software, algorithm | Photoshop | Adobe | https://www.adobe.com |
Additional files
-
Supplementary file 1
U. maydis strains used in this study.
- https://doi.org/10.7554/eLife.48943.039
-
Transparent reporting form
- https://doi.org/10.7554/eLife.48943.040





