Antigen presentation and tumor immunogenicity in cancer immunotherapy response prediction
Figures
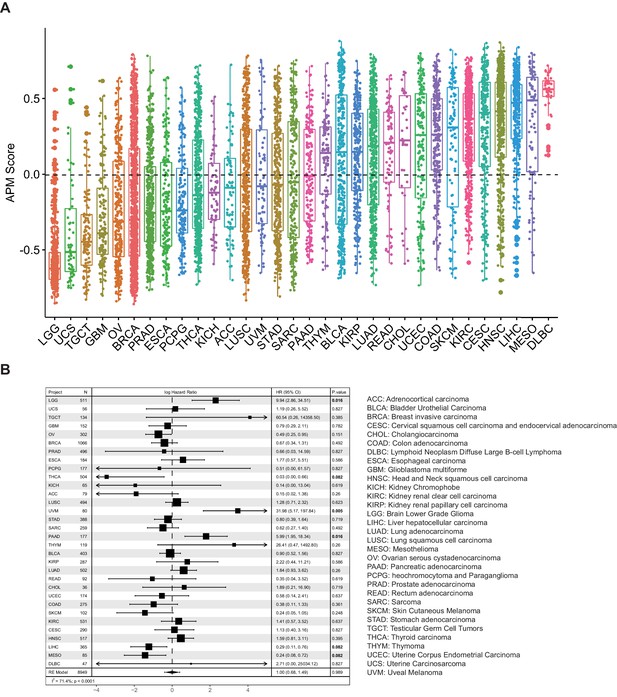
Analysis of antigen processing and presenting machinery (APM) score in 32 cancer types.
(A) APM scores were calculated with GSVA in 32 TCGA cancer types. (B) Results of Cox proportional hazards regression analysis using APM score for all solid cancers. Forest plots showing loge hazard ratio (95% confidence interval). Cox p-values are adjusted the with false discovery rate (FDR) method, p-values less than 0.1 are in bold. The pooled hazard ratio and p-value are generated by the random effect model. The statistical test for heterogeneity is also shown in the last column. Tumor types are ordered by median APM scores.
-
Figure 1—source data 1
APM gene list for GSVA.
- https://doi.org/10.7554/eLife.49020.005
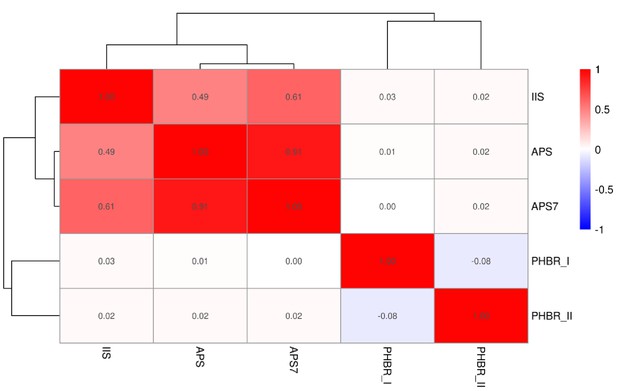
Correlations between immune infiltration score (IIS), APS, 7-APM genes, PHBR I and PHBR II in the TCGA pan-cancer dataset.
https://doi.org/10.7554/eLife.49020.004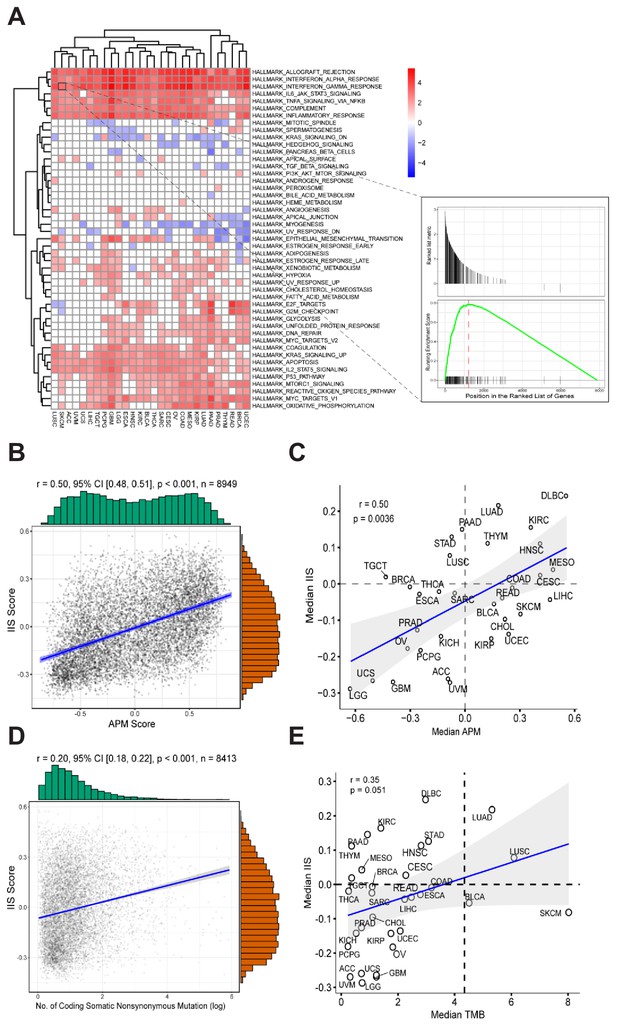
Gene expression signatures associated with high APM score.
(A) Gene sets enriched in patients with high APM score. (B) Significant correlation between APM score and IIS in 8949 cancer samples. (C) Significant correlation between APM score and IIS in different cancer types. (D) Correlation between TMB and IIS in 8413 cancer samples. (E) Correlation between TMB and IIS in different cancer types.
-
Figure 2—source data 1
Immune cell types and corresponding signature gene lists for GSVA.
- https://doi.org/10.7554/eLife.49020.011
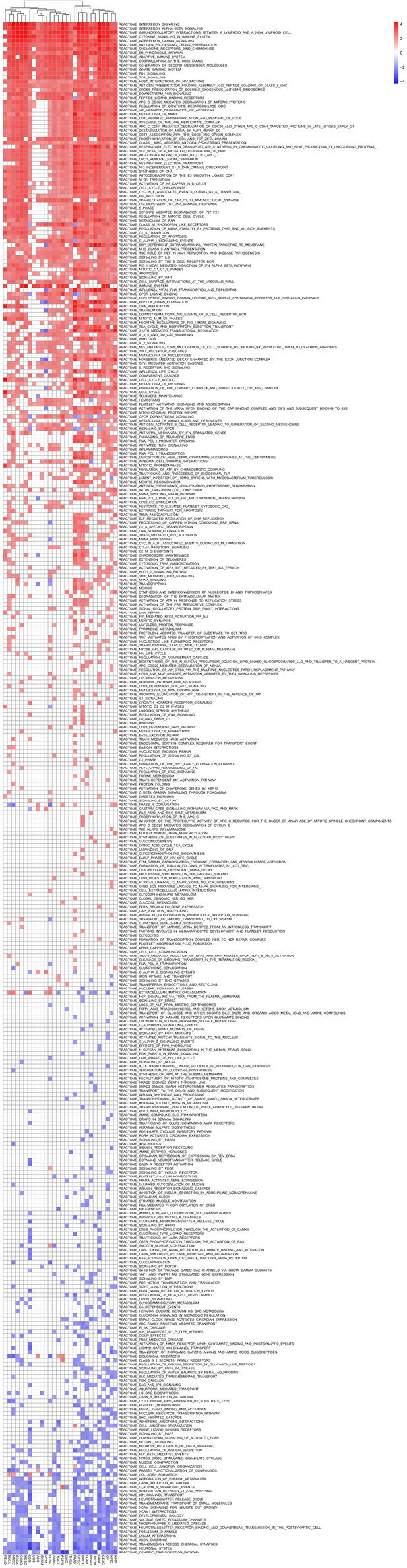
Gene sets that are enriched in 30 types of TCGA cancer patients with high APM score.
https://doi.org/10.7554/eLife.49020.007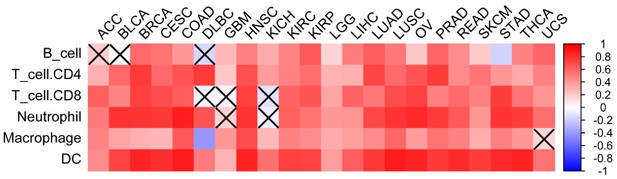
Correlation between IIS of GSVA and TIMER analysis (B_cell, etc.) results in 30 TCGA cancer types.
https://doi.org/10.7554/eLife.49020.008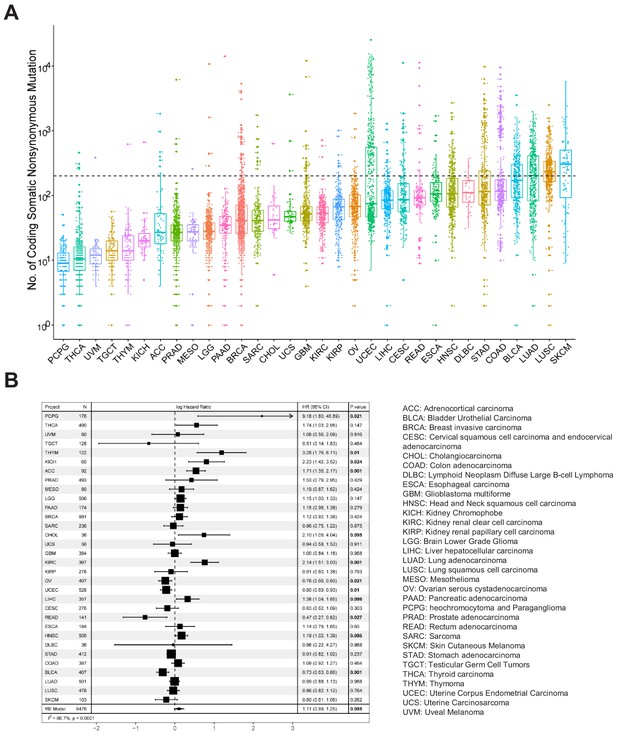
Analysis of tumor mutational burden (TMB) in 32 TCGA cancer types.
(A) Number of whole-exome non-synonymous mutation in 32 TCGA cancer types. (B) Results of Cox proportional hazards regression analysis using TMB for all solid cancers. Forest plots showing loge hazard ratio (95% confidence interval). Cox p-values are adjusted with the FDR method. p-values less than 0.1 are in bold. The pooled hazard ratio and p-value are generated by the random effect model. The statistical test for heterogeneity is also shown in the last column. Tumor types are ordered by median TMB values.
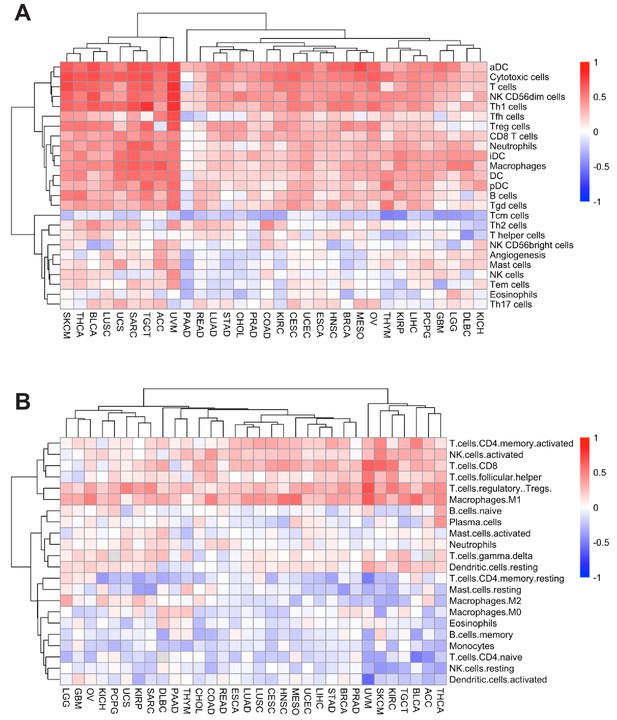
Immune cell subsets associated with APS were analyzed with IIS (A) or the CIBERSORT (B) method.
https://doi.org/10.7554/eLife.49020.010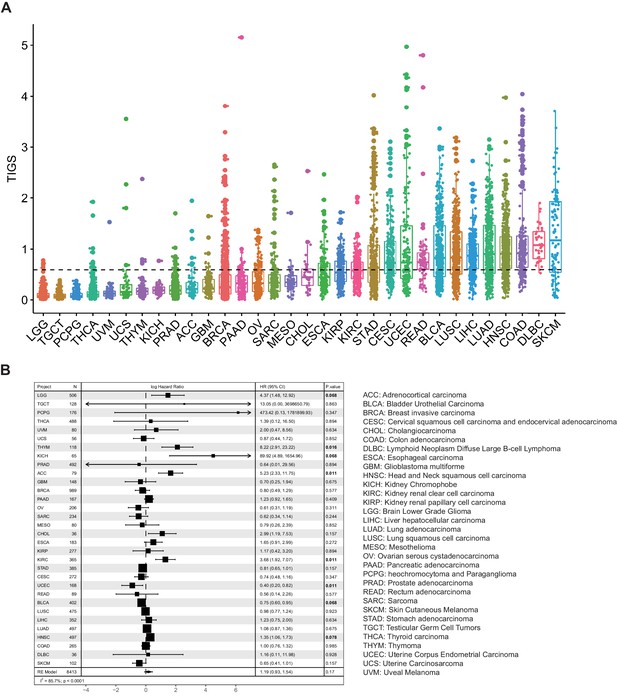
Tumor immunogenicity score (TIGS) analysis in 32 cancer types.
(A) Analysis of TIGS in 32 cancer types. (B) Results of Cox proportional hazards regression analysis using TIGS for all solid cancers. Forest plots showing loge hazard ratio (95% confidence interval). Cox p-values are adjusted with the FDR method. p-values less than 0.1 are in bold. The pooled hazard ratios and the p-values were generated using the random effect model. The statistical test for heterogeneity is also shown in the last column. Tumor types are ordered by median TIGS score.
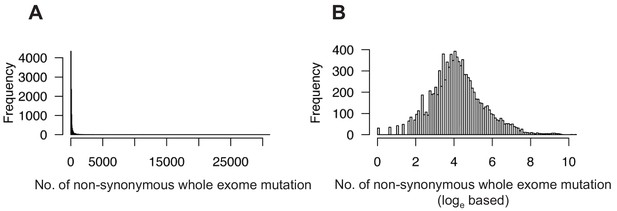
Pan-cancer distribution pattern of TMB in 9613 TCGA cancer samples.
(A) The distribution of non-synonymous whole-exome mutation counts of 9613 TCGA cancer samples. (B) Loge-based TMB values show a Gaussian distribution.
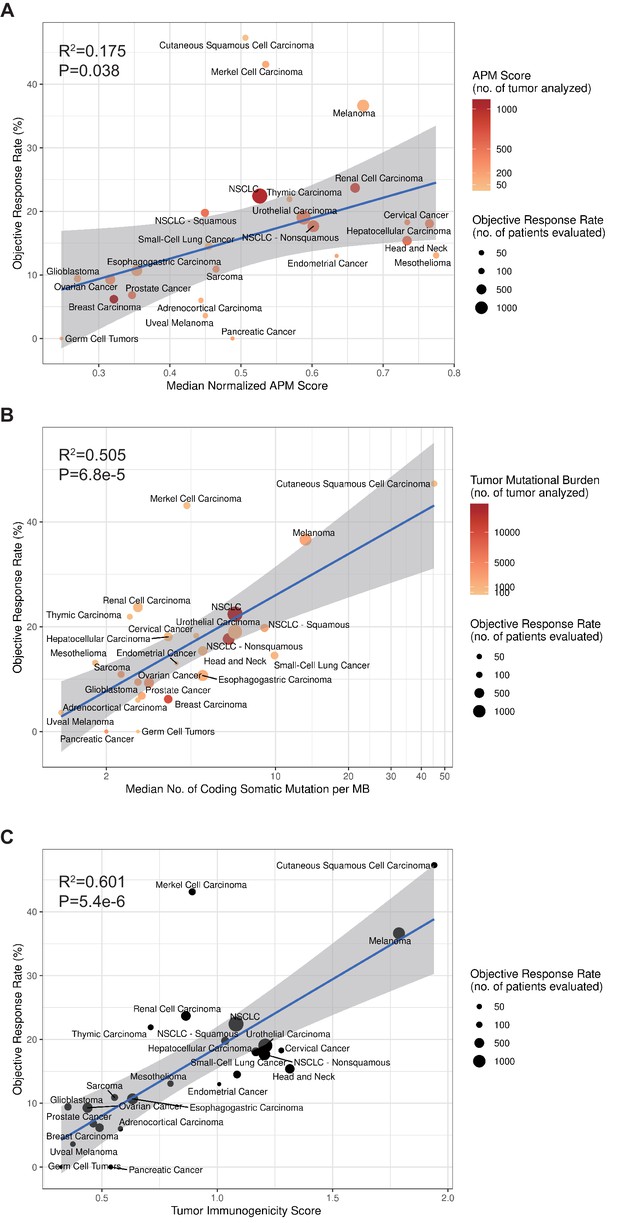
TIGS and predicted pan-cancer response rates to PD-1 inhibition.
Correlation between (A) APS, (B) TMB, (C) TIGS and objective response rate (ORR) with anti-PD-1 or anti-PD-L1 therapy in 25 cancer types. Shown are median normalized APS (A), median number of TMB (non-synonymous mutation/MB) in log scale (B) and TIGS in 25 tumor types or subtypes among patients who received inhibitors of PD-1 or PD-L1 (C), as described in published studies for which data regarding the ORR are available. The number of patients who were evaluated for the ORR is shown for each tumor type (size of the circle), along with the number of tumor samples that were analyzed to calculate the APS, TMB or TIGS (degree of shading of the circle).
-
Figure 4—source data 1
List of citations for individual studies used in pooled analysis of objective response rate.
- https://doi.org/10.7554/eLife.49020.016
-
Figure 4—source data 2
Summary of pooled ORR, median TMB and median APS by tumor type or subtype.
- https://doi.org/10.7554/eLife.49020.017
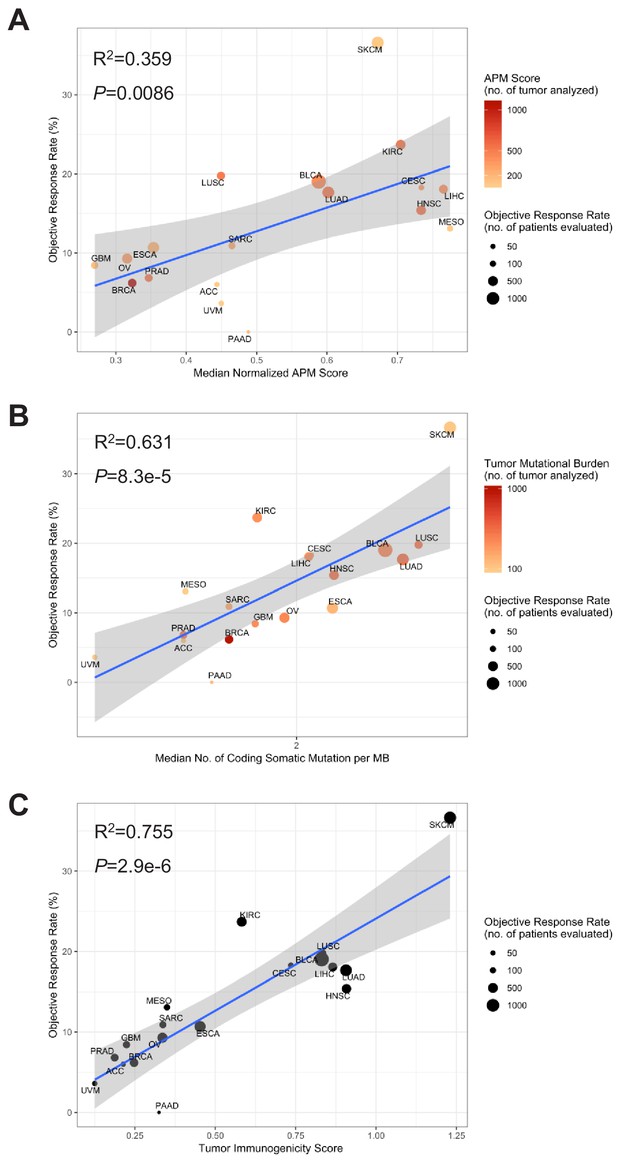
TIGS and predicted pan-cancer response rates to PD-1 inhibition.
Correlation between (A) APS, (B) TMB, (C) TIGS and objective response rate (ORR) with anti-PD-1 or anti-PD-L1 therapy in 18 cancer types. Shown are median normalized APS, median number of TMB (non-synonymous mutation/MB) in log scale and TIGS in 18 TCGA tumor types. The number of patients who were evaluated for the objective response rate is shown for each tumor type (size of the circle), along with the number of tumor samples that were analyzed to calculate the APS, TMB or TIGS (degree of shading of the circle). This analysis is similar to main Figure 4, except that APS, TMB and TIGA are all calculated for TCGA datasets.
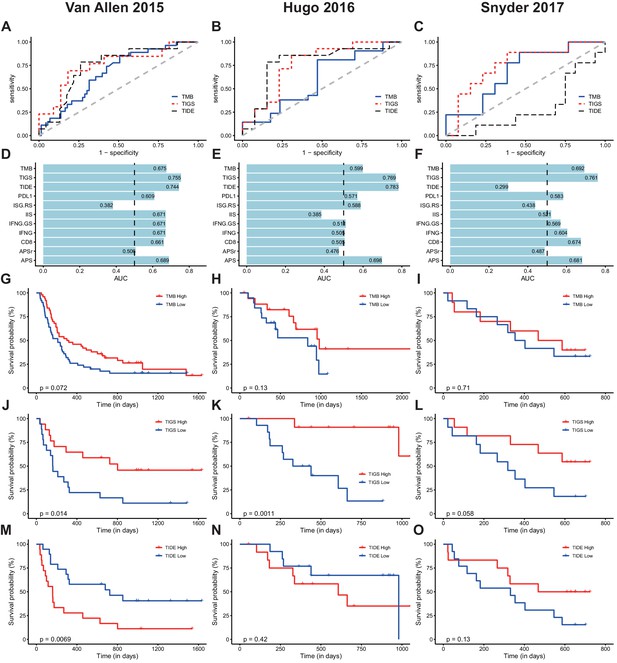
TIGS predicts clinical response to ICI immunotherapy.
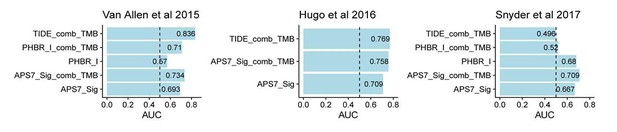
(A) ROC curves for the performance of TMB, TIDE and TIGS in predicting anti-CTLA4 Immunotherapy response in 35 melanoma patients (dataset from Van Allen et al., 2015). (B) ROC curves for the performance of TMB, TIDE and TIGS in predicting anti-PD-1 immunotherapy response in 27 melanoma patients (dataset from Hugo et al., 2016). (C) ROC curves for the performance of TMB, TIDE and TIGS in predicting anti-PD-L1 immunotherapy response in 22 urothelial cancer patients (dataset from Snyder et al., 2017). (D–F) AUC values of TMB, TIGS, TIDE, PDL1, immune infiltration score (IIS), interferon gamma gene expression signature (IFNG), CD8, APS and random genes as negative control for APS quantification (APSr) in the Van Allen et al. (2015) dataset (D), the Hugo et al. (2016) dataset (E) and the Snyder et al. (2017) dataset (F). The performance of a random predictor (AUC = 0.5) is represented by the dashed line. (G,J,M) Patients were grouped on the basis of TMB (G), TIGS (J) or TIDE (M) status. The Kaplan–Meier (KM) overall survival curves were compared between TMB-High and TMB-Low (100 patients), between TIGS-High vs TIGS-Low (35 patients) or between TIDE-High and TIDE-Low (37 patients) in the Van Allen et al. (2015) dataset. (H,K,N) Patients were grouped on the basis of TMB (H), TIGS (K) or TIDE (N) status. The KM overall survival curves were compared between TMB-High and TMB-Low (37 patients), between TIGS-High and TIGS-Low (26 patients) or between TIDE-High and TIDE-Low (26 patients) in the Hugo et al. (2016) dataset. (I,L,O) Patients were grouped on the basis of TMB (I), TIGS (L) or TIDE (O) status. The KM overall survival curves were compared between TMB-High and TMB-Low (22 patients), TIGS-High and TIGS-Low (22 patients) or TIDE-High and TIDE-Low (25 patients) in the Snyder et al. (2017) dataset.
-
Figure 5—source data 1
List of genes in the lists used for CD8, IFNG, ISG.RS and IFNG.GS signature calculation.
- https://doi.org/10.7554/eLife.49020.021
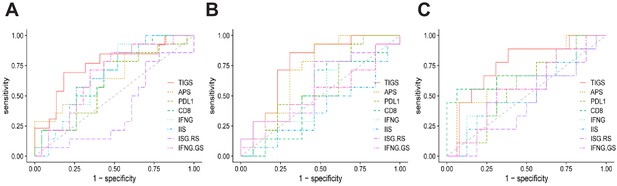
ROC curves for the performance of APS, CD8, IFNG, IIS, PDL1 and TIGS in predicting immunotherapy response in the Van Allen et al. (2015) melanoma dataset (A), the Hugo et al. (2016) melanoma dataset (B) and the Snyder et al. (2017) urothelial cancer dataset (C).
https://doi.org/10.7554/eLife.49020.019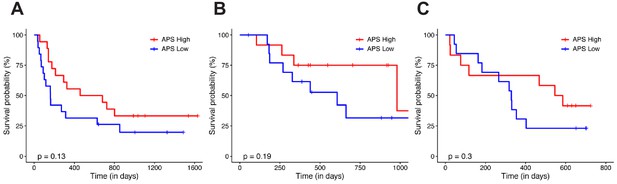
APS in predicting the clinical response to immunotherapy.
Patients were grouped on the basis of APS status. The Kaplan–Meier (KM) overall survival curves were compared between APS-High and APS-Low in the Van Allen et al. (2015) melanoma dataset (A), in the Hugo et al. (2016) melanoma dataset (B), and in the Snyder et al. (2017) urothelial cancer dataset (C). Log-rank test p-values are shown.
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.49020.022





