Discrete viral E2 lysine residues and scavenger receptor MARCO are required for clearance of circulating alphaviruses
Figures
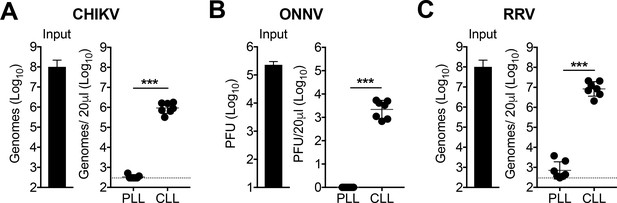
Phagocytic cells efficiently clear multiple alphaviruses from the circulation.
(A–C) WT C57BL/6 mice were treated intravenously (i.v.) with PBS- (PLL) or clodronate-loaded liposomes (CLL). At 42 hr post-treatment, mice were inoculated i.v. with CHIKV (A), ONNV (B) or RRV (C), and viral genomes in the inoculum (input) and serum at 45 min post-inoculation were quantified by RT-qPCR (A and C) or plaque assay (B). Mean ± SD. N = 7, two experiments. Mann-Whitney test; ***p<0.001. Figure 1—figure supplement 1 shows that viral RNA is undetectable in the clotted fraction of the blood.
-
Figure 1—source data 1
Raw data for Figure 1A-C.
- https://doi.org/10.7554/eLife.49163.005
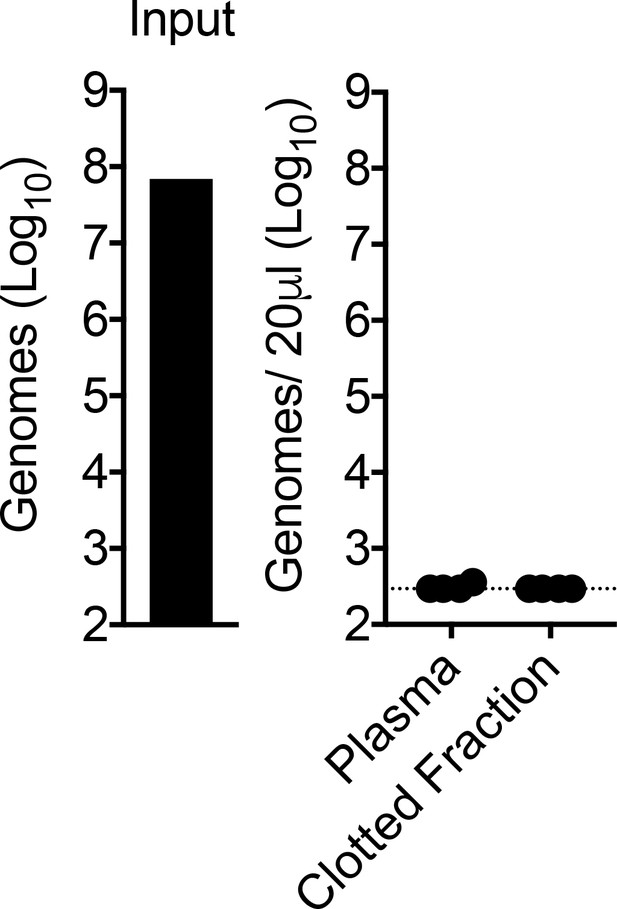
CHIKV RNA is undetectable in the clotted fraction of the blood at 45 min post-inoculation.
WT C57BL/6 mice were inoculated i.v. with CHIKV, and viral genomes in the inoculum (input), plasma, and clotted fraction of the blood at 45 min post-inoculation were quantified by RT-qPCR. Mean ± SD. N = 4, one experiment. Two-tailed unpaired t-test; p>0.05.
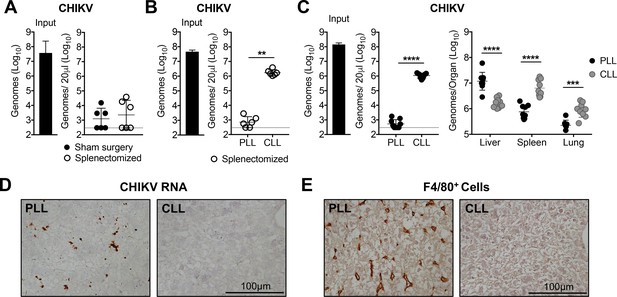
Liver Kupffer cells clear CHIKV from the circulation.
(A) WT C57BL/6 mice that underwent a sham or splenectomy surgery were inoculated i.v. with CHIKV and viral genomes in the inoculum and serum at 45 min post-inoculation were quantified by RT-qPCR. Mean ± SD. N = 6, two experiments. Mann-Whitney test; p>0.05. (B) Splenectomized WT C57BL/6 mice were treated i.v. with PLL or CLL. At 42 hr post-treatment, mice were inoculated i.v. with CHIKV and viral genomes in the inoculum (input) and serum at 45 min post-inoculation were quantified by RT-qPCR. Mean ± SD. N = 6, two experiments. Mann-Whitney test; **p<0.01. (C) WT C57BL/6 mice were treated and inoculated as in (B), and viral genomes present at 45 min post-inoculation in the serum or indicated tissues were quantified by RT-qPCR. Mean ± SD. N = 9, two experiments. Mann-Whitney test or Two-tailed t-test; ***p<0.001, ****p<0.0001. (D and E) WT C57BL/6 mice were treated as in (B) and inoculated with CHIKV at 42 hr post-treatment. Livers were collected at 45 min post-inoculation, and RNA Scope in situ hybridization (D) or IHC (E) were performed to visualize viral RNA localization or F4/80+ macrophages, respectively. Brown staining is indicative of viral RNA (D) or F4/80+ macrophages (E). N = 6–7, two experiments. Figure 2—figure supplement 1 shows representative images of CHIKV RNA Scope in situ hybridization and F4/80 IHC for all biological replicates, and Figure 2—figure supplement 2 shows flow cytometry analysis of liver macrophages and dendritic cells (DCs) in PLL- and CLL-treated mice.
-
Figure 2—source data 1
Raw data for Figure 2A-C.
- https://doi.org/10.7554/eLife.49163.009
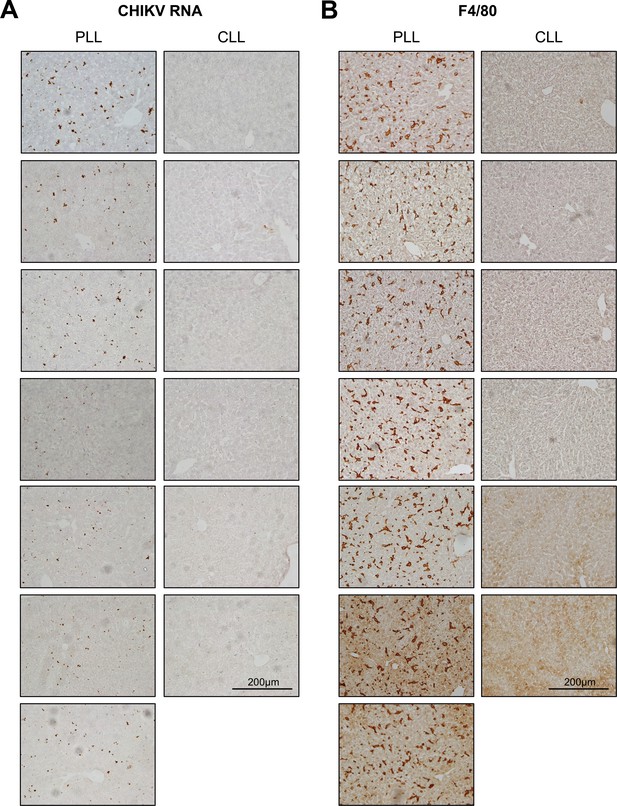
CHIKV RNA and F4/80+ cells are detectable in the livers of PLL- but not CLL-treated mice.
CHIKV-specific RNA scope in situ hybridization (A) and F4/80 antigen-specific immunohistochemistry (B) of liver sections from each mouse in the experiment shown in Figure 1D and E to visualize CHIKV RNA and F4/80+ cells (brown staining). N = 6–7, two experiments.
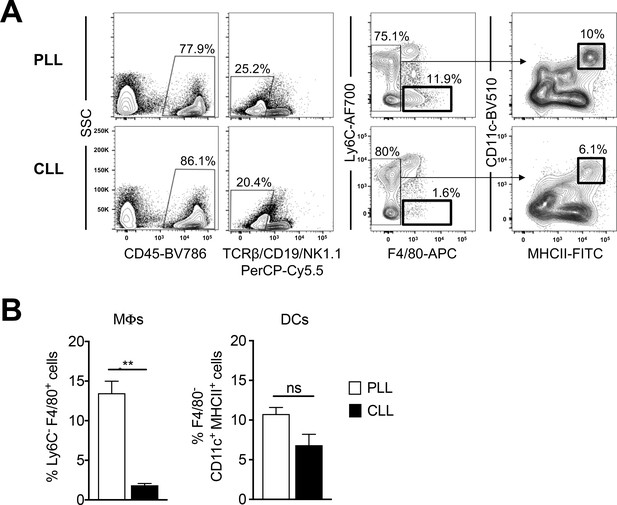
Clodronate liposome treatment depletes liver macrophages, but has minimal impact on DCs.
Livers were collected from splenectomized WT C57BL/6 mice in Figure 2B at 45 min post-inoculation and macrophage and DC populations were evaluated by flow cytometry. The representative flow plots (A) and percentage of cells (B) are shown. Mean ± SD. N = 3, one experiment (representative of two additional experiments in WT C57BL/6 mice that had not undergone surgery). Two-tailed, unpaired t-test. **p<0.01.
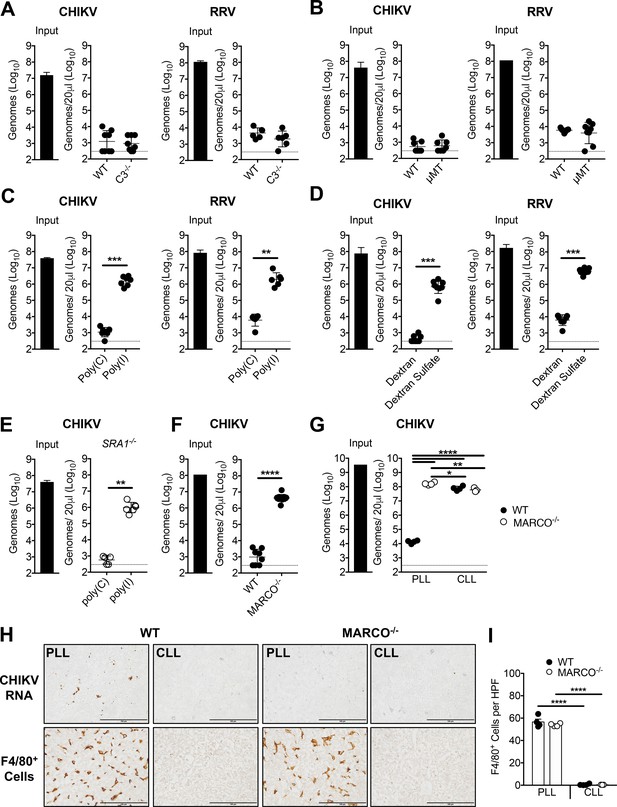
The scavenger receptor MARCO is required for clearance of CHIKV and RRV.
(A) WT or C3-/- C57BL/6 mice were inoculated i.v. with the indicated virus, and viral genomes in the inoculum (input) and serum at 45 min post-inoculation were determined by RT-qPCR. Mean ± SD. N = 6–8, two experiments. Mann-Whitney test; p>0.05. (B) WT or μMT C57BL/6 mice were inoculated i.v. with the indicated virus, and analyzed as in (A). Mean ± SD. N = 7–8, two experiments. Mann-Whitney test; p>0.05. (C) WT C57BL/6 mice were treated i.v. with 200 μg of poly(I) or poly(C) 5 min prior to inoculation with the indicated virus. Viral genomes in the serum at 45 min post-inoculation were quantified by RT-qPCR. Mean ± SD. N = 6–7, two experiments. Mann-Whitney test or Two-tailed unpaired t-test; **p<0.01, ***p<0.001. (D) WT C57BL/6 mice were treated i.v. with 200 μg of dextran or dextran sulfate 5 min prior to inoculation with the indicated virus, and analyzed as in (C). Mean ± SD. N = 7–8, two experiments. Mann-Whitney test or Two-tailed unpaired t-test; ***p<0.001. (E) SR-A1-/- mice were treated, incoculated, and evaluated as in (C). Mean ± SD. N = 5–6, two experiments. Mann-Whitney test; **p<0.01. (F) WT or MARCO-/- C57BL/6 mice were inoculated and evaluated as in (A). Mean ± SD. N = 8–11, two experiments. Mann-Whitney test; ****p<0.0001. (G–I) WT and MARCO-/- C57BL/6 mice were treated i.v. with PBS- (PLL) or clodronate-loaded liposomes (CLL). At 42 hr post-treatment, mice were inoculated i.v. with CHIKV. (G) Viral genomes in the inoculum (input) and serum at 45 min post-inoculation were quantified by RT-qPCR. Mean ± SD. N = 3–4. One-way ANOVA with Tukey’s multiple comparisons test; *p<0.05, **p<0.01, ****p<0.0001. (H) Livers were collected at 45 min post-inoculation, and RNA Scope in situ hybridization or IHC were performed to visualize viral RNA localization or F4/80+ macrophages, respectively. Brown staining is indicative of viral RNA or F4/80+ macrophages. (I) F4/80 positive cells in 10 randomly selected high power fields (HPF) per section were counted in a blinded manner and used to calculate the average number of F4/80 positive cells per field. Mean ± SD. N = 3–4. One-way ANOVA with Tukey’s multiple comparisons test; ****p<0.0001. Figure 3—figure supplement 1 shows the mRNA expression of various scavenger receptors in murine liver cell subsets.
-
Figure 3—source data 1
Raw data for Figure 3A-G, I.
- https://doi.org/10.7554/eLife.49163.012
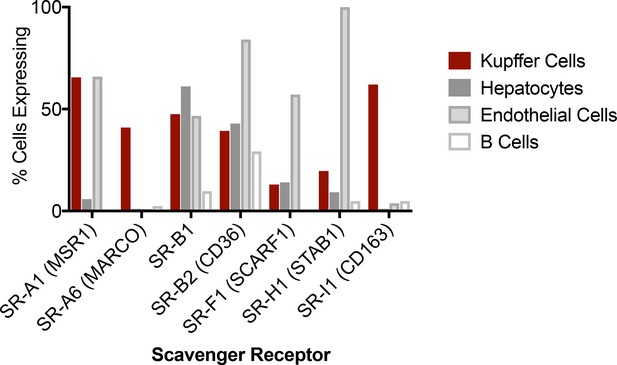
Expression of various scavenger receptors by murine liver cell subsets.
Transcriptome data were mined from the Tabula Muris (Tabula Muris Consortium et al., 2018), which FACS sorted single liver cells into a 384-well plate, evaluated cellular RNA by Illumina sequencing, and aligned reads to the mm10plus genome. Clustering and marker gene expression were used to identify Kupffer cells (Emr1+, Clec4F+, Cd68+, Irf7+) , hepatocytes (Alb+, Ttr+, Apoa1+, Serpina1c+), endothelial cells (Pecam1+, Nrp1+, Kdr+, Oit3+), and B cells (Cd79a+, Cd79b+, Cd74+, Cd19+). The percent of analyzed cells expressing a given SR is displayed.
CHIKV E2 K200R evades phagocytic cell-mediated clearance.
(A) WT C57BL/6 mice were inoculated i.v. with CHIKV or CHIKV E2 K200R, and viral genomes in the inoculum and serum at the indicated times post-inoculation were evaluated by RT-qPCR. Mean ± SD. N = 6 per time point, two experiments. Two-way ANOVA with Bonferroni’s correction; ****p<0.0001. (B) WT C57BL/6 mice were inoculated with human fibroblast-derived CHIKV or CHIKV E2 K200R, and viral genomes in the inoculum and in the serum at 45 min post-inoculation were quantified by RT-qPCR. Mean ± SD. N = 6, two experiments. Two-tailed, unpaired t-test; ****p<0.0001. (C) WT C57BL/6 mice were treated i.v. with PBS liposomes or clodronate liposomes 42 hr prior to i.v. inoculation with CHIKV or CHIKV E2 K200R. Viral genomes in the inoculum and serum at 45 min post-inoculation were determined by RT-qPCR. Mean ± SD. N = 8, two experiments. Kruskal-Wallis; **p<0.01, ***p<0.001 (D) WT C57BL/6 mice were inoculated i.v. with the indicated viruses, and viral genomes in the inoculum and in the serum at 45 min post-inoculation were quantified by RT-qPCR (99659, SL15649), focus forming assay (37997), or plaque assay (ONNV SG650). Mean ± SD. N = 3, one experiment. Mann-Whitney test; p>0.05. Figure 4—figure supplement 1 shows that mutation of E2 glycosylation sites does not influence CHIKV clearance from the circulation. Figure 4—figure supplement 2 shows that the E2 K200R mutation does not alter the thermostability of CHIKV particles in serum (A), and that the E2 K200R mutation allows CHIKV particles to evade clearance from the circulation in multiple distinct mouse strains (B).
-
Figure 4—source data 1
Raw data for Figure 4A-D.
- https://doi.org/10.7554/eLife.49163.016
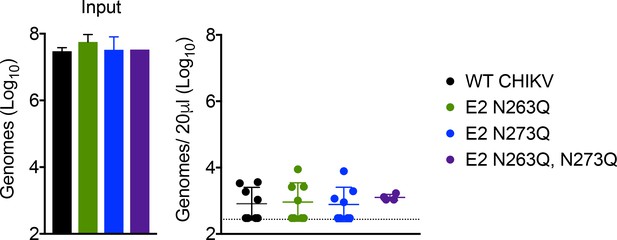
Mutation of putative CHIKV glycosylation sites has no impact on viral clearance from the circulation.
WT C57BL/6 mice were inoculated with WT CHIKV or the indicated mutants and viral genomes in the serum at 45 min post-inoculation were quantified by RT-qPCR. Mean ± SD. N = 8 with two experiments for WT CHIKV, E2 N263Q, and E2 N273Q; N = 4 with one experiment for E2 N263Q, N273Q. Kruskal-Wallis with Dunn’s multiple comparison test; p>0.05 for all comparisons.
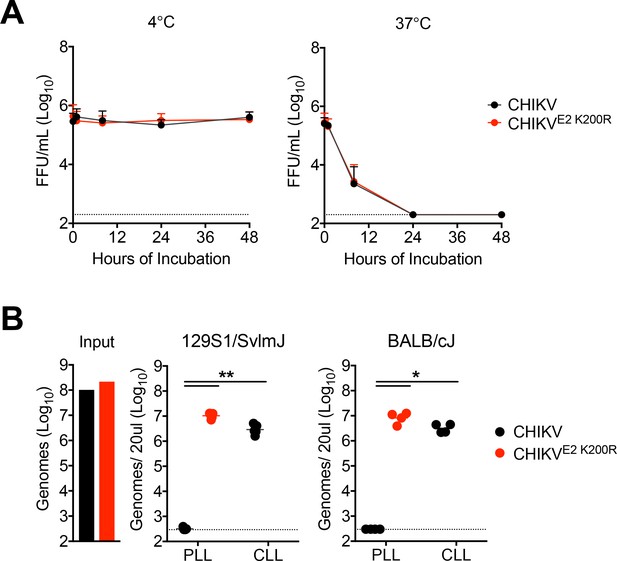
CHIKV E2 K200R does not impact virus stability and evades clearance in multiple mouse strains.
(A) CHIKV or CHIKV E2 K200R particles were incubated in serum at 37°C or 4°C. At the indicated times, infectious virus in the serum was quantified by focus formation assay. Mean ± SD. N = 6, two experiments. Two-way ANOVA with Bonferroni’s correction. (B) WT 129S1/SvlmJ or BALB/cJ mice were were inoculated i.v. with CHIKV or CHIKV E2 K200R, and viral genomes in the inoculum or in the serum at 45 min post-inoculation were evaluated by RT-qPCR. Mean ± SD. N = 4, one experiment. Mann-Whitney test relative to PLL treated CHIKV inoculated group; *p<0.05, **p<0.01.
An E2 K200R mutation or deletion of MARCO allows for enhanced viremia and more rapid viral dissemination.
(A) WT C57BL/6 mice were inoculated in the left footpad with 1,000 PFU of the indicated virus. At 24 hpi, infectious virus in the ipsilateral left ankle, contralateral right ankle, and serum were quantified by focus formation assay (CHIKV) or plaque assay (ONNV). Mean ± SD. N = 5–10, two experiments. Mann-Whitney test; **p<0.01, ****p<0.0001. (B) WT or MARCO-/- C57BL/6 mice were inoculated as in (A) with CHIKV or CHIKV E2 K200R. At 24 hpi, infectious virus in the ipsilateral left ankle, contralateral right ankle, right quadriceps, and serum were quantified by focus formation assay. Mean ± SD. N = 9, two experiments. Kruskal-Wallis or One-way ANOVA; **p<0.01, ***p<0.001, ****p<0.0001.
-
Figure 5—source data 1
Raw data for Figure 5A-B.
- https://doi.org/10.7554/eLife.49163.018
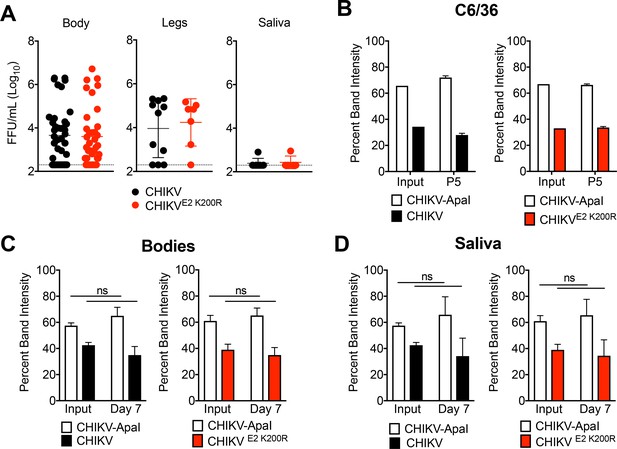
CHIKV E2 K200R has no impact on vector competence or viral fitness in mosquitoes.
(A) Ae. aegypti mosquitoes were fed a blood meal containing 1.1 × 106 PFU/mL of CHIKV or CHIKV E2 K200R, and the head, legs, and saliva were collected at three dpi. Samples initially found to be positive for virus were evaluated by focus formation assay to quantify infectious virus. Mean ± SD. N = 50, one experiment. Mann-Whitney test; p>0.05. (B) Ae. albopictus C6/36 cells were infected in triplicate at an MOI of 1 PFU/cell with a 1:1 mixture of CHIKV marked with an ApaI restriction site (CHIKV-ApaI) and WT CHIKV or CHIKV E2 K200R, and 1/10th of the supernatant was serially passaged onto new C6/36 cells every 24 hr. RNA was extracted from the inoculum and supernatant of passage 5, cDNA was generated, and PCR amplified. Digestion of the PCR product was used to identify ratios of ApaI marked to unmarked virus, and the percent band intensity is displayed. Mean ± SD. N = 3, one experiment. (C and D) Ae. aegypti mosquitoes were microinjected with 138 PFU of 1:1 mixtures of CHIKV-ApaI and CHIKV, or CHIKV-ApaI and CHIKV E2 K200R. Bodies (C) and saliva (D) were collected at seven dpi. Ratios of each virus present in the input and in samples at day seven were evaluated as described in (B). Mean ± SD. N = 20, one experiment. Two-way ANOVA with Bonferroni’s correction; p>0.05 for all comparisons.
-
Figure 6—source data 1
Raw data for Figure 6A-D.
- https://doi.org/10.7554/eLife.49163.020
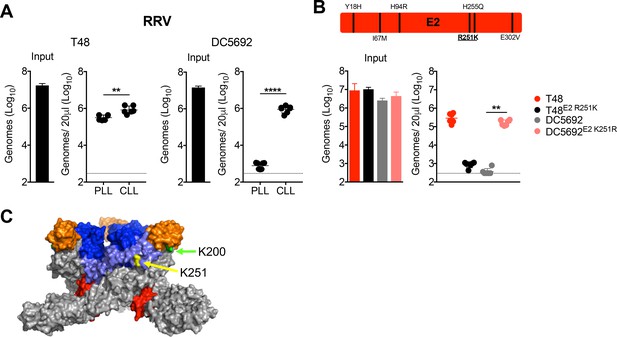
RRV clearance from the circulation is mediated by a distinct lysine residue, E2 K251.
(A) WT C57BL/6 mice were treated i.v. with PLL or CLL 42 hr prior to i.v. inoculation with RRV strains T48 or DC5692. Viral genomes in the inoculum and in the serum at 45 min post-inoculation were quantified by RT-qPCR. Mean ± SD. N = 6, two experiments. Two-tailed unpaired t-test; **p<0.01, ****p<0.0001. (B) Schematic representation of the six amino acid differences between T48 and DC5649 within E2 shown, with position 251 underlined. WT C57BL/6 mice were inoculated i.v. with the indicated viruses and mutants. Viral genomes in inoculum and in the serum at 45 min post-inoculation were quantified as in (A). Mean ± SD. N = 6, two experiments. Kruskal-Wallis; **p<0.01. (C) CHIKV trimeric spike (PDB 3J2W), with E2 K200 highlighted in green, and E2 K251 (RRV numbering) highlighted in yellow. The E1 glycoprotein is shown in gray, E2 domain A is shown in blue, E2 domain B is shown in orange, E2 domain C is shown in red, and the β-ribbon connector is shown in light blue. Figure 7—figure supplement 1 shows evaluation of the clearance of RRV T48 and RRV DC5692 structural gene chimeras (A), demonstrates that the E2 K251R mutation does not alter the thermostability of RRV particles in serum (B), and that the clearance phenotypes of RRV T48 and RRV T48 E2 R251K viral particles are maintained when purified viral particles are used for mouse inoculation studies (C).
-
Figure 7—source data 1
Raw data for Figure 7A-C.
- https://doi.org/10.7554/eLife.49163.023
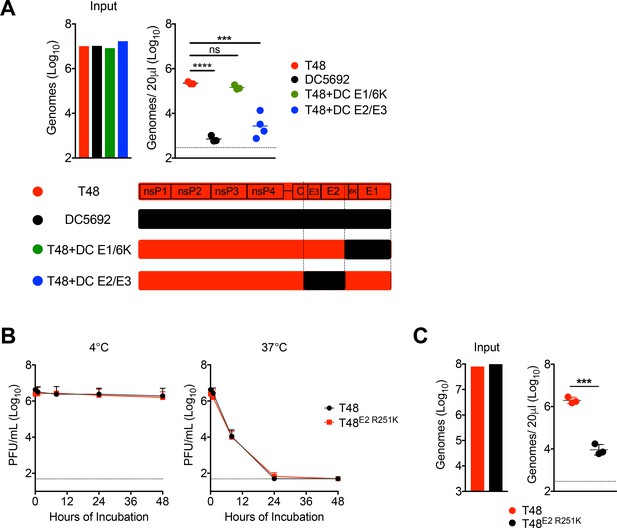
Chimeric analysis of T48 and DC5692, stability of T48 versus T48 E2 R251K, and serum clearance of purified viral particles.
(A) WT C57BL/6 mice were inoculated i.v. with RRV strains T48, DC5692, or chimeras between T48 and DC5692 (DC), as shown in the schematic. Viral genomes in the inoculum and in the serum were quantified by RT-qPCR. Mean ± SD. N = 4, representative of two experiments. One-way ANOVA with Tukey’s correction; ***p<0.001, ****p<0.0001. (B) RRV T48 or T48 E2 R251K particles were incubated in serum at 37°C or 4°C. At the indicated times, infectious virus in the serum was quantified by plaque assay. Mean ± SD. N = 6, two experiments. Two-way ANOVA with Bonferroni’s correction. (C) WT C57BL/6 mice were inoculated i.v. with purified particles of RRV T48 or T48 E2 R251K derived in serum free media, and viral genomes in inoculum and in the serum at 45 min post-inoculation were quantified by RT-qPCR. Mean ± SD. N = 3, one experiment. Two-tailed, unpaired t-test; ***p<0.001.
Lysine residues at E2 200 or E2 251 are essential for CHIKV and RRV clearance, respectively.
(A) Properties of the amino acid side chains selected for substitution analysis at E2 K200 or K251. Amino acid structures generated using PepDraw. (B) WT C57BL/6 mice were inoculated i.v. with a panel of CHIKV mutants with different amino acid substitutions at position E2 200. Viral genomes in the inoculum and in the serum at 45 min post-inoculation were quantified by RT-qPCR. Mean ± SD. N = 4, one experiment. Kruskal-Wallis, comparing each group to WT virus containing K; ***p<0.001. (C) WT C57BL/6 mice were inoculated i.v. with a panel of RRV mutants with different amino acid substitutions at position E2 251. Viral genomes in the inoculum and in the serum at 45 min post-inoculation were quantified by RT-qPCR. Mean ± SD. N = 4, one experiment. One-way ANOVA, comparing each group to virus containing K; ****p<0.0001. (D) WT C57BL/6 mice were inoculated i.v. with CHIKV or CHIKV E2 K252R, and viral genomes in inoculum and in the serum at 45 min post-inoculation were quantified by RT-qPCR. Mean ± SD. N = 8, two experiments. Mann-Whitney test; p>0.05.
-
Figure 8—source data 1
Raw data for Figure 8B-D.
- https://doi.org/10.7554/eLife.49163.025
Additional files
-
Supplementary file 1
Primers used to generate mutant viruses through site-directed mutagenesis.
- https://doi.org/10.7554/eLife.49163.026
-
Supplementary file 2
Key Resources Table.
- https://doi.org/10.7554/eLife.49163.027
-
Transparent reporting form
- https://doi.org/10.7554/eLife.49163.028





