The yellow gene influences Drosophila male mating success through sex comb melanization
Figures
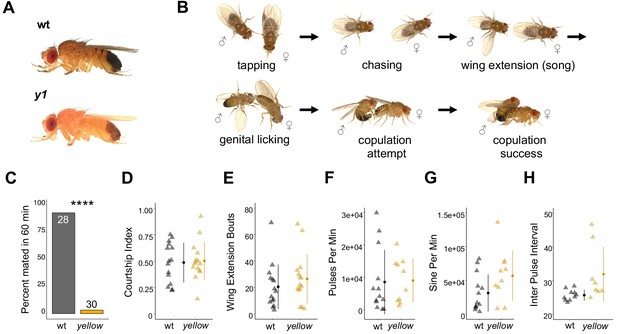
The Drosophila melanogaster yellow gene is required for male mating success.
(A) Photographs comparing wild-type and yellow (y1) body pigmentation [Reprinted from Atlas of Drosophila Morphology, 1 st Edition, Sylwester Chyb and Nicolas Gompel, Body Markers, pp.173, 175, Copyright (2013), with permission from Elsevier] This panel is not covered under the CC-BY 4.0 licence. (B) Snapshots from videos illustrating D. melanogaster courtship behaviors. (C) y1 males (yellow) showed significantly lower mating success levels compared to wild-type males (black) in non-competitive, one-hour trials. Sample sizes are shown at the top of each barplot. (D–H) y1 males showed similar levels of courtship activity and song compared to wild-type males. (D) Courtship index: the proportion of time a male engages in courtship activity divided by the total observation period. (E) Wing extension bouts: the number of unilateral wing extensions during the observation period. (F) Pulses per minute. (G) Sine per minute. (H) Inter pulse interval. (D–H) Show individual points that represent single fly replicates. Circles represent means and lines SD. Significance was measured using Fisher’s exact test in (C) and Welch’s Two Sample t-test in (D–H). Comparisons that were statistically (p<0.05) are indicated (****p<0.0001).
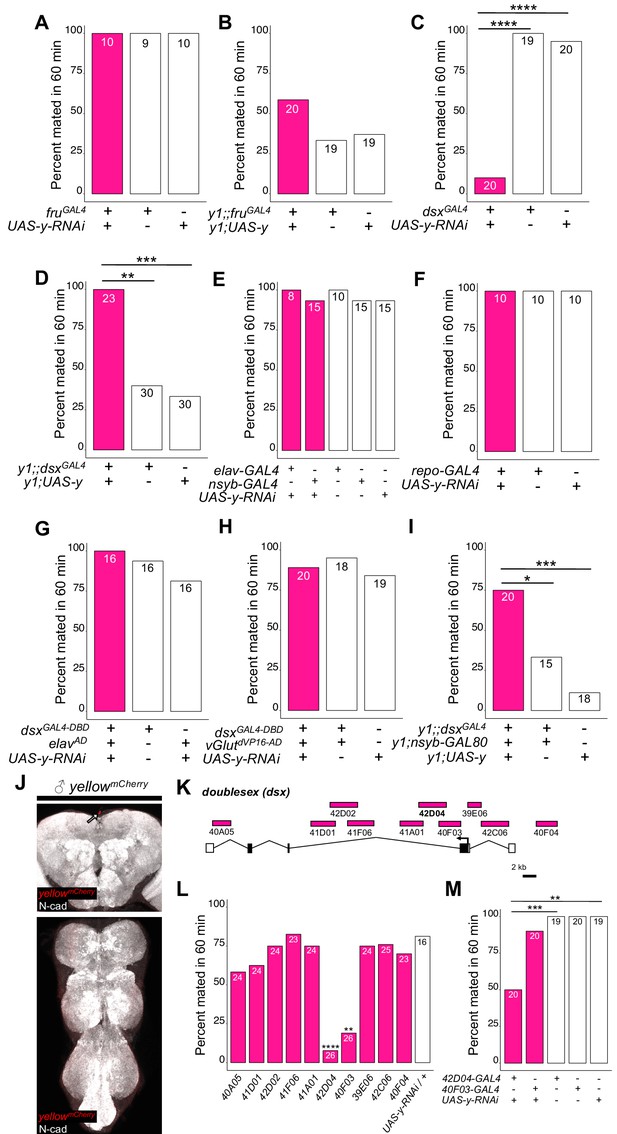
yellow expression in non-neuronal doublesex-expressing cells, but not fruitless-expressing cells, is necessary and sufficient for male mating success.
(A,B) Neither expressing yellow-RNAi nor yellow-cDNA in fru-expressing cells using fruGAL4 (Stockinger et al., 2005) affected male copulation. (C) Expressing yellow-RNAi in dsx-expressing cells using dsxGAL4 (Robinett et al., 2010) significantly inhibited male mating success. (D) Expressing yellow in dsx-expressing cells using dsxGAL4 in a y1 mutant background was sufficient to restore male mating success. (E,F) Expressing yellow-RNAi using pan-neuronal (elav-GAL4 and nsyb-GAL4) and pan-glia (repo-GAL4) drivers did not affect male mating success. (G) Restricting yellow-RNAi expression to dsx-expressing neurons using the split-GAL4 technique, combining dsxGAL4-DBD (Pavlou et al., 2016) with elavVP16-AD (Luan et al., 2006), did not affect male mating success. (H) Restricting yellow-RNAi expression to dsx-expressing glutamatergic neurons using the split-GAL4 technique, combining dsxGAL4-DBD (Pavlou et al., 2016) with vGlutdVP16-AD (Gao et al., 2008) did not affect male mating success. (I) Expressing yellow in dsx-expressing cells restricted outside the CNS using dsxGAL4 and nsyb-GAL80 (courtesy of Julie Simpson) in a y1 mutant background significantly increased male mating success. (J) Brain and ventral nerve cord of adult male ymCherry flies stained with anti-N-Cadherin (N-cad) antibody labeling neuropil (white) and anti-DsRed antibody labeling Yellow::mCherry (red). We observed sparse, inconsistent signal outside the CNS at the top of the brain in males (white arrow), but we were unable to confirm a previous report that ymCherry is expressed in the adult brain (Hinaux et al., 2018). (K) Diagram of the male exon structure of the dsx locus highlighting 10 genomic fragments between 1.7 and 4 kb used to clone Janelia enhancer trap GAL4 drivers (Pfeiffer et al., 2008). Black boxes indicate coding exons. White boxes indicate 5’ and 3’ UTRs, and the arrow in exon two denotes the transcription start site. (L) Expressing yellow-RNAi using each Janelia dsx-GAL4 driver identified 42D04-GAL4 and 40 F03-GAL4 as affecting male mating success when compared with the yellow-RNAi control. (M) A replicate experiment comparing 42D04-GAL4 and 40F03-GAL4 effects on male mating success with both GAL4 and UAS parental controls confirmed the significant effect of 42D04-GAL4 but not 40F03-GAL4. We attribute differences in the 40F03-GAL4 effect between (L) and (M) to between experiment variability in the levels of male mating success; each common genotype tested in (L), for example, mated at higher levels in (M), but 42D04-GAL4 consistently showed a significant effect relative to controls. Sample sizes are shown at the top of each barplot. Significance was measured using Fisher’s exact tests with Bonferroni corrections for multiple comparisons. Comparisons that were statistically (p<0.05) are indicated (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001).
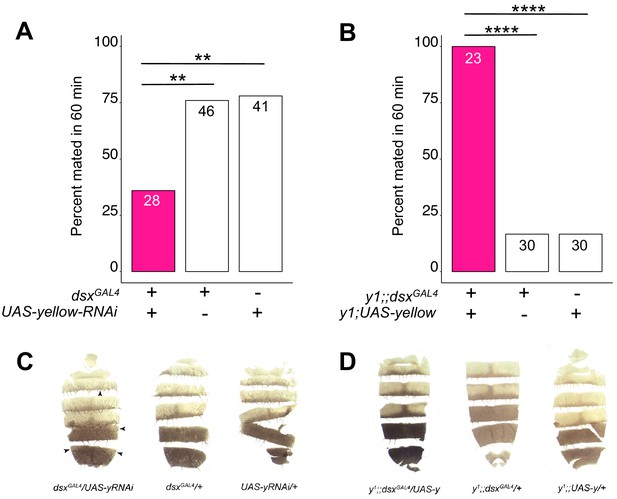
yellow expression in dsx-expressing cells is necessary and sufficient for male mating success.
(A) Expressing yellow-RNAi in dsx-expressing cells using dsxGAL4 (Rideout et al., 2010) significantly inhibited male mating success. (B) Expressing yellow in dsx-expressing cells using dsxGAL4 in a y1 mutant background was sufficient to restore male mating success. (C) Expressing yellow-RNAi using dsxGAL4 (Rideout et al., 2010) partially reduced black melanin levels in the male A5 and A6 abdominal tergites (black arrowheads), consistent with prior work (Williams et al., 2008; Rogers et al., 2014; Kalay et al., 2016). (D) Expressing yellow using dsxGAL4 partially elevated black melanin levels in the male A5 and A6 abdominal tergites. Sample sizes are shown at the top of each barplot. Significance was measured using Fisher’s exact tests with Bonferroni corrections for multiple comparisons. Comparisons that were statistically (p<0.05) are indicated (**p<0.01, ****p<0.0001).

The mating regulatory sequence (MRS) from Drapeau et al. (2006) does not affect male mating success.
(A) Diagram of the yellow locus highlighting the putative ‘mating regulatory sequence’ (MRS) (pink) region mapped in Drapeau et al. (2006) and a predicted dsx binding site (yellow) identified by ChIP-seq in Clough et al. (2014). The predicted binding site was identified based on in vivo Doublesex occupancy data (PWM score = 88.7) localized between 356,273 and 356,286 bp on the X chromosome (see Supplementary Table S2 in Clough et al., 2014). The wing-body enhancer region is indicated in blue, which was cloned upstream of GAL4 in Gilbert et al. (2006) to make the wing-body-GAL4 line. (B) Expressing yellow-RNAi using wing-body-GAL4 reduced black melanin to y1 levels, and expressing yellow in a y1 mutant background using wing-body-GAL4 restores black melanin synthesis to wild-type (wt) levels. (C) Expressing yellow-RNAi using wing-body-GAL4 did not inhibit male mating success. (D) Expressing yellow using wing-body-GAL4 in a y1 mutant background did not restore male mating success. (E) Brain and VNC of adult male and female flies stained with anti-GFP (green) antibody for myrGFP expressed using wing-body-GAL4 and counterstained with anti-nC82 (magenta) for neuropil. (F) Diagram illustrating the CRISPR/Cas9-facilitaed homology-directed repair (HDR) strategy used to excise and replace the MRS (pink) with pHD-DsRed-attP (red) (Gratz et al., 2014). Two sgRNAs (pink letters) were designed towards target PAM sites (blue letters) at the most 5’ and 3’ bounds of the MRS (scissors). Sanger sequencing chromatograms illustrate the location of each cut site (black arrows) relative to the transcription start site. DsRed was removed using Cre-lox recombinase (Siegal and Hartl, 1996). (G) PCR validation of DsRed removal and MRS deletion. (H) Excising the putative MRS did not inhibit male male mating success. Sample sizes are shown at the top of each barplot. Significance was measured using Fisher’s exact tests with Bonferroni corrections for multiple comparisons. Comparisons that were statistically (p<0.05) are indicated (****p<0.0001).

Expressing yellow-RNAi in subsets of CNS tissue does not affect male mating success.
(A,B) Expressing yellow-RNAi using a series of CNS, dopaminergic, and serotonergic GAL4 drivers did not affect male mating success. Significance was measured using Chi-square tests with Bonferroni corrections for multiple comparisons. Sample sizes are shown at the top of each barplot. Significance was measured using Fisher’s exact tests with Bonferroni corrections for multiple comparisons. None of the comparisons were statistically significant.

ymCherry expression in adult female central nervous system.
Brain and ventral nerve cord of adult female ymCherry flies stained with anti-N-Cadherin (N-cad) antibody labeling neuropil (white) and anti-DsRed antibody labeling Yellow::mCherry (red). We observed sparse, inconsistent signal outside the CNS at the top of the brain (white arrow), but we were unable to confirm a previous report that ymCherry is expressed in the adult brain (Hinaux et al., 2018).
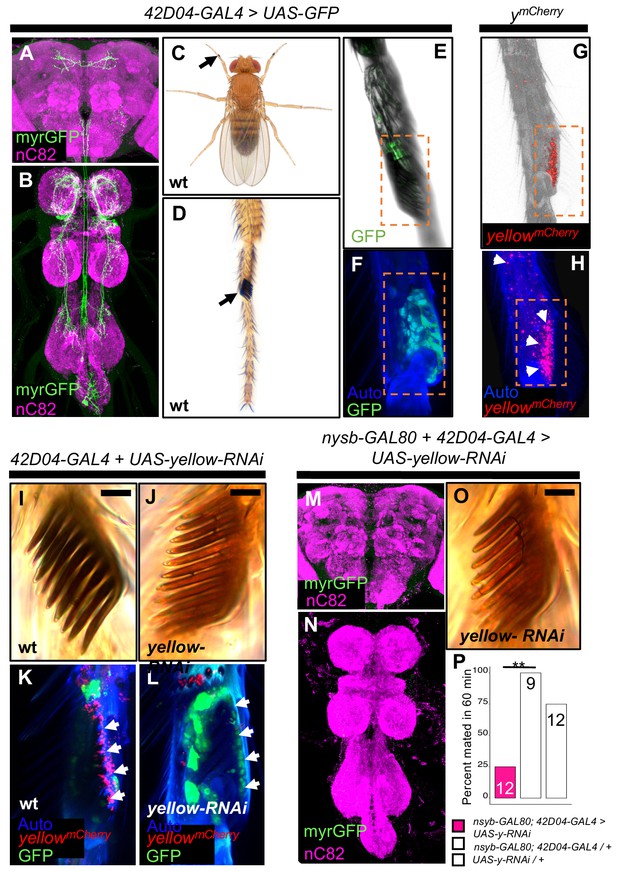
yellow expression in non-neuronal 42D04-GAL4 expressing cells is necessary for sex comb melanization and male mating success.
(A,B) Brain and ventral nerve cord of adult male fly stained with anti-GFP (green) antibody for myrGFP expressed using 42D04-GAL4 and counterstained with anti-nC82 (magenta) for neuropil. (C) Wild-type (wt) D. melanogaster adult male fly highlighting the location of sex combs (Nicolas Gompel). (D) Close up of a wild-type (wt) sex comb on the first tarsal segment (ts1) of the front leg (courtesy of Nicolas Gompel). (E) Bright field illumination of a male front leg expressing cytGFP (green) in sex-comb cells using 42D04-GAL4. (F) Confocal image of the sex comb cells expressing cytGFP (green) with 42D04-GAL4 and leg cuticle autofluorescence (blue). (G) Confocal image of a ymCherry male leg highlighting native ymCherry sex comb expression (red). (H) Zoomed in confocal image shown in (G) with leg cuticle autofluorescence (blue) and native ymCherry sex comb expression (red). (I) Wild-type (wt) sex comb. (J) Loss of black melanin in sex combs in males expressing yellow-RNAi using 42D04-GAL4. (K) Co-localization of ymCherry (red) at the base of the sex comb cells expressing cytGFP (green) with 42D04-GAL4. (L) Loss of ymCherry (red) at the base of the sex comb cells expressing cytGFP (green) and yellow-RNAi using 42D04-GAL4. (M,N) Brain and ventral nerve cord of adult male expressing nsyb-GAL80 to block GAL4 activity in the CNS, stained with anti-GFP (green) antibody for myrGFP expressed using 42D04-GAL4, and counterstained with anti-nC82 (magenta) for neuropil. (O) Loss of black melanin in sex combs in nsyb-GAL80 males expressing yellow-RNAi using 42D04-GAL4. (P) Expressing yellow-RNAi using 42D04-GAL4 in males expressing nsyb-GAL80 significantly inhibited male mating success. Scale bars in (I), (J), and (O) measure 12.5 μm. Sample sizes are shown at the top of each barplot. Significance was measured using Fisher’s exact tests with Bonferroni corrections for multiple comparisons. Comparisons that were statistically (p<0.05) are indicated (**p<0.01).
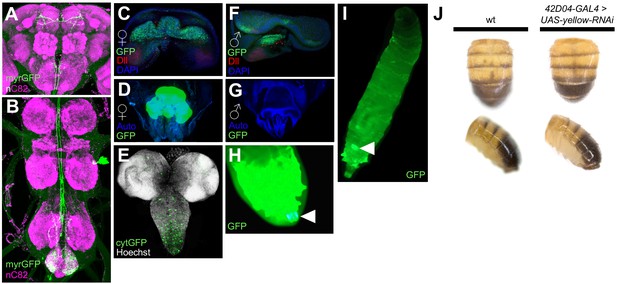
Expression pattern of 42D04-GAL4.
(A,B) Brain and VNC of adult female fly stained with anti-GFP (green) antibody for myrGFP expressed using 42D04-GAL4 and counterstained with anti-nC82 (magenta) for neuropil. (C) L3 larval female genital disc stained with anti-GFP (green) antibody for cytGFP expressed using 42D04-GAL4, anti-Dll (red) for Distal-less expression, and counterstained with DAPI (blue) for DNA (courtesy of Janelia Fly Light). (D) Adult female genitalia native cytGFP (green) expressed using 42D04-GAL4. (E) L3 CNS native cytGFP (green) expressed using 42D04 (F) L3 larval male genital disc stained with anti-GFP (green) antibody for cytGFP expressed using 42D04-GAL4, anti-Dll (red) for Distal-less expression, and counterstained with DAPI (blue) for DNA (courtesty of Janelia Fly Light). (G) Adult male genitalia did not show native cytGFP expression using 42D04-GAL4. (H) L3 larval posterior spiracle (white arrowhead) native cytGFP (green) expression. (I) L3 larva whole body highlighting native cytGFP (green) expression in the genital disc (white arrowhead). (J) Expressing yellow-RNAi using 42D04-GAL4 does not affect body pigmentation relative to wild-type (wt) flies.
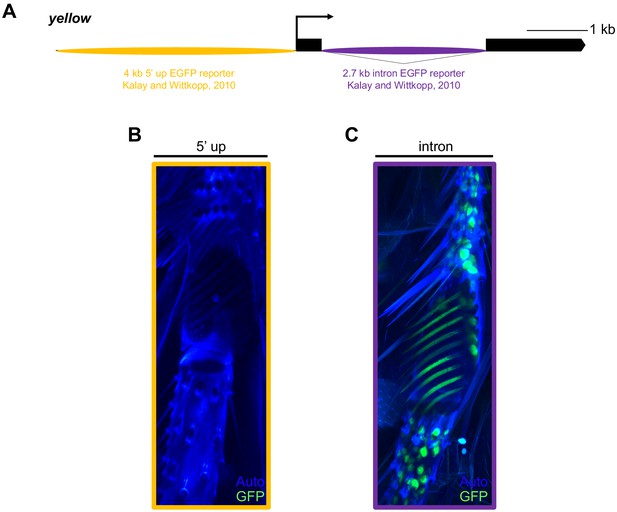
yellow EGFP reporters localize yellow sex comb expression to the intronic bristle enhancer.
(A) Diagram of the yellow locus highlighting two D. melanogaster enhancer regions (5’ up including the wing, body, and putative MRS enhancers reported in Geyer and Corces, 1987, Martin et al., 1989, and Drapeau et al., 2006; and intron, including the bristle and putative sex comb enhancer reported in Geyer and Corces, 1987 and Martin et al., 1989) that were cloned upstream of an EGFP reporter in Kalay and Wittkopp (2010). (B) Confocal image of a 96 hr old after pupal formation (APF) pupal sex comb expressing cytGFP under the control of the 5’ up enhancer region. (C) Confocal image of a 96 hr APF pupal sex comb expressing cytGFP under the control of the intronic enhancer region, highlighting expression in bristle sockets, sex comb sockets, and sex comb teeth.
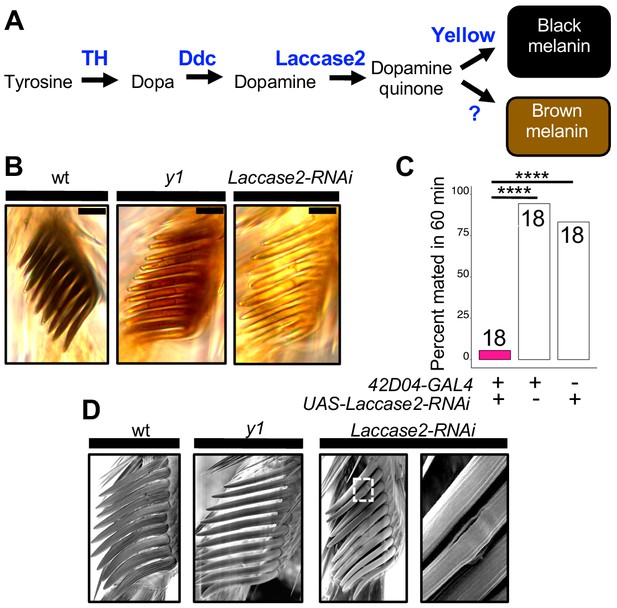
Sex comb melanization is specifically required for male mating success.
(A) Simplified version of the insect melanin synthesis pathway. (B) Light microscopy images of sex combs from wild-type (wt), y1, and 42D04-GAL4; UAS-Laccase2-RNAi males. Expressing Laccase2-RNAi in sex combs completely blocked melanin synthesis. (C) Expressing Laccase2-RNAi using 42D04-GAL4 in males significantly inhibited male mating success. (D) Scanning Electron Microscopy (SEM) of sex combs from wild-type (wt), y1, and Laccase2-RNAi males (expressed using 42D04-GAL4). Compared to wild-type, sex comb teeth in y1 mutants appeared thinner and smoother, whereas Laccase2-RNAi sex comb teeth appeared even smoother than y1 mutants, and one comb tooth had a visible crack in the cuticle (white rectangle, enlarged on the right). Scale bars in (B) measure 12.5 μm. Sample sizes are shown at the top of each barplot. Significance in was measured using Fisher’s exact tests with Bonferroni corrections for multiple comparisons. Comparisons that were statistically (p<0.05) are indicated (****p<0.0001).
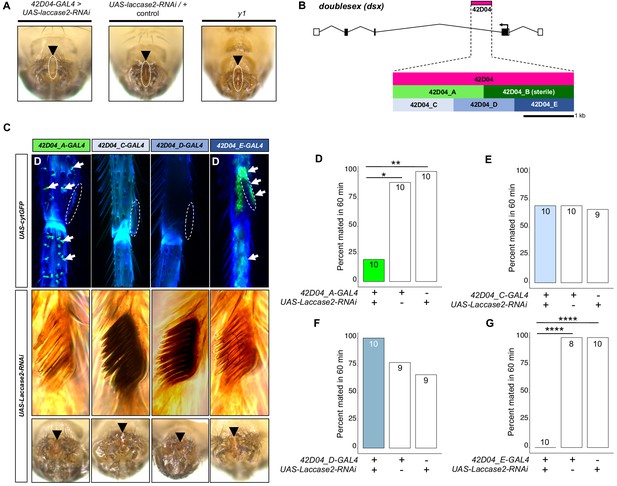
Genetic dissection of the 42D04-GAL4 enhancer confirms the specific role of sex comb melanization, and not the aedeagus, in male mating success.
(A) Expressing Laccase2-RNAi using 42D04-GAL4 blocked melanin synthesis in the aedeagus, however, brown melanin is still visible in y1 mutants. (B) Diagram of the male exon structure of the dsx locus highlighting the strategy used to dissect the 42D04-GAL4 expression pattern. Five new GAL4 lines were created by synthesizing different sized sub-fragments of the 42D04-GAL4 enhancer fragment and cloning them upstream of GAL4 (see Supplementary Materials and methods). Note, 42D04_B-GAL4 could not be maintained, since female flies expressing GAL4 using this enhancer region were all sterile and showed necrotic growths on their genitalia. (C) Expression pattern of 42D04_A,C,D, and E-GAL4 lines. Expressing cytGFP using 42D04_A-GAL4 showed GFP (green) localized to bristle sockets, and 42D04_E-GAL4 shows bright GFP in the sex comb and lower leg region. 42D04_C-GAL4 and 42D04_D-GAL4 did not show GFP expression in the legs. Expressing Laccase2-RNAi using 42D04_A-GAL4 and 42D04_E-GAL4 blocked melanin synthesis in the sex combs but not the aedeagus. (D) Expressing Laccase2-RNAi using 42D04_A-GAL4 and 42D04_E-GAL4 inhibited male mating success. Sample sizes are shown at the top of each barplot. Significance was measured using Fisher’s exact tests with Bonferroni corrections for multiple comparisons. Comparisons that were statistically (p<0.05) are indicated (*p<0.05, **p<0.01, ****p<0.0001).
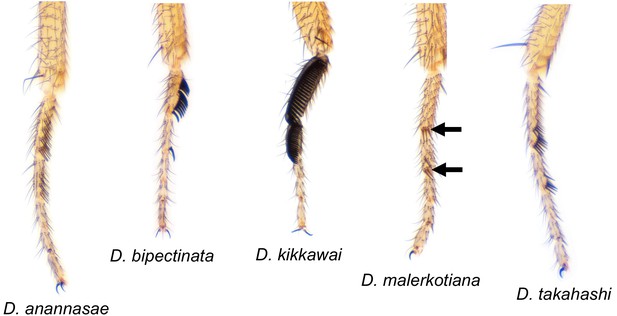
Drosophila species with varying sex comb morphology used for high-speed video assays.
D. anannasae, D. bipectinata, D. kikkawai, D. malerkotiana, and D. takahahi male front forelegs, highlighting variation in sex comb morphology (Nicolas Gompel).

Sex comb melanization is required for male mating success with y1 females.
(A) y1 males showed increased male mating success with y1 females. (B) Expressing Laccase2-RNAi using 42D04-GAL4 in males significantly inhibited male mating success with y1 females. Significance was measured using Fisher’s exact tests with Bonferroni corrections for multiple comparisons. Comparisons that were statistically (p<0.05) are indicated (**p<0.01, ***p<0.001).

Videos
Wild-type courtship and copulation.
https://doi.org/10.7554/eLife.49388.004y1 courtship with wild-type female.
https://doi.org/10.7554/eLife.49388.005Wild-type copulation.
https://doi.org/10.7554/eLife.49388.006Copulation attempts between y1 male and wild-type female after 3 hr of courtship.
https://doi.org/10.7554/eLife.49388.007Copulation attempts between male expressing yellow-RNAi in dsxGAL4-expressing cells and wild-type female.
https://doi.org/10.7554/eLife.49388.013Copulation attempts between male expressing yellow-RNAi in 42D04-GAL4-expressing cells and wild-type female.
https://doi.org/10.7554/eLife.49388.014High-speed (1000 fps) video capture of copulation attempts between y1 male and wild-type female.
https://doi.org/10.7554/eLife.49388.018High-speed (1000 fps) video capture of wild-type copulation.
https://doi.org/10.7554/eLife.49388.019Copulation attempts between male expressing Laccase2-RNAi in 42D04-GAL4-expressing cells and wild-type female.
https://doi.org/10.7554/eLife.49388.024High-speed (1000 fps) video capture of copulation attempts between male expressing Laccase2-RNAi in 42D04-GAL4-expressing cells and wild-type female.
https://doi.org/10.7554/eLife.49388.025Drosophila anannasae wild-type copulation.
https://doi.org/10.7554/eLife.49388.026Drosophila bipectinata wild-type copulation.
https://doi.org/10.7554/eLife.49388.027Drosophila kikkawai wild-type copulation.
https://doi.org/10.7554/eLife.49388.028Drosophila malerkotiana wild-type copulation.
https://doi.org/10.7554/eLife.49388.029Drosophila takahashi wild-type copulation.
https://doi.org/10.7554/eLife.49388.030Drosophila willistoni wild-type copulation.
https://doi.org/10.7554/eLife.49388.031Additional files
-
Source code 1
R code used for statistical analyses.
- https://doi.org/10.7554/eLife.49388.032
-
Supplementary file 1
Sequences used for cloning.
- https://doi.org/10.7554/eLife.49388.033
-
Supplementary file 2
Summary table containing data for all mating trials.
- https://doi.org/10.7554/eLife.49388.034
-
Supplementary file 3
Summary of all statistical tests reported in the paper.
- https://doi.org/10.7554/eLife.49388.035
-
Supplementary file 4
Key Resources Table.
- https://doi.org/10.7554/eLife.49388.036
-
Transparent reporting form
- https://doi.org/10.7554/eLife.49388.037





