Decoding WW domain tandem-mediated target recognitions in tissue growth and cell polarity
Figures
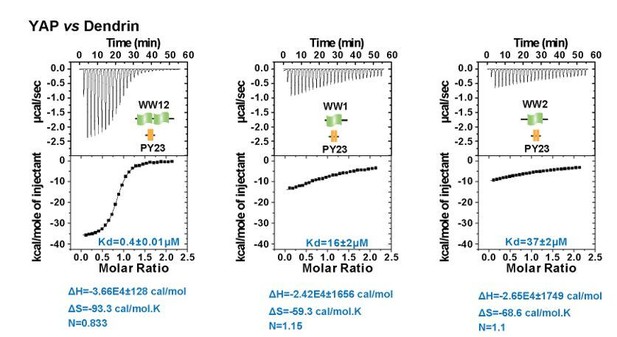
Super strong bindings of the KIBRA WW tandem to PY-motif containing proteins in cell growth control.
(A) Schematic diagram showing the domain organization of selected WW-tandem containing proteins and multiple PY-motifs containing proteins in cell growth and polarity. (B) ITC-derived binding affinities of WW tandems from KIBRA and YAP in binding to PY motifs from PTPN14, Dendrin, AMOT and LATS1. (C–D) Example ITC titration curves showing the bindings of KIBRA WW12 to PTPN14 PY12 (C) and to Dendrin PY23 (D). Due to the very tight direct bindings between the WW tandem and each of the PY-motifs, we used a competition-based binding assay (i.e. by first saturating each PY-motifs with YAP WW12 which has a modest binding to the PY-motif) to obtain reliable dissociation constants. (E–G) ITC-based measurements of the bindings between WW tandems of KIBRA and YAP and PY motifs of AMOT and Expanded. Both PY motifs of AMOT and Expanded are separated by long and flexible linkers. For simplicity, we have only listed Kd values (showed as values ± fitting errors) for the ITC experiments in the figure. Other thermodynamic parameters (i.e. Kd, ΔH, TΔS, and N value) of each ITC titration experiment covered in this study are compiled and presented in Supplementary file 2. The exact sequence of the WW tandems and its binding targets investigated in this study is compiled in Supplementary file 4.
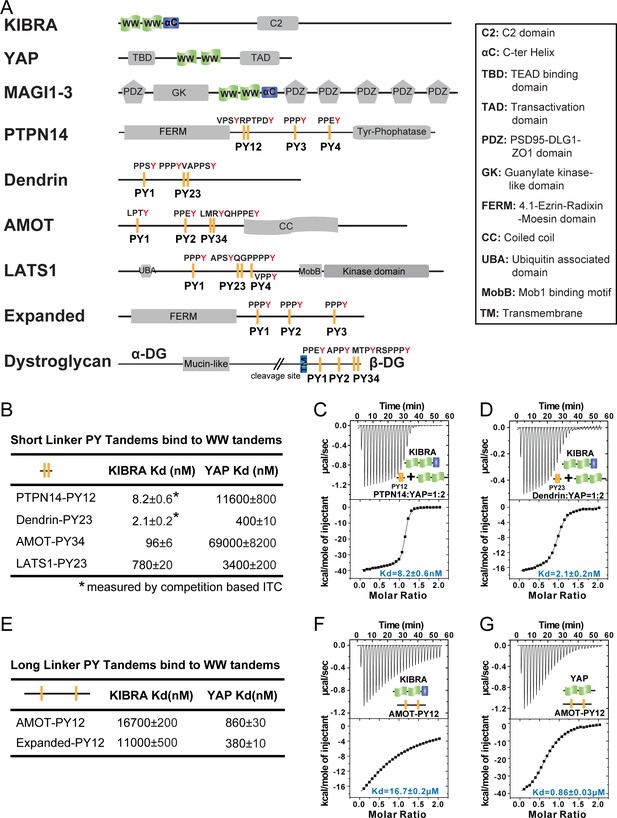
Super strong bindings of the KIBRA WW tandem to PY-motif containing proteins functioning in cell growth control.
(A) Amino acid sequence alignment of the closely spaced PY motifs from PTPN14, AMOTs and LATS1. (B) ITC-derived binding affinities of isolated WWs and WW tandems from KIBRA and YAP in binding to closely connected PY motif. (C and D) Superimposition of ITC curves of isolated WWs and WW tandems from KIBRA (C) and YAP (D) in binding to closely spaced PY motif. (E and F) ITC-based measurements of the bindings between WW tandems from KIBRA and YAP, and various PY motifs of AMOT (E) and LATS1 (F). Due to the existence of multiple binding sites on AMOT PY1-4, the binding affinity between KIBRA WW tandem and AMOT PY1-4 could not be properly analyzed by curve fitting, and thus no Kd value was reported.
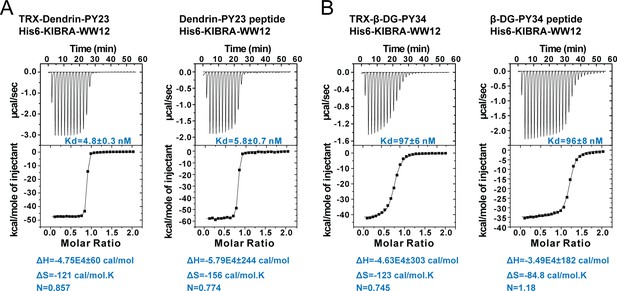
ITC experiments showing the bindings of Dendrin PY23 or β-DG PY34 to KIBRA WW12, showing that the TRX-tag had minimal impact on the bindings of each of the two peptides to the KIBRA WW12 tandem.
In this figure, TRX-Dendrin PY23 or TRX-β-DG PY34 represents each peptide was fused to the C-terminal end of the TRX tag. Dendrin PY23 peptide or β-DG PY34 peptide represents synthetic peptide encompassing each of the minimal PY motifs.
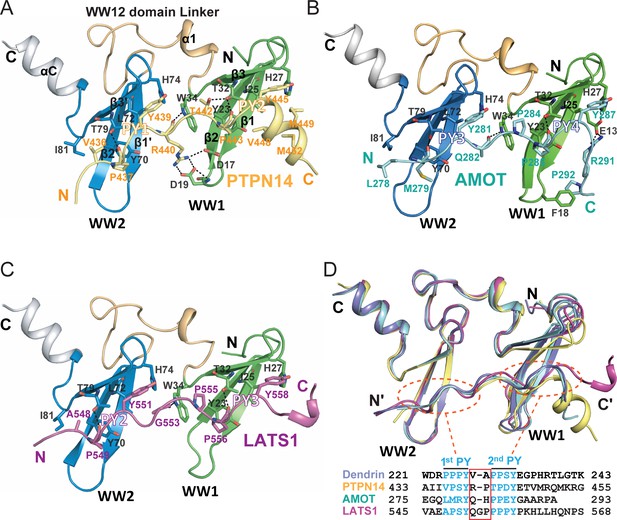
Structures of the KIBRA WW tandem in Complex with various closely spaced PY motifs.
(A–C) Combined ribbon and stick model showing the structures and detailed interactions of the KIBRA WW tandem in complex with PY motifs from PTPN14 (A), AMOT (B), and LATS1 (C). In the crystals of the KIBRA/AMOT complex, WW12 adopts a domain-swapped dimer similar to the crystals of the KIBRA/Dendrin complex described earlier (Ji et al., 2019), so the KIBRA/AMOT supramolecular complex is formed from two different polypeptides in this structure. However, in solution KIBRA and AMOT form a 1:1 complex. (D) Superimposition of the structures of the KIBRA WW tandem in complex with PY motifs from PTPN14, AMOT, LATS1, and Dendrin. The amino acid sequences of the PY motifs are numbered and aligned at the bottom of the panel, and the red box highlights the 2–3 residue linking sequences of the PY motifs. The color-coding scheme for the structures is used throughout the manuscript.
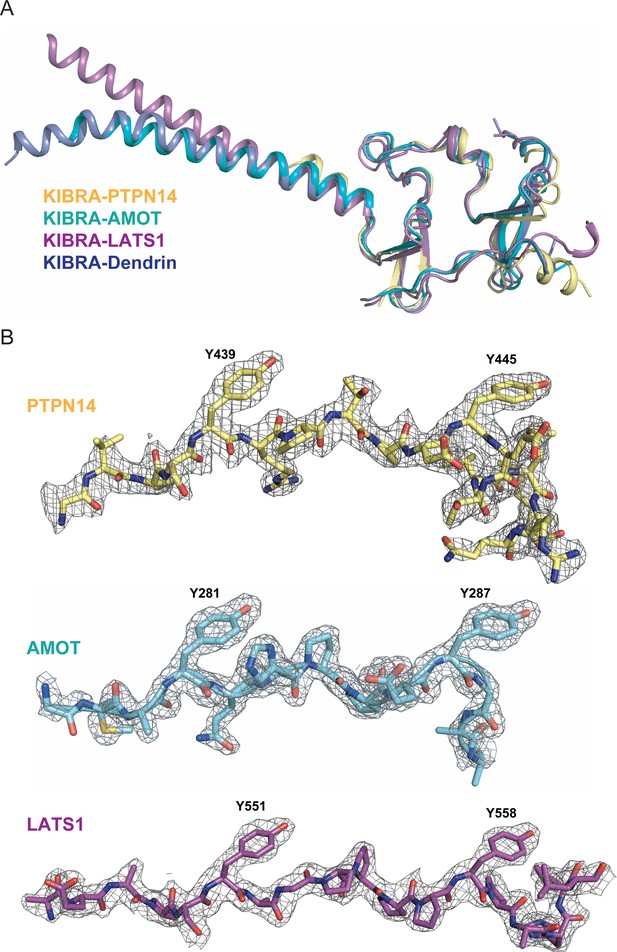
Structures of KIBRA WW12 in Complex with various closely connected PY motifs.
(A) Superimposition of the overall structures of the KIBRA WW tandem in complex with PY motifs from PTPN14, AMOT, LATS1, and Dendrin. (B) The electron density maps of PY motifs from PTPN14, AMOT and LATS1 are shown and are colored grey and contoured at 1.5δ.
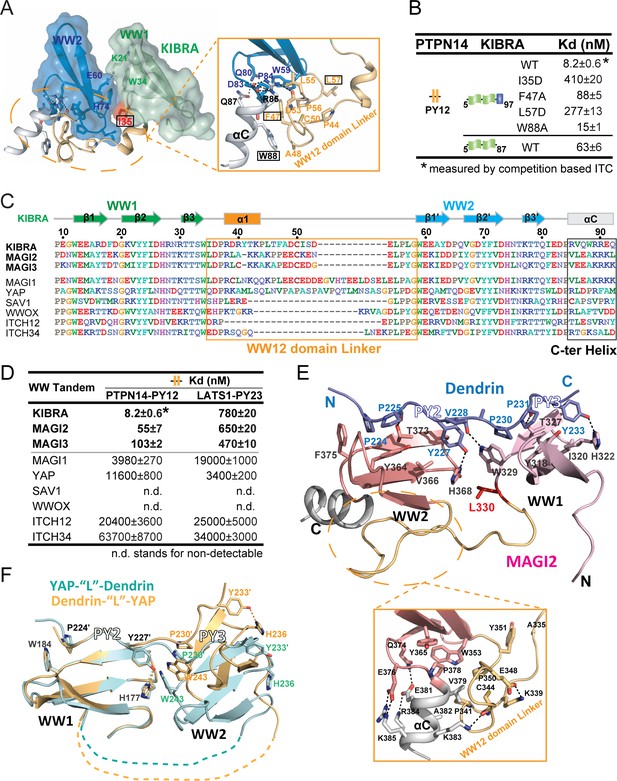
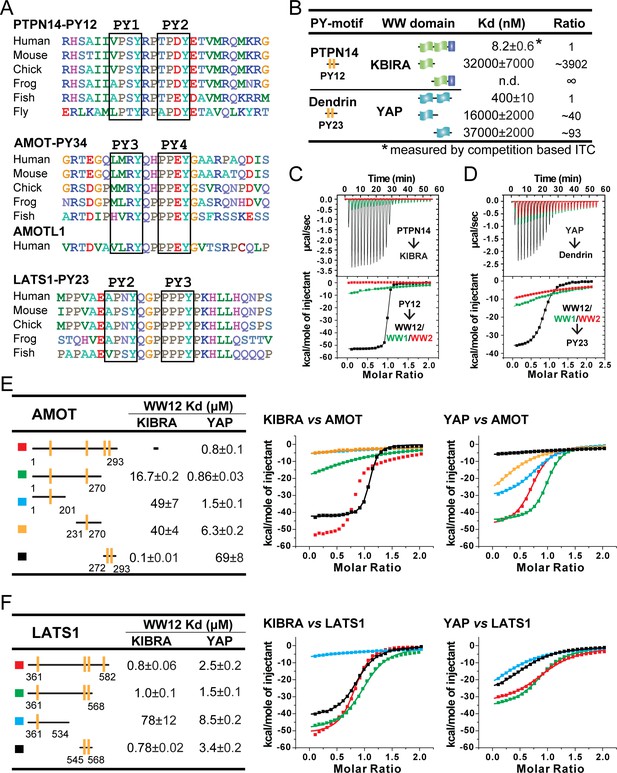
Structural basis governing the strong bindings of various WW tandems to their PY-motif containing targets.
(A) Combined surface and ribbon-stick model showing the structural basis for the formation of the KIBRA WW tandem supramodule. The detailed interactions between the inter-domain linker and C-terminal α-helix (αC) are shown in an expanded insert at right. The residues chosen for mutational analysis in Panel B are boxed. (B) ITC-based measurements comparing the binding affinities of the KIBRA WW tandem and its various mutants to PTPN14 PY12. (C) Amino acid sequence alignment of the WW tandems from KIBRA, MAGI1, MAGI2, MAGI3, YAP, SAV1, WWOX and ITCH-WW12/WW34, showing that the WW tandems of KIBRA, MAGI2, and MGAI3 are more similar to each other. (D) ITC-derived affinities of various WW tandems binding to PTPN14 PY12 and LATS1 PY23. (E) Combined ribbon and stick diagram showing the structure and detailed interaction of the MAGI2 WW tandem in complex with Dendrin PY23. The detailed interactions between the inter-domain linker and C-terminal α-helix are shown in an expanded box below the structure. (F) Ribbon and stick model showing the superimposition of two versions of the structures of YAP WW tandem in complex with Dendrin PY23. The structure colored in brown was obtained by fusing Dendrin-PY23 to the N-terminus of the YAP WW tandem, and the one colored in cyan was obtained by fusing Dendrin-PY23 to the C-terminus of the YAP WW tandem. The structures are superimposed by overlaying the WW1, showing different orientations of WW2 between the two structures. The dotted lines denote the linking sequence between WW1 and WW2, which could not be traced in the two crystal structures.
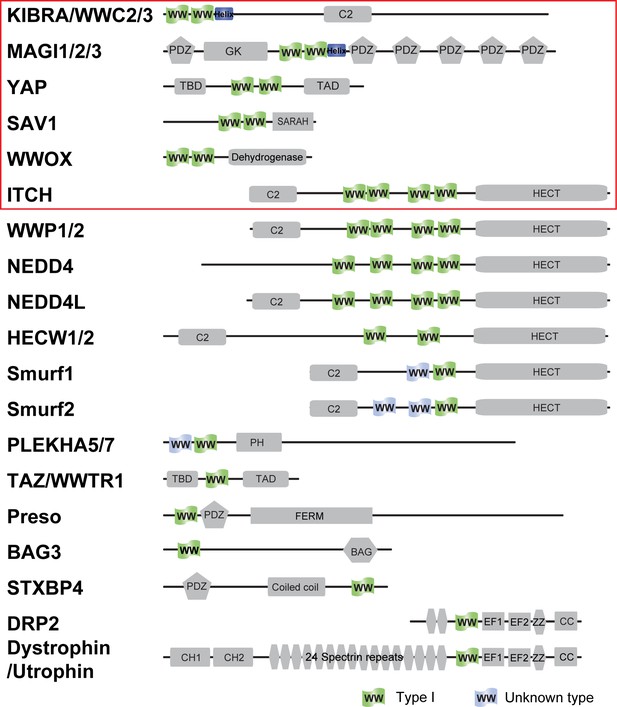
Domain organization of Type I WW containing human proteins.
The proteins tested in this study are labeled with a red box.
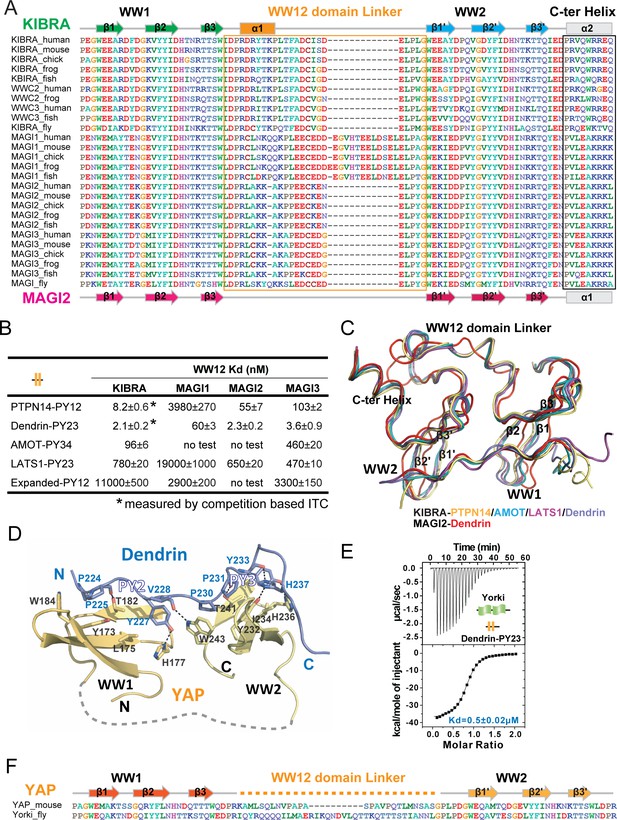
Structural basis governing the strong bindings of various WW tandems to their PY-motif containing targets.
(A) Amino acid sequence alignment of the WW tandems of KIBRA/WWC and MAGI families from different species. (B) ITC-based measurements comparing the binding affinities between various closely spaced PY motifs and WW tandems from KIBRA and MAGI family. (C) Superimposition of the complex structures of KIBRA and MAGI2 WW tandems with their respective ligands. (D) Ribbon and stick model showing the detailed interactions of YAP WW tandem in complex with Dendrin PY23. (E) ITC-based measurement of the binding between Yorkie WW tandem and Dendrin PY23. (F) Amino acid sequence alignment of the WW tandems of mouse YAP and Drosophila Yorkie.
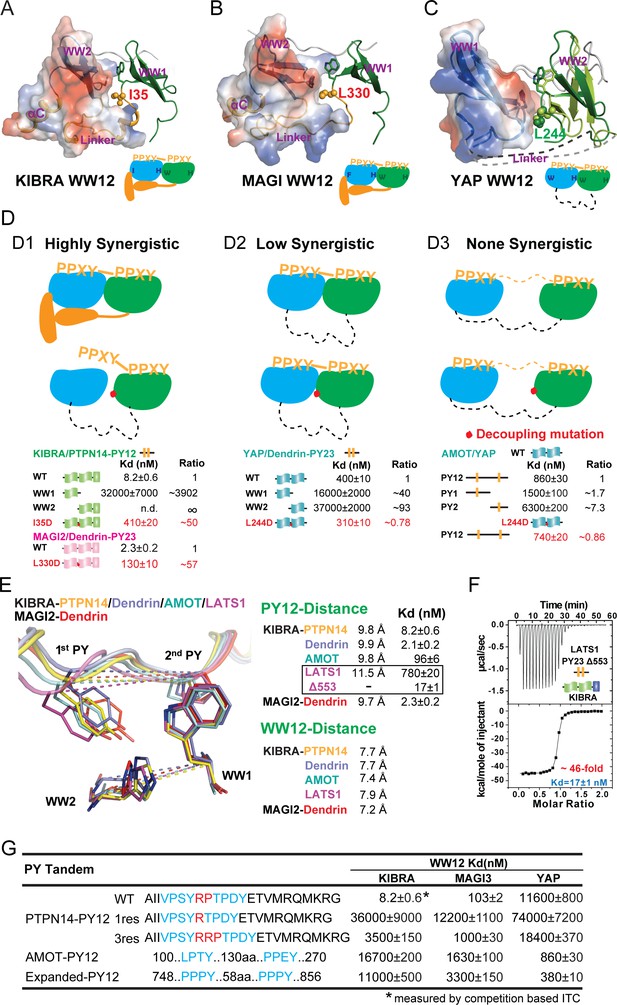
Binding modes of WW tandems to PY motifs.
(A–C) Combined surface and ribbon diagram showing the domain coupling of the WW tandems of KIBRA (A), MAGI2 (B), and YAP (C), each in complex with Dendrin PY23. The cartoon in each panel is used to show the coupling mechanism of the WW domain tandems in the complexes. (D) Three distinct modes of WW tandem-mediated bindings to various PY motif targets. (D1), Highly synergistic bindings of tightly coupled WW domains in the tandem with two PY motifs separated by two and only two residues. In this mode, isolated WW domains have very weak binding to PY motifs as shown by the quantitative binding affinity data below the model cartoon. Perturbations of the inter-domain couplings (indicated by a red dot in the model corresponding to the I35D in KIBRA and L330D in MAGI2) also severely impair the WW tandem/target bindings. (D2) Low synergistic binding of WW tandems with PY motif targets as illustrated by the two WW domains with no apparent direct coupling but binding to two PY motifs separated only by a very short linker. In this mode, isolated WW domains have very weak binding to PY motifs. Modest synergism can be brought to action by the two closely spaced PY motifs. Changes of WW domain surface outside the binding areas (i.e. L244D of YAP) do not affect WW tandem/target bindings. (D3) None-synergistic bindings of WW tandems to PY motifs separated by long linkers. In this mode, bindings of the two WW domains to PY-targets are simply additive in their Kd values. (E) Ribbon-stick model showing the distances of the two PY motifs as well as two WW domains in the five complex structures. The inter-WW domain distances are indicated by the signature Trp at the end of β3-strand of WW1 and His in the β2-β3 hairpin of WW2. The distances between the PY motifs are measured Tyr in the first PY motif and the first Pro in the second PY motif. The figure also shows that the Kd values of the binds are nicely correlated to the distances between the two PY motifs in the complexes. (F) ITC-based measurement showing that removal of one residue (Gly553) in the 3-residue linker (‘QGP’) between LATS1 PY23 converted the LATS1 mutant into a super strong KIBRA WW tandem binder, presumably by shortening the inter-PY motif distance from 11.5 Å to ~9.8 Å as shown in panel E. (G) ITC-derived binding affinities of various WW tandems to PTPN14 PY12 motif mutants with different inter-motif linker lengths, showing that shortening or lengthening of the 2-residue PY motif linker led to weakening of the binding. AMOT PY12 and Expanded PY12 are used as examples of PY motifs separated by longer linkers.
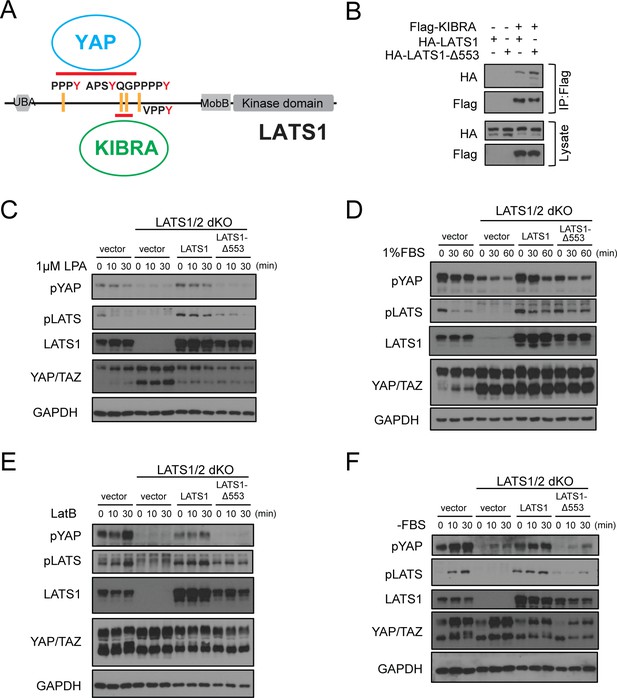
Deletion of Gly553 in LATS1 inhibits YAP phosphorylation in cells.
(A) Schematic diagram showing the interaction of LATS with KIBRA or YAP. (B) LATS1-△553, compared to WT LATS1, has an enhanced binding to KIBRA in HEK293A cells. (C, D) Comparison of YAP and LATS1 phosphorylations of wild type HEK293A cells, LATS1/2 dKO cells, LATS1/2 dKO cells stably expressing LATS1 or LATS1-△553. Cell were serum-starved for 4 hr and then treated with 1 μM LPA (C) or 1%FBS (D) for indicated times. (E, F) Comparison of YAP and LATS1 phosphorylations of wild type HEK293A cells, LATS1/2 dKO cells, LATS1/2 dKO cells stably expressing LATS1 or LATS1-△553 treated with 0.2 mg/ml LatB (E) or serum starvation (F) for indicated times.
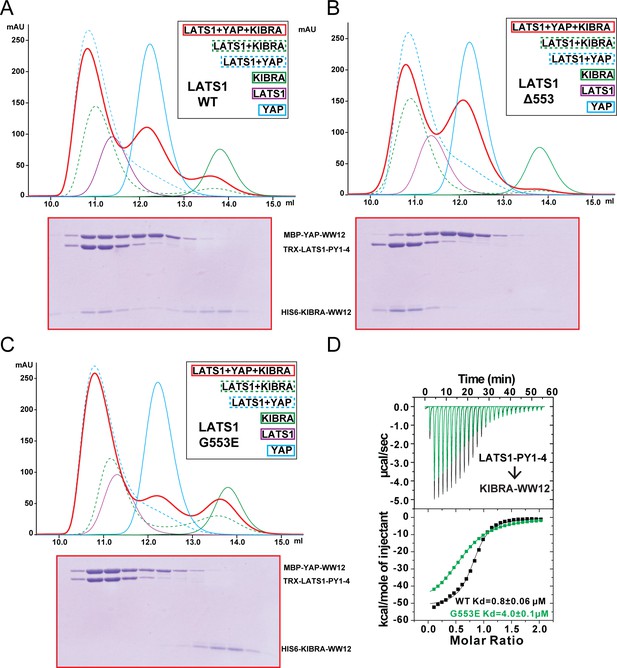
KIBRA and YAP compete for binding to LATS1.
(A) Analytical gel filtration chromatography showing that KIBRA WW12 and YAP WW12 compete with each other for binding to WT LATS1 PY1-4. The elution fractions of the KIBRA WW12, YAP WW12 and LATS1 PY1-4 (or its mutants in Panels B and C below) mixture at a 1:1:1 ratio (red curve) were analyzed by SDS-PAGE. The proteins used were: His6-KIBRA WW12 (aa 5–97), MBP-tagged YAP WW12 (aa 151–251), and Trx-tagged LATS1 PY1-4 (aa 361–582) or the corresponding Gly553 mutants below. (B) LATS1 PY1-4(Δ553) displayed enhanced binding to KIBRA WW12, as all KIBRA WW12 was shift to the complex fractions and YAP WW12 shifted to the free form. (C) LATS1 PY1-4(G553E) cannot bind to KIBRA WW12 as all KIBRA WW12 shifted to the free form, and LATS1 PY1-4(G553E) became preferentially bound to YAP WW12. (D) ITC-based measurements of the bindings of KIBRA WW12 to WT LATS1 PY1-4 or LATS1 PY1-4(G553E).
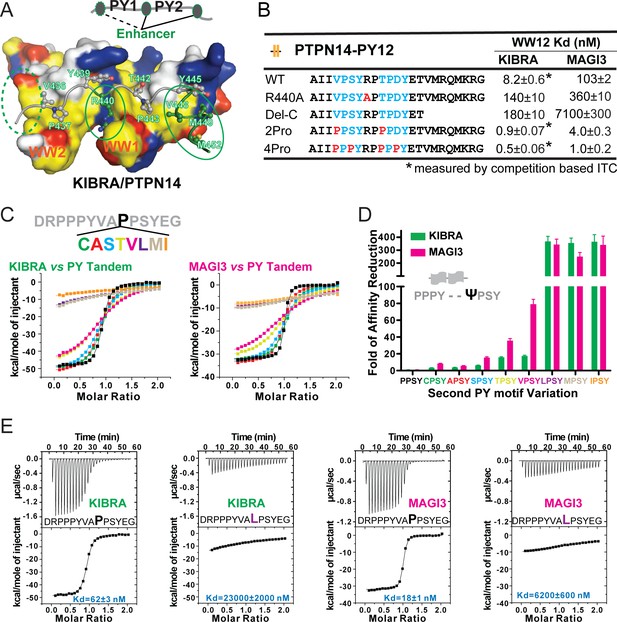
Specificity determinants of closely spaced PY motif to the WW tandems of KIBRA and MAGI2/3.
(A) Surface combined with ribbon-stick model showing the binding between the KIBRA WW tandem and PTPN14 PY12. The figure is to show that, in addition to the WW/PY motif interactions, the inter-PY motif linker and the extension sequence following PY2 (as indicated by the green ovals) also play critical roles for the super strong bindings between the WW tandem and the PY motifs. Additionally, residues preceding PY1 (indicated by the dotted green oval) may also enhance the binding. In the drawing, the hydrophobic, negatively charge, positively charged, and polar residues are colored in yellow, red, blue and white, respectively, in the surface model. (B) ITC-derived binding affinities of the KIBRA and MAGI3 WW tandem to various PTPN14 PY12 mutants. For bindings with nM or sub-nM Kd values, competition-based ITC assays were used as described in Figure 1C and D. (C) Mutational analysis of the contributions of different amino acids corresponding to the first Pro in the second PY motif of Dendrin PY23 in binding to KIBRA and MAGI3 WW tandems. The figure shows the superposition plot of ITC-based binding curves of binding reactions performed under the same experimental condition (also see Figure 6—figure supplement 1 for re-scaled ITC curve) (D) Comparison of relative binding affinity difference of various PY motif mutants tested in Panel C. In this comparison, the bindings of the KIBRA and MAGI3 WW tandems to the wild type PY motifs with the optimal ‘PPSY’ sequence are set at the base value of ‘1’. The fold of affinity reductions of each mutant are plotted. (E) The representative ITC curves of showing the binding of the KIBRA/MAGI3 WW tandems to the WT PY motif peptide or to one of the representative mutants.
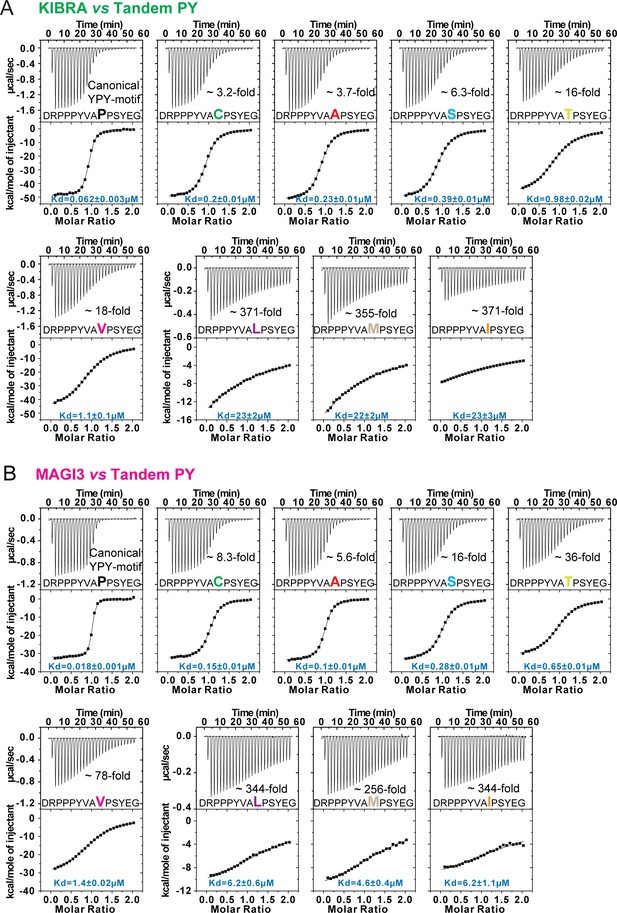
Bindings between WW tandems and closely spaced PY motifs with 2nd PY variation.
(A and B) ITC-based measurements comparing the binding of WW tandems from KIBRA (A) and MAGI3 (B) to Dendrin-PY23 with variations on 2nd PY motif.
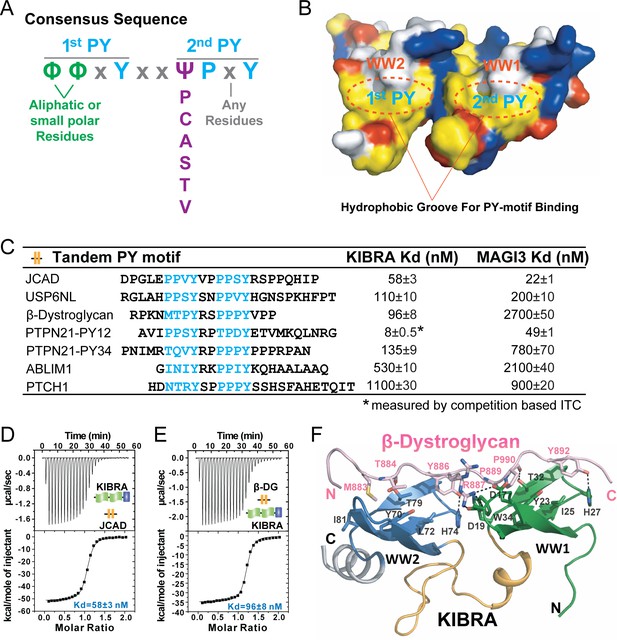
A consensus sequence of closely-spaced PY motifs capable of binding to KIBRA, MAGI2, and MAGI3 WW tandems with high affinities.
(A) The consensus sequence pattern of two PY motif-containing sequences that can synergistically bind to tightly coupled WW tandems such as those from KIBRA, MAGI2, and MAGI3. (B) Surface diagram showing two hydrophobic PY motif binding pockets of WW tandem that are precisely spaced due to the tight inter-domain coupling. The color coding is the same as in Figure 5A. (C) ITC-based measurements of the binding affinities of selected targets with PY motifs fitting the consensus sequence indicated in panel A to WW tandems from KIBRA and MAGI3. (D and E) Representative ITC binding curves of the bindings described in panel C, showing the bindings of KIBRA WW tandem to PY motifs JCAD (D) and β-Dystroglycan (E). (F) Ribbon-stick model showing the structure and detailed interaction of the KIBRA WW tandem in complex with the PY motifs from β-Dystroglycan.
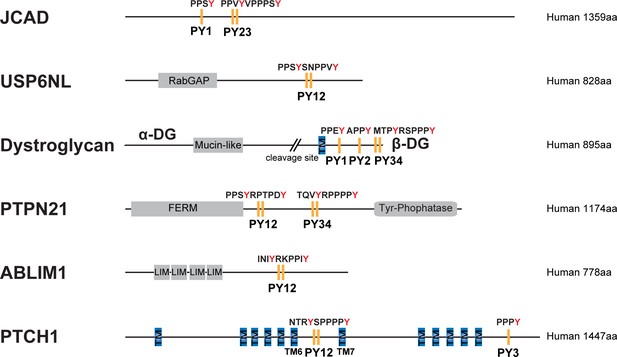
Domain organization of selected proteins with PY motifs fitting the consensus sequence indicated in panel Figure 7A.
https://doi.org/10.7554/eLife.49439.016Additional files
-
Supplementary file 1
Statistics of Data Collection and Model Refinement of the crystal structures.
- https://doi.org/10.7554/eLife.49439.017
-
Supplementary file 2
Summary of thermodynamic parameters for the binding of each WW domain or tandem to motifs derived by ITC experiments.
- https://doi.org/10.7554/eLife.49439.018
-
Supplementary file 3
List of human proteins containing the ‘ΦΦxYxxΨPxY’ motif.
- https://doi.org/10.7554/eLife.49439.019
-
Supplementary file 4
Key Resources Table.
- https://doi.org/10.7554/eLife.49439.020
-
Transparent reporting form
- https://doi.org/10.7554/eLife.49439.021