The mammalian LINC complex component SUN1 regulates muscle regeneration by modulating drosha activity
Figures
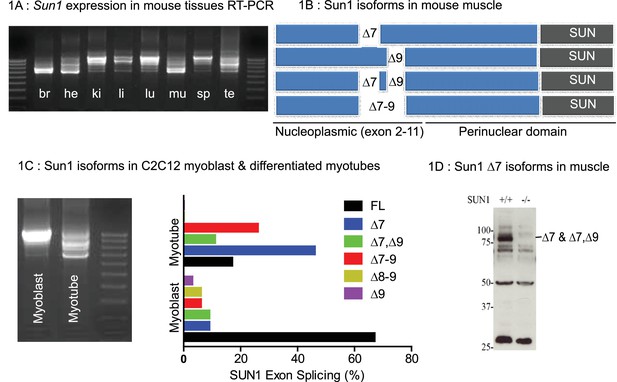
Tissue specific Splice Isoforms of Sun1.
(A) Sun1 cDNAs encoding the nucleoplasmic domain were amplified by RT-PCR from murine tissues; brain (br), heart (he), kidney (ki), liver (li), lung (lu), hind-limb muscle (mu), spleen (sp), and testis (te). The outermost lanes are the 100 bp DNA markers. (B) A diagram shows four different Sun1 cDNAs expressed in muscle due to alternative splicing between exons 7 to 9 (n = 34 clones sequenced). (C) Amplified Sun1 cDNAs from C2C12 myoblasts and differentiated myotubes showing major differences in expression of the spliced variants. These cDNAs were cloned and sequenced (n = 34 clones) with the Sun1 isoform percentages being presented in the chart, together with the full-length transcript. (D) Western blot showing the Sun1 Δ7 antibody specifically recognizes ~75 kD SUN1 protein in skeletal muscle lysates and is not detected in Sun1–/– muscle.

Amino acid sequence alignment of mouse SUN1 and human SUN1Δ6.
The mouse SUN1 exon 7 to 9 sequences are color coded and are not conserved in the human SUN1 gene.
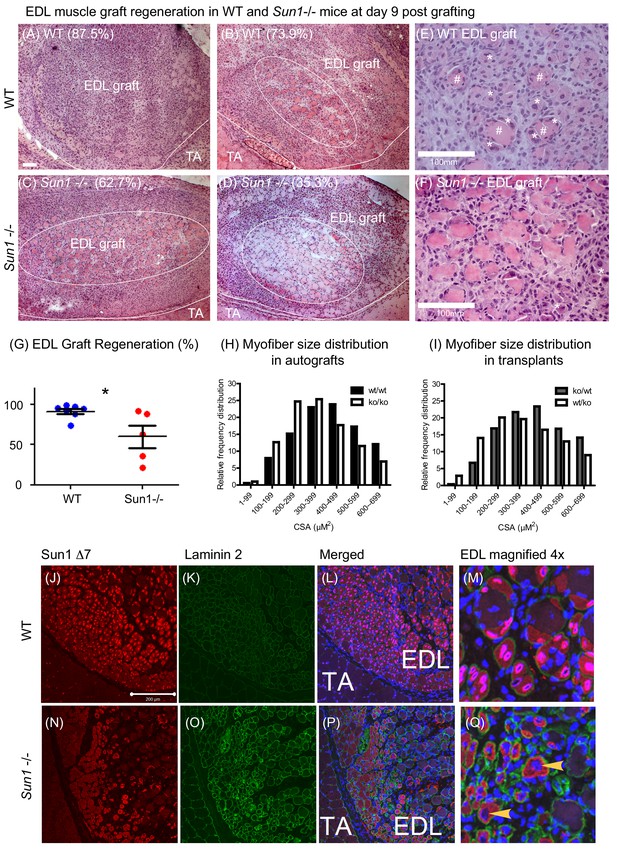
Loss of SUN1 retards muscle regeneration.
(A) Haematoxylin and eosin (H and E) stained images of EDL grafts at day nine post engraftment. In the WT grafts (A and B) and Sun1–/– grafts (C and D) the central necrotic muscles are demarcated by a white oval, with the boundary of the EDL graft and TA muscle being marked with a white line. Scale bar, 100 μm. (E) Magnified view of necrotic muscle fibers (#), with regenerating myofibers (*) clustered around the necrotic muscle and clusters of regenerated muscle fibers (*) in the WT EDL graft. (F) Myogenesis had initiated at the Sun1–/– /– periphery (*), however the core area was occupied with necrotic muscle fibers. (G) Comparison of muscle regeneration between WT (n = 7) and Sun1–/– (n = 5) mice. The numbers are expressed as a percentage of newly regenerated myofibers over total muscle numbers in the graft, with mean ± SEM and *p<0.05. Percentage of regeneration is included (in parenthesis) in the H and E images. (H and I) The cross sectional area (CSA) of the regenerated myofibers in both autografts and reciprocal grafts were quantified and presented in CSA distribution curves. (J) SUN1 Δ7 and - Δ7, Δ9 isoforms localize to the NE of regenerating EDL myofibers in vivo. Images (red immunostaining) of (top panel) SUN1 Δ7 at the nuclear envelope (NE) of WT muscle fibers; (K) laminin 2 (green) staining defines the boundary of individual myofibers and (L) with DAPI staining to mark the nuclei. (M) Sun1 Δ7 and - Δ7, Δ9 isoforms (red immunostaining) are expressed in regenerated muscle myofibers NEs (defined by the green laminin staining), with the isoforms being absent in surrounding non muscle cell nuclei (blue). (Q) In the Sun1–/– grafts the Sun1 Δ7 antibody did not stain the NE of regenerated muscle myofibers (as defined by the green laminin staining and yellow arrowheads), (bottom panel). The red cytoplasmic signal in myofibers is non-specific probably due to cross-reactivity by the polyclonal antibodies. Scale bar, 200 μm.
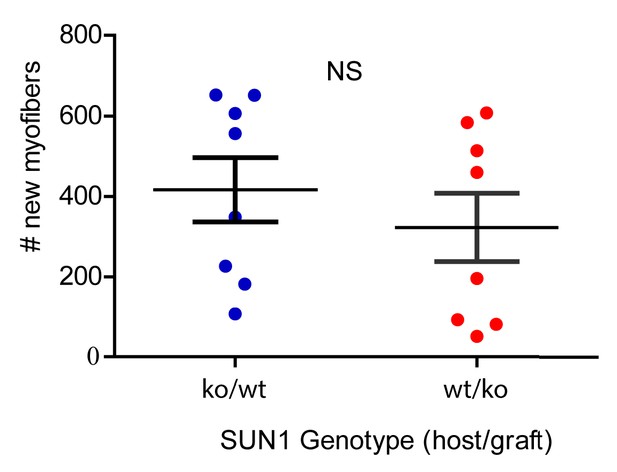
Number of regenerated myofibers in reciprocal graft transplants; WT EDL into Sun1–/– host (ko/wt) and Sun1–/– EDL into WT host (wt/ko) reveals no significant differences.
https://doi.org/10.7554/eLife.49485.005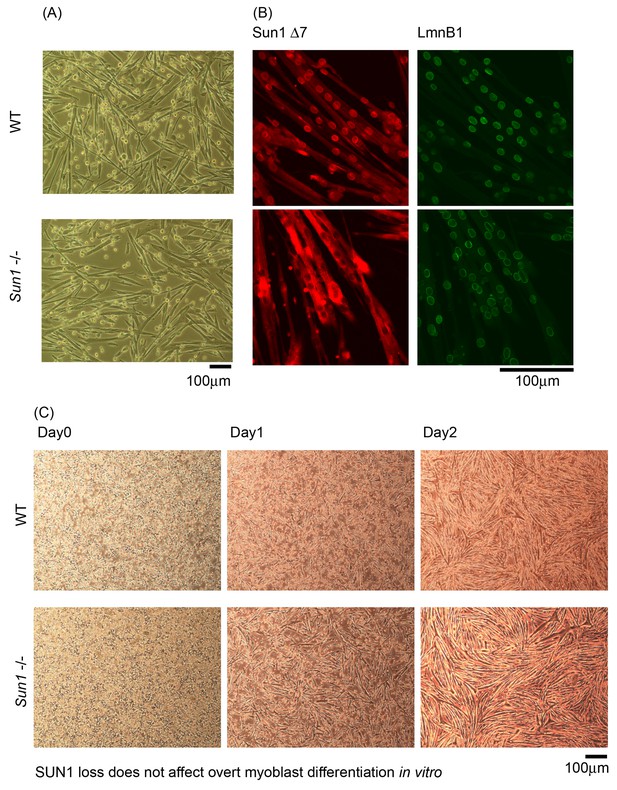
Sun1 expression in differentiating myoblast cultures.
(A) The in vitro differentiation of primary myoblasts derived from Sun1–/– and WT hind-limb muscle is similar. (B) Confocal immunostaining with SUN1Δ7 and Lamin B1 antibodies; SUN1Δ7 isoforms localize to the NEs of WT and are absent in the Sun1–/– myotubes. (C) In vitro differentiation of primary myoblasts at three time-points showing no overt differences in myotube formation.
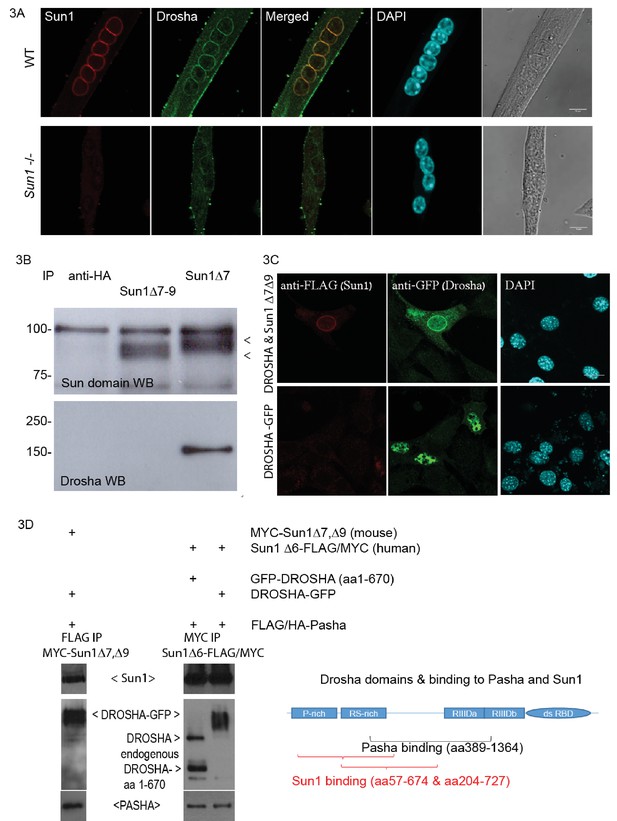
Sun1 interacts with Drosha.
(A) Confocal images of Drosha and SUN1 isoforms (12.10F antibody recognizing all SUN1 isoforms) localized to the NE of WT myotube nuclei, but not in the Sun1–/– myotubes. (B) Immuno-precipitation of SUN1 isoforms from C2C12 myotubes with two SUN1 antibodies (Δ7 and Δ7–9). SUN1 immuno-precipitates were detected with an anti-SUN domain antibody. Drosha was co-precipitated with SUN1 Δ7, but not with the Sun1 Δ7–9 isoform or anti-HA control antibodies. (C) Transient transfection of tagged cDNAs of Drosha and SUN1 Δ7 isoform in NIH3T3 cells, reveals enriched GFP tagged Drosha to the NE in nuclei co-expressing FLAG tagged SUN1 Δ7, Δ9 (top panel). Drosha expression in the absence of SUN1 Δ7,Δ9 localized throughout the nucleoplasm (bottom panel). (D) Transient transfection of tagged cDNAs in HEK293 cells for co-immuno-precipitation analysis. The Drosha schematic diagram shows the proline rich-, arginine/serine rich domains, the catalytic domains (RIIID) and RNA binding domain. The Pasha- and Sun1 binding domains partially overlap.
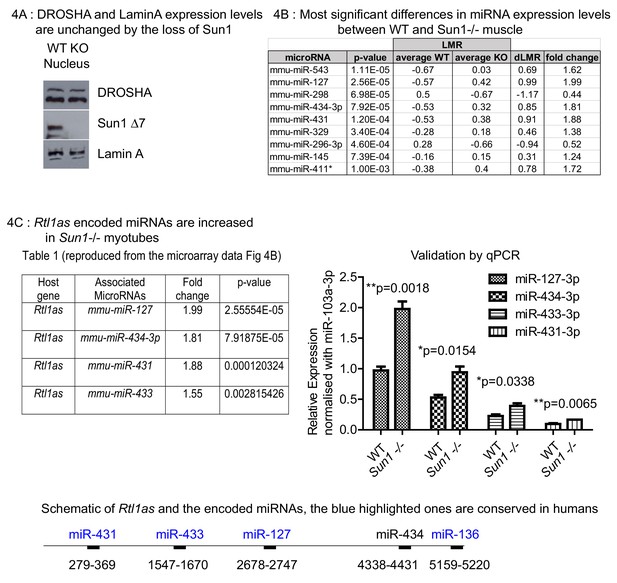
Effect of Sun1 loss on DROSHA expression and on the regenerating myofibre miRNA profile.
(A) Equal Drosha and Lamin A protein levels were present in the nuclear fractions from Sun1–/– and WT myotube cultures. WT and Sun1–/– primary myoblasts were differentiated into myotubes and then processed into cytoplasmic and nuclear fractions. Sun1 Δ7 antibody was the control for the WT lysate, Lamin A antibody was the positive control for the nuclear fraction. (B) Table lists the miRNAs that show significant fold differences in expression levels between WT and Sun1–/– myotubes. The p-values are based on the t-test. The top five miRNAs pass the Bonferroni correction for multiple testing (0.000161). Average expression (LMR) are shown for both groups, including the difference (dLMR) and the conversion into fold change. (C) The four miRNAs are all encoded by the Rtl1as transcript (see underlying schematic of the Rtl1as transcript and the position of the miRNAs). These miRNAs were expressed at significantly higher levels in Sun1–/– myotube cultures compared to WT cultures (4C Right panel). The relative abundance of each miRNA is also shown – (miR-127–3p>miR-434–3 p>miR-433–3p>miR431-3 p) and was determined by the CT values (qRT-PCR).
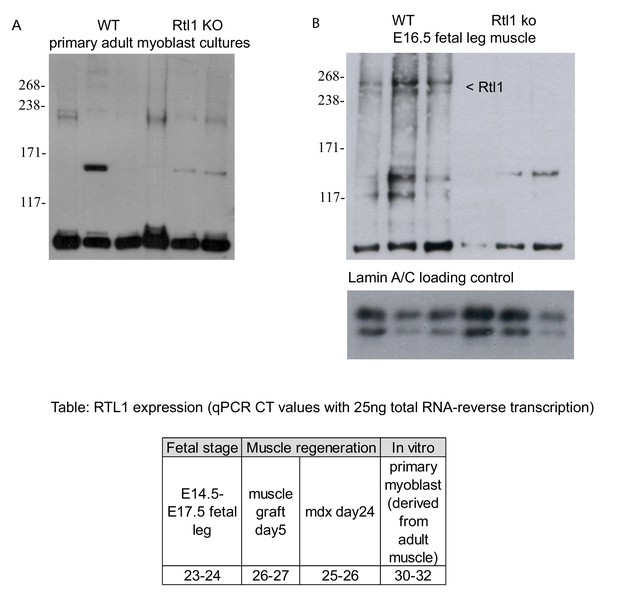
RTL1 protein was undetectable in WT primary myoblasts derived from hindlimb muscle, Rtl1 null derived primary myoblasts were a negative control (Left panel).
In comparison, WT fetal leg muscle expressed RTL1 protein with Rtl1 null fetal leg muscle as a negative control (Right panel). The table shows the CT values derived from qPCR experiments (with 25 ng total RNA) comparing Rtl1 RNA transcript levels during fetal development stages (E14.5-E17.5), in WT muscle grafts and mdx muscle representing muscle regeneration and in WT primary myoblast cultures. Higher levels of Rtl1 expression correlate with lower CT values.
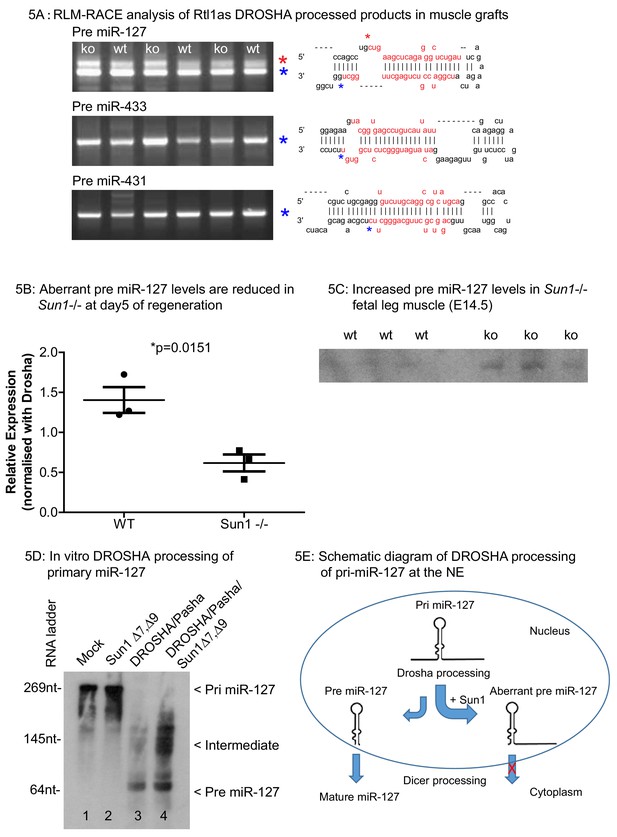
RLM-RACE analysis of Rtl1as DROSHA processed products in muscle grafts.
(A) RLM-RACE was performed on muscle graft samples from D5 of regeneration to identify Drosha mediated cleavage of Rtl1as RNA. PCR bands were cloned and sequenced to map Drosha cleavage sites. Sequences of pre-miRNAs in the hairpin configuration and the mature miRNA sequences are highlighted in red, mapped Drosha cleavage sites are marked by asterisks. (B) Quantitation from D5 grafts of WT and Sun1–/– muscle grafts showing aberrant pre-miR-127 levels are reduced in the Sun1–/– muscle. (C) Total RNA from WT and Sun1–/– muscle (5 ug) were resolved with 6% TBE-urea gel for detection of pre-miR-127. (D) Drosha, Pasha and SUN1 Δ7, Δ9 proteins were immunoprecipitated from HEK293 cells and incubated with primary miR-127 RNA in vitro. Background non-specific breakdown of primary miR-127 RNA by contaminating RNase from the HEK293 lysate (lanes 1 and 2). Drosha/Pasha cleaved the pri miR-127 to pre miR-127 (lane 3). In the presence of SUN1, Drosha cleaved pri miR-127 into intermediate miR-127 RNA and pre miR-127 (lane 4). (E) Schematic of pri miR-127 and Drosha processing into pre miR-127; the expected product of microprocessor cleavage in nucleus. Pre miR-127 is then exported out of nucleus into cytoplasm for further processing into mature miRNA by Dicer. Sun1 recruits Drosha to the NE and this might disrupts Drosha activity resulting in aberrant processing of pri-miR-127. Aberrant pre-miR-127 might not be exported out in the nucleus resulting in a reduction of mature functional miR-127 RNA.
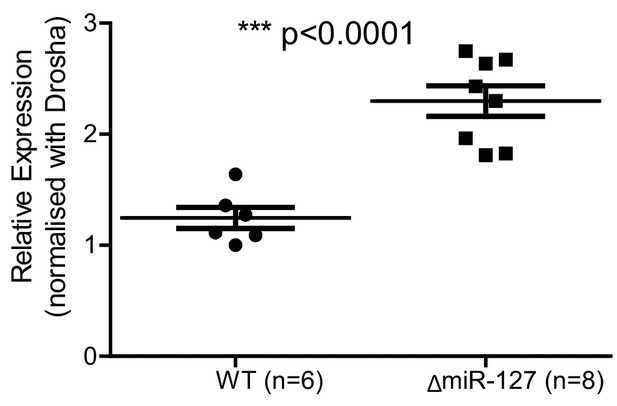
Rtl1 expression levels were quantified by RT-qPCR in miR-127 Δ and WT hind-limb muscle from D24 mice.
Rtl1 levels were significantly increased in the miR-127Δ hind-limb muscle at D24.
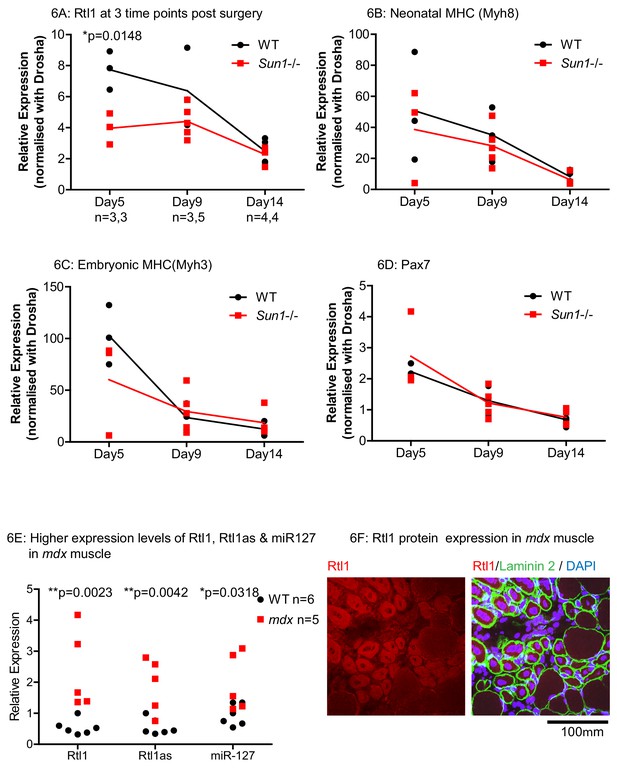
Effects of Sun1 loss on Rtl1 and other muscle specific gene expression levels during regeneration.
(A) Rtl1 expression increases in regenerating WT myofibers but at lower levels in Sun1–/– grafts. Both the grafted muscle (left hind-limb) and control muscle (right hind-limb) were harvested at three time-points (Days 5, 9 and 14 post-grafting) for qRT-PCR. The relative expression levels in the muscle grafts over control muscle in each sample with (n) representing sample size is presented at each time-point. WT grafts showed significantly higher Rtl1 expression compared to Sun1–/– grafts at D5. (B–D) High expression levels of Mhy8, Mhy3 and Pax7 were noted in these grafts at D5 indicating effective regeneration. (E) Rtl1, Rtl1as and miR-127 showed significantly increased levels in the hind-limb muscle of mdx mice compared to WT littermates at D24 (postnatal). Statistical analysis with unpaired t test; two-tailed p values *p<0.05 and **p<0.005. (F) Confocal image analysis with the anti-Rtl1 antibody revealed RTL1 protein localizes to the cytoplasm of regenerating mdx myofibers that have a centrally located nucleus (left panel). Fiber location was determined by co-staining with an anti laminin2 antibody and DAPI (right panel).
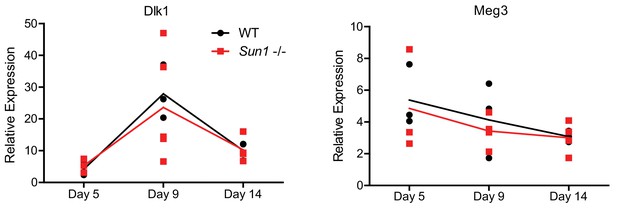
The Dlk1 and Meg3 imprinted genes in autografts of WT and Sun1–/– mice were quantified by RT-qPCR and did not show any significant differences.
https://doi.org/10.7554/eLife.49485.013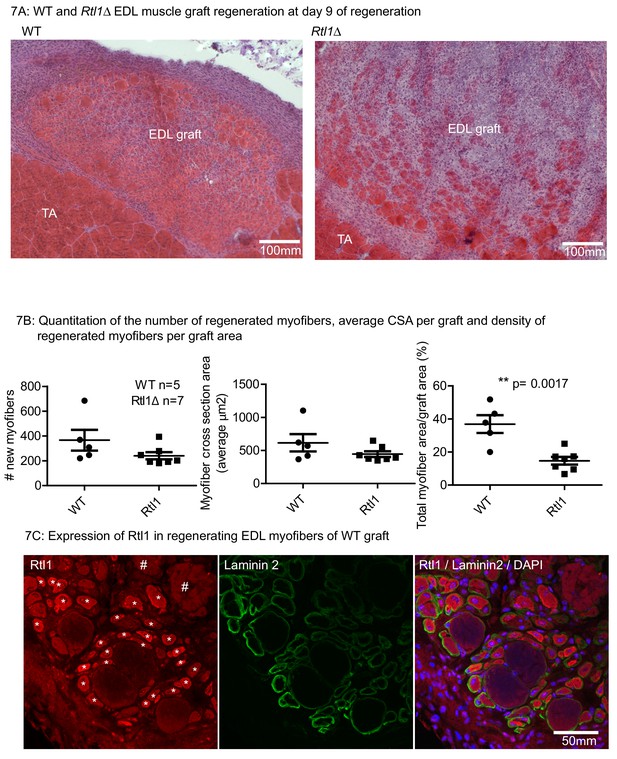
Loss of Rtl1 impairs muscle regeneration.
(A) H and E images of Rtl1 Δ and WT EDL autografts at D9 post grafting. (B) The Rtl1 Δ EDL grafts have fewer regenerated myofibers (p=0.0017) and the central region of the graft is occupied by non-muscle cells, while myogenesis was almost completed in the WT graft. (C) RTL1 protein expression was present in regenerating myofibers (*) as shown by co-staining of anti-RTL1, anti-laminin2 antibody and DAPI. Necrotic muscles are marked with # (C left panel).
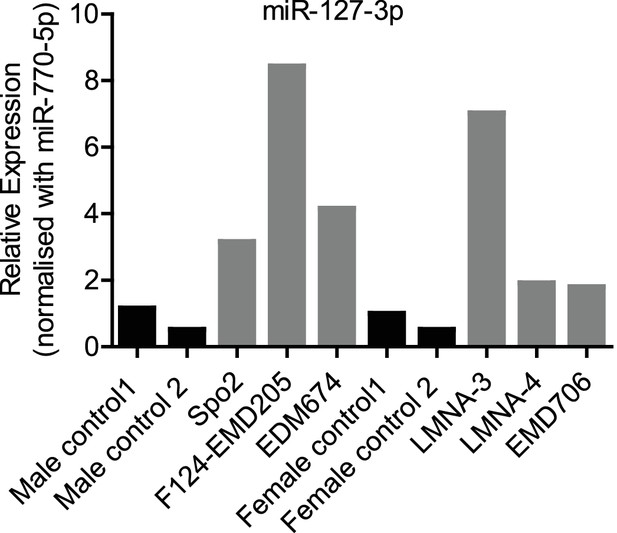
miR-127–3 p levels were quantified in healthy human biopsy samples and patients diagnosed with different LMNA mutations.
Refer to Supplementary file 2 for the list of LMNA patient mutations and their associated pathologies.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (M. musculus, males and females) | SUN1+/- | PMID: 19211677 | RRID: MGI:3838371 | mixed strain, (C57BL/6J × 129/J); SUN1+ /- males and females for breeding |
| Strain, strain background (M. musculus, males and females) | miR-127 Δ | PMID: 26138477 | RRID: MGI:5789800 | C57BL/6J; miR-127 Δ females mated with WT C57BL/6 males for breeding |
| Strain, strain background (M. musculus, males) | Rtl1Δ | PMID: 26138477 | RRID: MGI:5789800 | mixed strain, (C57BL/6J × 129/J); miR-127 Δ males mated with WT 129Sv females to get Rtl1Δ pups |
| Strain, strain background (M. musculus, males) | mdx | Jackson Laboratory | RRID: IMSR_JAX:001801 | mixed strain, (C57BL/6) |
| Biological sample (M. musculus) | Primary myoblast cell (Wildtype and SUN1 -/-) | This paper | Derived from WT and SUN1 - /- littermates, males | |
| Cell line (H. sapiens) | Hek-293 | ATCC | CRL-1573TM | |
| Cell line (M. musculus) | NIH/3T3 | ATCC | CRL-1658TM | |
| Cell line (M. musculus) | C2C12 | ATCC | CRL-1772TM | |
| Transfected construct (M. musculus) | pCMV6- FLAG-SUN1 Δ7 Δ9 | This paper | Mammalian expression construct | |
| Transfected construct (M. musculus) | pCMV6- MYC-SUN1 Δ7 Δ9 | This paper | Mammalian expression construct | |
| Transfected construct (H. sapiens) | pCMV6- SUN1 Δ6 -MYC/FLAG | Origene | RC226167 | Mammalian expression construct |
| Transfected construct (H. sapiens) | pCMV6- Drosha-GFP | This paper | Mammalian expression construct | |
| Transfected construct (H. sapiens) | pFLAG/HA- DGCR8 (Pasha) | PMID: 15589161 | RRID: Addgene_10921 | |
| Recombinant DNA reagent | pSPT18 -primary miR127 | This paper | In vitro transcription of pri-miR127 for Drosha cleavage assay | |
| Recombinant DNA reagent | pSPT18-pre- miR127 | This paper | In vitro transcription, with DIG-labelling of probe | |
| Antibody | anti-Drosha (rabbit polyclonal) | Abcam | Cat# ab12286 | IF(1:100), WB (1:500) |
| Antibody | anti-Lamin B (goat polyclonal) | Santa Cruz Biotechnology | Cat# sc-6217 | IF(1:100) |
| Antibody | anti-Lamin A/C (rabbit polyclonal) | Cell Signaling Technology | Cat# 2032 | WB(1:500) |
| Antibody | anti-MYC (rabbit monoclonal) | Cell Signaling Technology | Cat# 2278 | WB(1:2000) |
| Antibody | anti-FLAG (rabbit monoclonal) | Cell Signaling Technology | Cat# 14793 | IF(1:100), WB(1:2000) |
| Antibody | anti-laminin 2 (rat monoclonal) | Enzo life sciences | 4H8-2 | IF(1:200) |
| Antibody | anti-SUN1 12F10 (mouse monoclonal) | This paper | RRID: AB_2813863 | this antibody recognises all mouse SUN1 isoforms |
| Antibody | anti-SUN1 Δ7 (rabbit polyclonal) | PMID: 26417726 | Raised with peptide RDRTLKPPHLGHC by YenZym Antibodies | |
| Antibody | anti-SUN1 Δ7–9 (rabbit polyclonal) | This paper | Raised with peptide CGGDRTLKPRDLLVQ by YenZym Antibodies | |
| Antibody | anti-SUN1 (rabbit polyclonal) | PMID: 19211677 | this antibody recognises all mouse SUN domain | |
| Antibody | anti-Rtl1 (rabbit polyclonal) | This paper | RRID: AB_2813865 | antibody produced by YenZym Antibodies |
| Antibody | anti-GFP 14F5 (mouse monoclonal supernatant) | This paper | provided by Brian Burke, A*STAR |
Additional files
-
Source data 1
Raw data for graphs.
- https://doi.org/10.7554/eLife.49485.016
-
Supplementary file 1
SUN1∆7, ∆9 Y2H interaction candidates and libraries screened.
- https://doi.org/10.7554/eLife.49485.017
-
Supplementary file 2
Clinical phenotypes and genotypes of LMNA mutated patients.
LGMD1B: Limb girdle muscular dystrophy type 1B; DCM: Dilated cardiomyopathy; CD: Conduction disease; CD-ARRH: conduction defects with arrhythmias, EDMD; Emery-Dreifuss muscular dystrophy; ICD: implantable cardiac defibrillator, PLD: Partial lipodystrophy.
- https://doi.org/10.7554/eLife.49485.018
-
Supplementary file 3
List of primer sequences.
- https://doi.org/10.7554/eLife.49485.019
-
Transparent reporting form
- https://doi.org/10.7554/eLife.49485.020





