Chromosome territory formation attenuates the translocation potential of cells
Figures
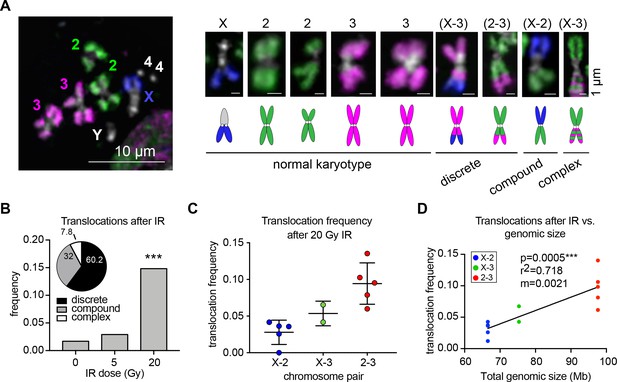
Whole-chromosome Oligopaints can efficiently detect IR-induced translocations.
(A) Left: representative metaphase spread with chromosome paints in control BG3 cells. DNA is stained with Hoechst and is shown in white. Right: representative chromosomes 48 hr after irradiation. Both normal and rearranged chromosomes are shown, with cartoon schematics of the chromosomes directly below. The chromosomes involved in the rearrangement (if any) are listed above, and the classification of each translocation type is listed below. (B) Total translocation frequency after varying doses of IR for 3–5 biological replicates. n = 592, 368, and 442 spreads counted for no IR, 5 Gy, and 20 Gy, respectively. Inset: Pie graph depicting the relative translocation types identified after 20 Gy of IR as a percent of total translocations. (C) Dot plot showing translocation frequencies for 2–5 populations of cells 48 hr after 20 Gy IR treatment. Only two replicates were included for X-3 due to pre-existing translocations in those cell sub-populations. (D) Scatterplot showing the translocation frequency after 20Gy IR (Y-axis) versus total genomic size of the chromosome pair (X-axis). The data shown represent five biological replicates. m = slope of line of best fit. P-value was calculated by linear regression.
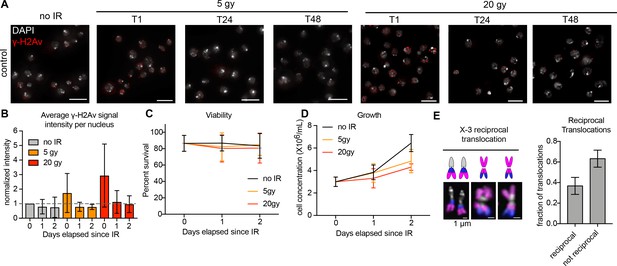
Additional data related to Figure 1.
(A) Representative images showing anti-γ-H2Av staining on BG3 cells before IR (no IR), and 1 hr (T1), 24 hr (T24), and 48 hr (T48) after IR with either 5 or 20 Gy treatments. Scale bar equals 10 μm. (B) Quantification of γ-H2Av fluorescence intensity as shown in (A). (C) Line graph showing average cell viability before and after IR, measured by trypan blue staining. Error bars show standard deviation between biological replicates. (D) Line graph showing average cell growth rates before and after IR over a 48 hr period. Error bars show standard deviation between biological replicates. (E) Left: Schematic and representative mitotic chromosome spread showing reciprocal translocations between chromosomes X and 3. Right: bar graph showing the average fraction of translocations after 20 Gy IR that were reciprocal for 2–5 biological replicates (n = 101 cells quantified total). Error bars show SEM between biological replicates.
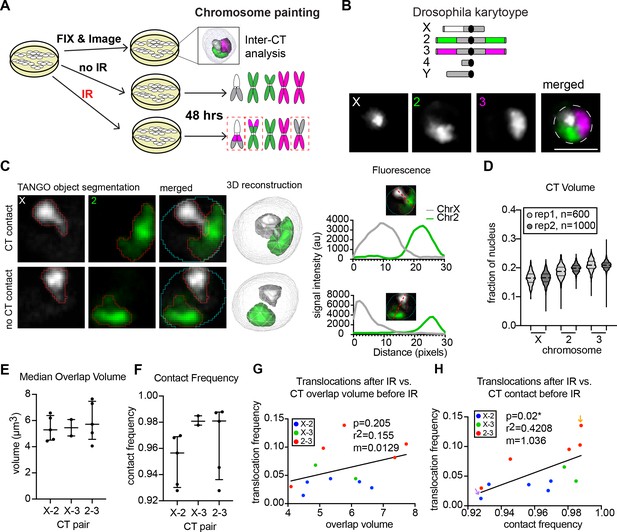
Inter-CT contacts correlate with translocation frequency in Drosophila.
(A) Schematic of experimental design in which cells are split into three groups: one of which is harvested immediately for interphase FISH analysis, the second is subjected to IR treatment, and the third is used as a ‘no IR’ control. The latter two groups are allowed to recover for two additional days before karyotype analysis to identify translocations (dashed red boxes). (B) Top: Cartoon depiction of BG3 cell karyotype and chromosome paints. Unlabeled heterochromatin is shown in gray. Bottom: representative nucleus with Oligopaints labeling chromosome X (white), 2 (green), and 3 (magenta). Dotted line in merged image represents the nuclear edge. Scale bar equals 5 μm. (C) Left: Representative image showing chromosome X paint (white) and chromosome two paint (green) in two representative nuclei illustrating CT contact and no CT contact. CT segmentation is shown as a red outline. Final 3D rendering is shown on the right. Right: Line plots of fluorescence intensity from the two cells depicted on left. In the top graph, voxel colocalization is observed while there is no voxel colocalization in the bottom graph. (D) Violin plot of CT volume for chromosomes X, 2, and three as a fraction of nuclear volume in BG3 cells. Each violin represents a single cell population, and two biological replicates are shown, each with >500 nuclei being measured. (E) Dot plot showing the median CT overlap volume between chromosome pairs defined by the X-axis, for 2–5 cell populations, where each dot represents the median of a cell population of n > 500 cells. (F) Dot plot showing the fraction of cells with CT contact between chromosome pairs defined by the X-axis, for 2–5 cell populations, where each dot represents the average of a cell population of n > 500 cells. (G) Scatterplot showing the translocation frequency after 20Gy IR (Y-axis) versus median CT overlap volumes prior to IR (X-axis). The data shown represent 3–5 biological replicates. m = slope of line of best fit. P-value was calculated by linear regression. (H) Scatterplot showing the translocation frequency after 20Gy IR (Y-axis) versus inter-CT contact frequency prior to IR (X-axis). The data shown represent 3–5 biological replicates. m = slope of line of best fit. P-value was calculated by linear regression. Arrows depict populations with the lowest (purple) and highest (yellow) translocation events.
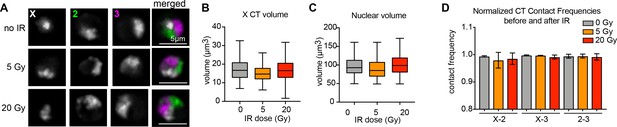
Additional data related to Figure 2.
(A) Representative nuclei with Oligopaints labeling chromosome X in white, chromosome two in green, and chromosome three in magenta under control conditions before IR (top), after 5 Gy IR (middle), and after 20 Gy IR (bottom). Scale bar equals 5 μm. (B) Tukey box plot showing chromosome X volume for a single biological replicate before and after IR. n = 550, 700, and 780 cells for no 0, 5, and 20 Gy respectively in B-D. (C) Tukey box plot showing nuclear volume for a single biological replicate before and after IR. (D) Bar graph showing normalized CT contact frequencies (between CT pairs labeled on the x-axis) for 2–5 biological replicates after varying doses of IR (no IR in gray, 5 Gy in orange, and 20 Gy in red). Error bars represent the standard deviation between replicates.
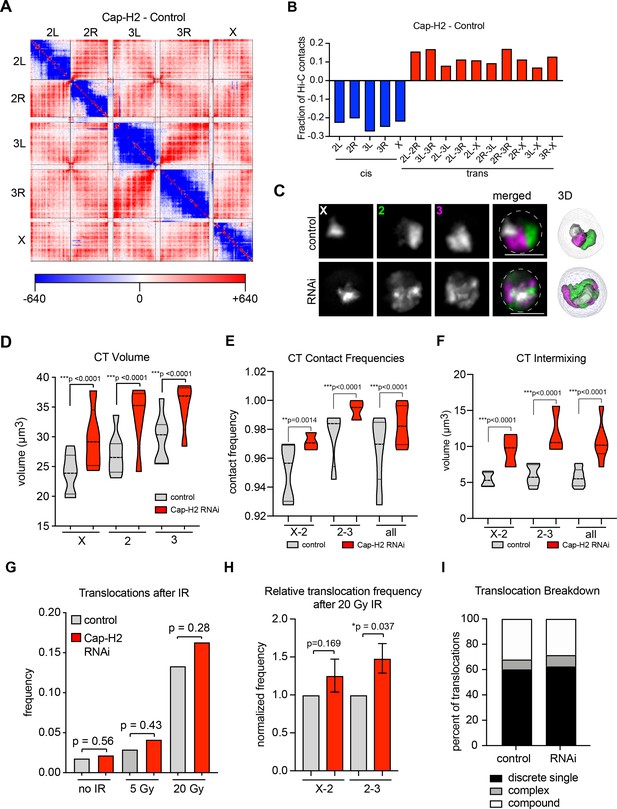
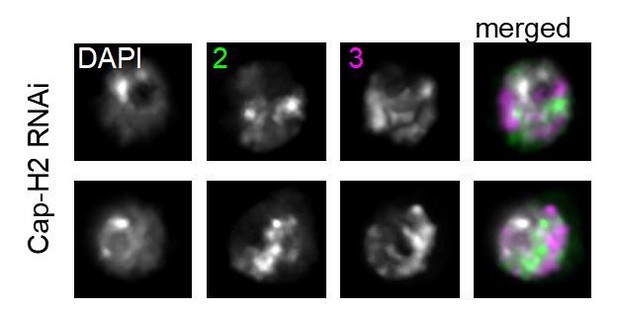
CT disruption following knockdown of Cap-H2 increases the translocation potential of long chromosomes.
(A) Whole genome heat map obtained by subtracting the two-dimensional contact matrix of Hi-C data from control and Cap-H2 depleted Kc167 cells. Hi-C data obtained from Li et al. (2015). (B) Bar graph showing the intra-chromosomal and inter-chromosomal changes for each chromosome pair calculated from the Cap-H2-Control Hi-C subtraction map. (C) Representative nucleus with Oligopaints labeling chromosome X (white), 2 (green), and 3 (magenta) in control conditions (top), or after Cap-H2 RNAi (RNAi; bottom). Dotted line in merged image represents the nuclear edge. Scale bar = 5 μm. Right: 3D rendering of segmented chromosome structures. (D) Violin plot showing average CT volumes across three biological replicates, both before and after IR, where n > 500 cells each, for control and Cap-H2 RNAi. p-values were determined by Student’s t-test. (E) Violin plot showing CT contact frequencies for X-2 and 2–3 CT pairs and combined across three biological replicates for control and Cap-H2 RNAi. p-values were determined by a Fisher’s Exact Test comparing contact and no contact for individual replicates. (F) Violin plot showing CT intermixing volumes for X-2 and 2–3 CT pairs and combined across three biological replicates for control and Cap-H2 RNAi. p-values were determined by Student’s t-test. (G) Bar graph showing the total translocation frequency after no IR, 5 Gy IR, 20 Gy IR, and combined (‘all’) for control and Cap-H2 RNAi cells. P-values were calculated by Fisher’s exact test comparing normal karyotypes to those with translocations for control and RNAi. (H) Bar graph showing fold-change in translocation frequencies of control and Cap-H2 RNAi cells after 20Gy IR. All data are normalized to controls, with controls shown in gray and Cap-H2 RNAi in red. p-values were calculated using Fisher’s exact test. (I) Stacked bar graphs showing the types and frequency of translocations in control and Cap-H2 RNAi cells after 20 Gy IR. n = 47 (control) and 56 (Cap-H2 RNAi) cells with translocations.
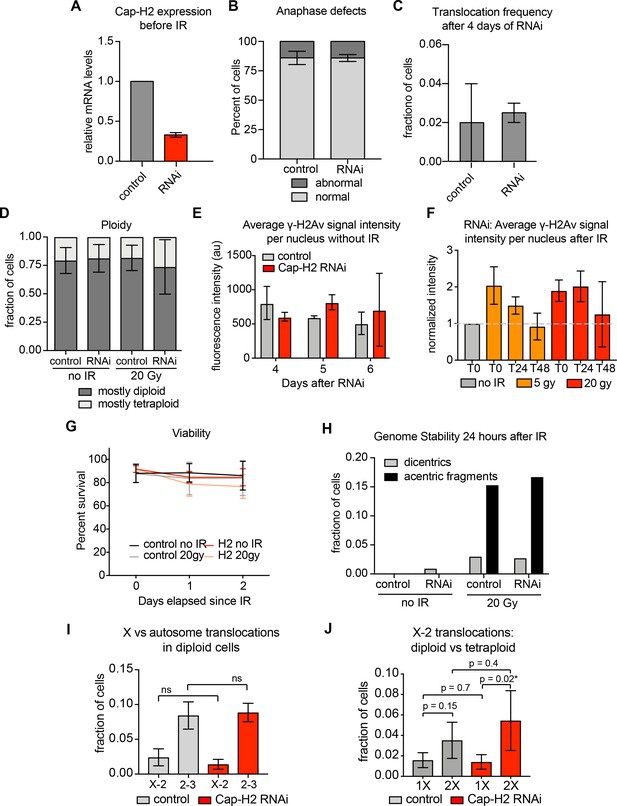
Additional data related to Figure 3.
(A) qPCR showing relative Cap-H2 mRNA levels in control or Cap-H2 RNAi cells 4 days after RNAi. Error bars show the standard deviation between technical qPCR replicates. (B) Bar graph showing percent of anaphase cells with defective chromosome segregation in control or RNAi cells. Error bars show standard deviation between biological replicates. p-values were determined by Fisher’s exact test comparing normal and abnormal anaphases in control versus RNAi cell populations. (C) Left: Bar graph showing the average translocation frequency after 4 days of control or Cap-H2 RNAi. Right: Bar graph showing the average translocation frequency after 17 days of control or Cap-H2 RNAi (four treatments). Error bars show SEM between biological replicates. (D) Bar graph showing percent of cells with mostly diploid (dark gray) or mostly tetaploid (light gray) karyotypes in control or Cap-H2 RNAi cells before and after 20 Gy IR. Error bars show standard deviation between biological replicates. (E) Quantification of anti-γ-H2Av fluorescence intensity per nucleus in Control and Cap-H2 RNAi cells after 4, 5, and 6 days of RNAi treatment. Error bars show standard deviation between biological replicates. (F) Quantification of anti-γ-H2Av fluorescence intensity in Cap-H2 RNAi cells before IR (T0), immediately after IR (T1), 24 hr (T24), and 48 hr (T48) either no IR (gray), 5 Gy IR (orange) or 20 Gy IR (red). Error bars show standard deviation between biological replicates. (G) Line graph showing average cell viability in control or Cap-H2 RNAi cells before and after 20 Gy IR, measured by trypan blue staining. Error bars show standard deviation between biological replicates. (H) Bar graph showing percent of cells with either dicentric chromosomes (gray bars) or acentric fragments (black bars) 24 hr after 20 Gy IR in control or Cap-H2 RNAi cells. (I) Bar graph showing fraction of diploid cells with translocations between chromosomes X-2 and 2–3 in control and Cap-H2 RNAi cells. (J) Bar graph showing fraction of Cap-H2-depleted cells harboring a translocation between chromosomes X and two in either diploid or tetraploid cells.
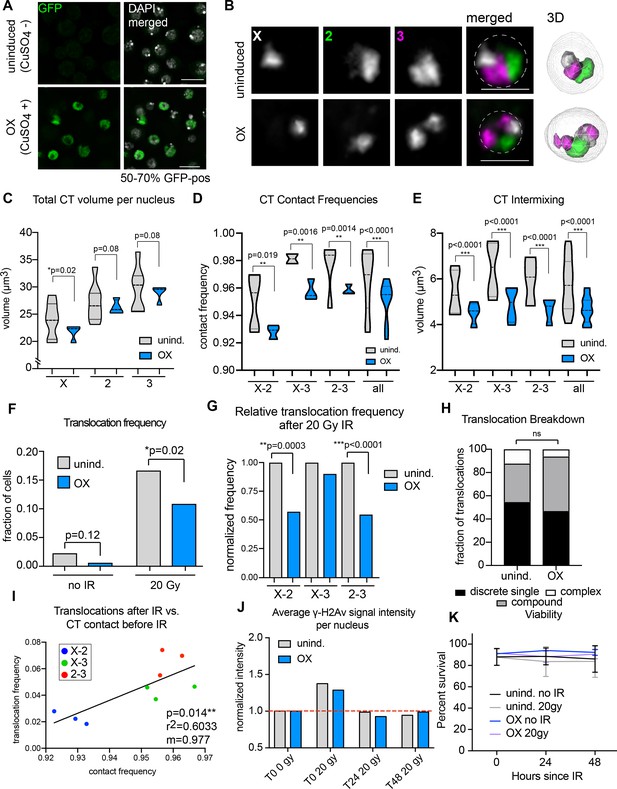
Increased Condensin II activity can attenuate the potential to form translocations in the presence of DNA damage.
(A) Immunofluorescence showing Cap-H2-GFP expression levels after induction (or uninduced, top row). DAPI is shown in gray. GFP is shown in green. Scale bar = 10 μm. (B) Left: representative nuclei with Oligopaints labeling chromosome X (white), 2 (green), and 3 (magenta) in control conditions (top), or after Cap-H2 overexpression (OX; bottom). Dotted line in merged image represents the nuclear edge. Scale bar = 5 μm. Right: 3D rendering of segmented chromosome structures. (C) Violin plot showing average CT volumes across three biological replicates, where n > 500 cells each, for control and Cap-H2 OX. P-values were calculated by Student’s t-test. (D) Violin plot showing CT contact frequencies for all CT pairs and combined across three biological replicates for control and Cap-H2 OX. p-values were determined by a Fisher’s Exact Test comparing contact and no contact for individual replicates. (E) Violin plot showing CT intermixing volumes for all CT pairs grouped together across three biological replicates for control and Cap-H2 OX. p-values were determined by Student’s t-test. (F) Total translocation frequency before or after 20Gy IR for control and Cap-H2 OX cells. p-values were calculated by Fisher’s exact test comparing normal karyotypes to those with translocations for control and OX. (G) Fold-change in translocation frequencies of control and Cap-H2 OX cells after 20Gy of IR. All data are normalized to controls, with uninduced controls being shown in gray and Cap-H2 OX shown in blue. P-values were calculated using Fisher’s exact test. (H) Stacked bar graphs showing the types and frequency of translocations in control and Cap-H2 OX cells after 20 Gy IR. n = 54 (control) and 63 (Cap-H2 OX) cells with translocations. p>0.5; calculated by Fisher’s exact test comparing control to OX for each category. (I) Scatterplot showing the translocation frequency of Cap-H2 OX cells after 20 Gy IR (Y-axis) versus CT contact frequencies before IR (X-axis). The data shown represent three biological replicates. m = slope of line of best fit. r2 and p values were calculated by linear regression. (J) Quantification of anti-γ-H2Av staining on control (gray) or Cap-H2 OX cells (blue) before IR (T0 0 Gy), immediately after 20 Gy IR (T0 20 Gy), 24 hr (T24 20 Gy), and 48 hr (T48 20 Gy). (K) Line graph showing average cell viability in control or Cap-H2 OX cells before and after 20 Gy IR, measured by trypan blue staining. Error bars show standard deviation between biological and technical replicates.
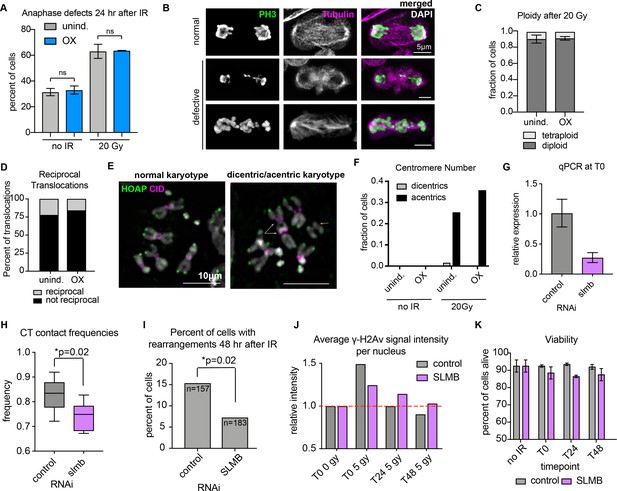
Additional data related to Figure 4.
(A) Bar graph showing percent of anaphase cells with defective chromosome segregation in control or OX cells before and 24 hr after IR with 20 Gy. Error bars show standard deviation between biological replicates. p>0.5; Fisher’s exact test comparing normal and abnormal anaphases in control and OX. (B) Representative IF images showing normal and abnormal anaphase cells. Anti-PH3S10 (mitotic marker) antibody is shown in green, and anti-tubulin in magenta. (C) Bar graph showing percent of cells with mostly diploid (dark gray) or mostly tetraploid (light gray) karyotypes in control or Cap-H2 OX cells. Error bars show standard deviation between biological replicates. (D) Stacked bar graphs showing the total fraction of translocations after 20 Gy IR that were reciprocal for 2 biological and 1 technical replicate (n=54-63 cells with translocations). (E) IF on metaphase chromosome spreads 24 hours after 20 Gy IR with anti-HOAP antibody (green) and anti-CID antibody (magenta), showing representative images of a normal karyotype (left) or karyotype with dicentric chromosomes (right; white arrows) and acentric fragments (right; yellow arrow). Scale bar equals 10 μm. (F) Quantification of IF shown in E, showing percent of cells with either dicentric chromosomes (gray bars) or acentric fragments (black bars) 24 hours after 20 Gy IR in control or Cap-H2 OX cells. (G) qPCR showing relative slmb mRNA levels in control or RNAi cells 4 days after RNAi treatment. Error bars show the standard deviation between technical qPCR replicates. (H) Tukey box plot showing CT contact frequencies for all CT pairs across 2 biological replicates for control and slmb RNAi. p-value was calculated by Mann-Whitney test to compare distributions. (I) Bar graph showing the total translocation frequency before or after 5Gy of irradiation for control and slmb RNAi cells. p-value was calculated by Fisher’s exact test comparing normal karyotypes to those with translocations for control and OX. (J) Quantification of anti-γ-H2Av fluorescence intensity on control or slmb RNAi cells before IR (T0 0 Gy), immediately after 5 Gy IR (T0 5 Gy), 24 hours (T24 5 Gy), and 48 hours (T48 5 Gy). These data represent one biological replicate, but were confirmed by a second biological replicate. (K) Bar graph showing average cell viability in control or slmb RNAi cells, measured by trypan blue staining. Error bars show standard deviation between 2 biological replicates.
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.49553.010