The Fml1-MHF complex suppresses inter-fork strand annealing in fission yeast
Figures
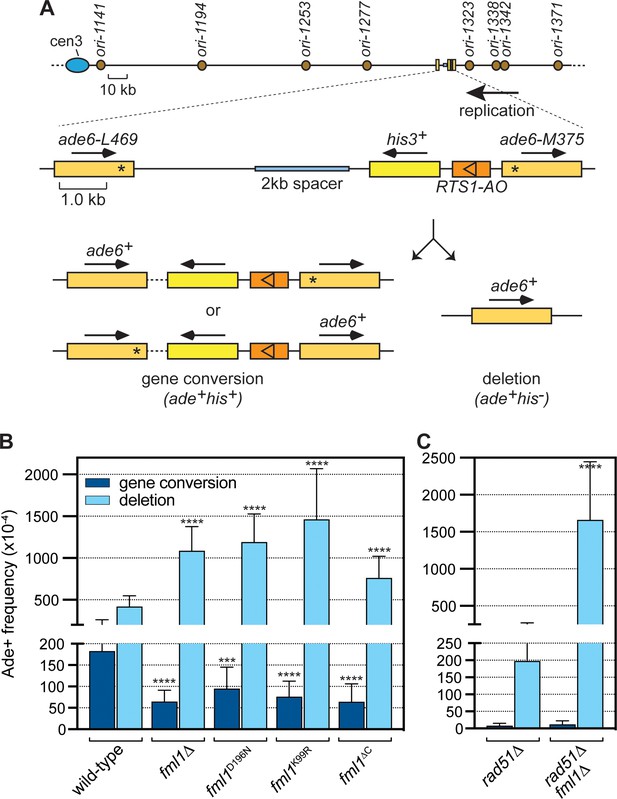
Comparison of SDD and gene conversion frequency in different fml1 mutant strains.
(A) Schematic of the direct repeat recombination reporter and its position on chromosome 3. Key features, including replication origins, prevailing direction of replication at the reporter, 2 kb spacer DNA and different types of Ade+ recombinant are indicated. Asterisks mark the position of point mutations in ade6-L469 and ade6-M375. (B) Ade+ recombinant frequencies for strains MCW8020 (wild-type), MCW8300 (fml1∆), MCW9268 (fml1D196N), MCW9281 (fml1K99R), and MCW9266 (fml1∆C). (C) Ade+ recombinant frequencies for strains MCW8296 (rad51∆) and MCW9496 (rad51∆ fml1∆). Data are mean values with error bars showing 1 SD. Significant differences relative to equivalent wild-type values are indicated by *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Ade+ recombinant frequencies with statistical analysis are also shown in Supplementary file 1.
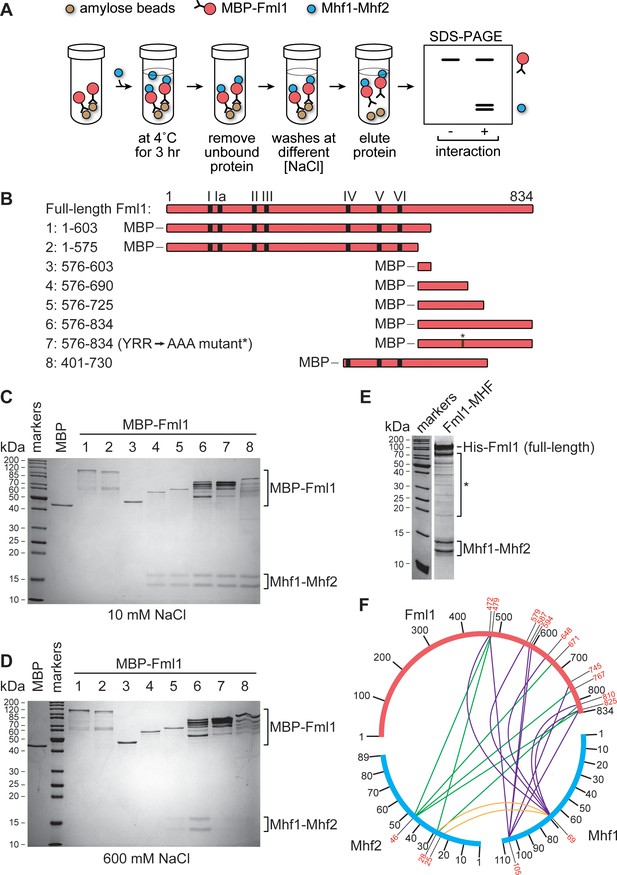
Mapping the region of Fml1 that interacts with Mhf1-Mhf2 (MHF).
(A) Experimental scheme for investigating the interaction between different MBP fused fragments of Fml1 and purified MHF. (B) Schematic of eight different Fml1 fragments tested for interaction with MHF. The terminal amino acids of each fragment are indicated by the numbers on the left-hand side of the panel. The position of the seven conserved helicase motifs (I – VI) in Fml1 and YRR to AAA mutation (*) are indicated. (C) Coomassie blue stained SDS-PAGE gel showing the results of pull-down experiments, with 10 mM NaCl washes, using the eight different MBP fused Fml1 fragments shown in B plus an MBP control. (D) The same as C except the washes were 600 mM NaCl. (E) Coomassie blue stained SDS-PAGE gel showing purified Fml1-MHF complex (4.3 µg). Truncated fragments of Fml1 and trace contaminant proteins are indicated by the asterisk. (F) Circos-like plot depicting crosslinking mass spectrometry analysis of the Fml1-MHF complex. Crosslinked lysines with a confidence score cut-off >15 are numbered in red.
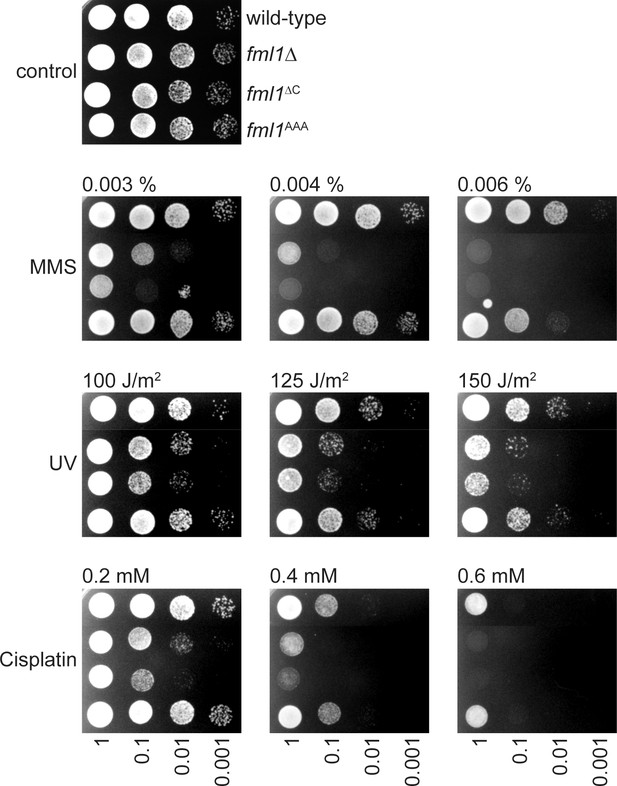
Spot assay comparing the genotoxin sensitivities of strains MCW1221 (wild-type), MCW2080 (fml1∆), MCW6923 (fml1∆C) and MCW6976 (fml1AAA).
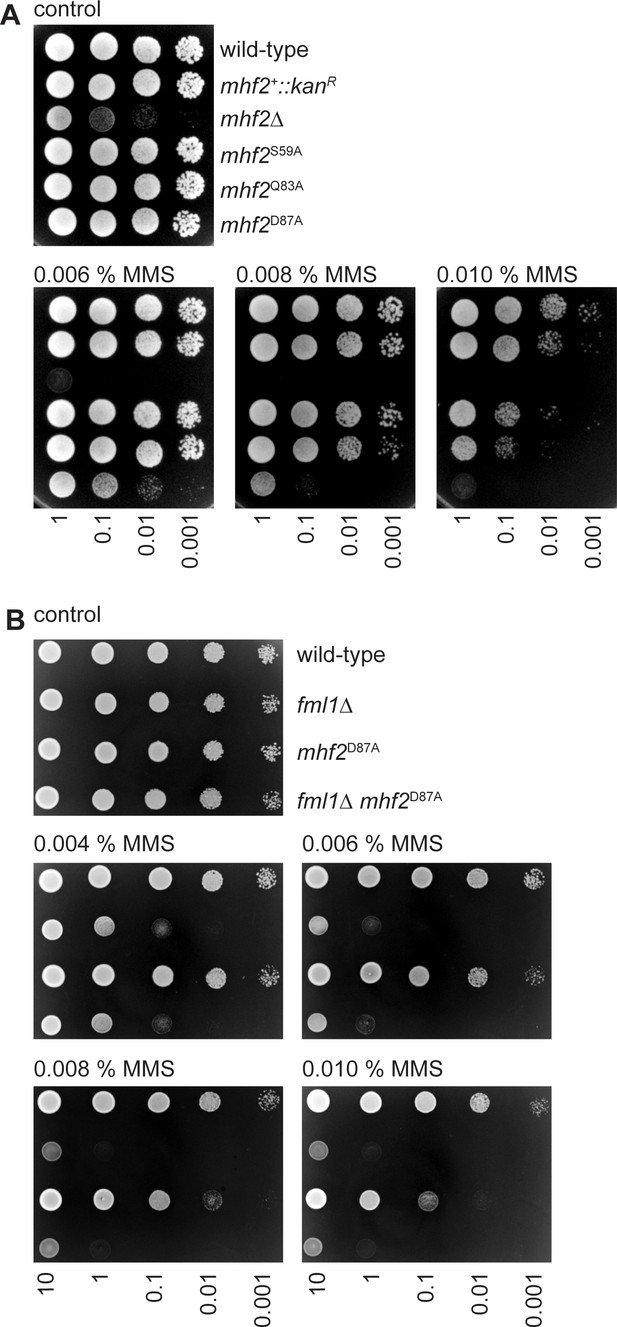
Spot assays comparing the MMS sensitivities of indicated mhf2 and fml1 mutant strains.
(A) The strains are MCW1221 (wild-type), MCW5677 (mhf2+::kanR), MCW5112 (mhf2∆), MCW6376 (mhf2S59A), MCW6379 (mhf2Q83A) and MCW6367 (mhf2D87A). (B) The strains are FO808 (wild-type), MCW4708 (fml1∆), MCW6319 (mhf2D87A) and MCW6570 (fml1∆ mhf2D87A). Note that the apparent difference in MMS sensitivity of the mhf2D87A strain in the experiments shown in panel A and B reflects differences in the potency of MMS between different batches of plates.
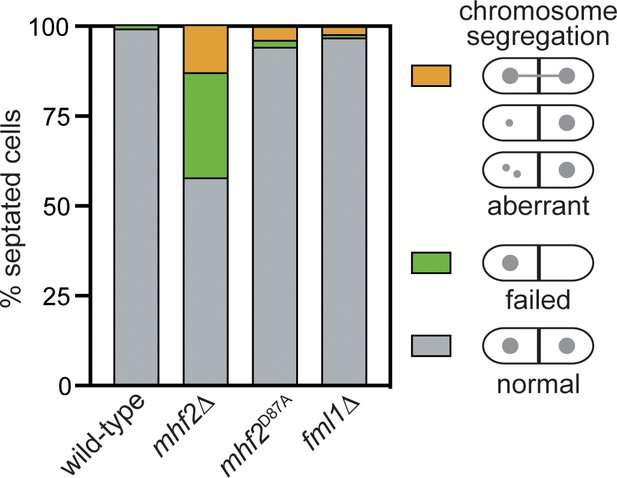
Analysis of chromosome segregation in septated cells.
Values are based on the analysis of approximately 300 septated cells from three independent asynchronously growing cultures incubated at 30 °C. The strains are FO808 (wild-type), MCW5112 (mhf2∆), MCW6319 (mhf2D87A) and MCW4708 (fml1∆).
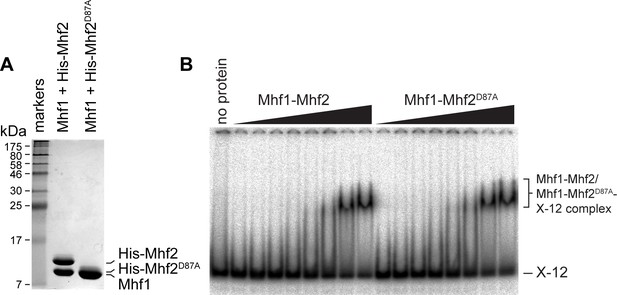
Comparison of DNA binding by Mhf1-Mhf2 and Mhf1-Mhf2D87A.
(A) Coomassie blue stained SDS-PAGE gel showing purified complex of Mhf1 plus His-tagged Mhf2 and Mhf1 plus His-tagged Mhf2D87A. Note that His-tagged Mhf2D87A co-migrates with Mhf1 unlike His-tagged Mhf2, which exhibits slower migration. Proteins were fully denatured by boiling in SDS loading dye containing ß-mercaptoethanol prior to running on the gel. Therefore, the difference in migration of His-tagged Mhf2D87A and His-tagged Mhf2 is not due to a difference in protein conformation. (B) EMSA showing binding of purified MHF complexes shown in A) to the synthetic Holliday junction X-12. Binding reactions contain 0.1 nM X-12 and increasing concentrations of MHF (100 nM, 200 nM, 500 nM, 1 µM, 2 µM, 4 µM, 8 µM and 10 µM).
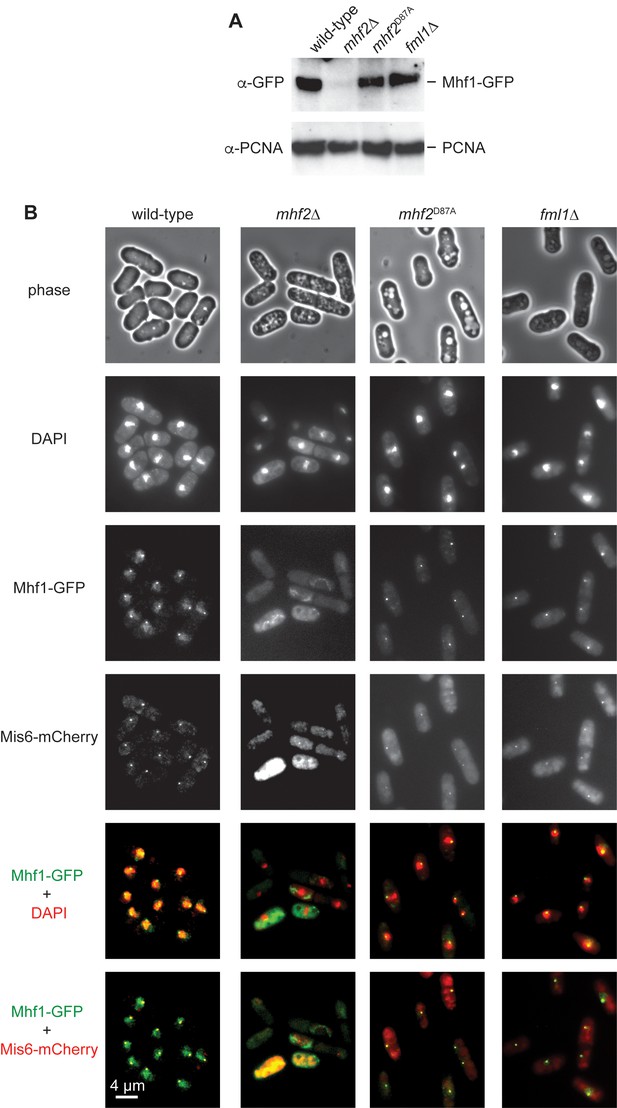
Analysis of Mhf1-GFP in wild-type, mhf2∆, mhf2D87A and fml1∆ cells.
(A) Western blot showing the relative amounts of Mhf1-GFP in whole cell lysates from strains MCW5846 (wild-type), MCW6845 (mhf2∆), MCW6884 (mhf2D87A) and MCW5963 (fml1∆). PCNA serves as a loading control. (B) Phase contrast and fluorescent microscopic images showing the nuclear localization of Mhf1-GFP in the same strains as in A. Note that the diffuse Mhf1-GFP fluorescence that extends beyond the DAPI staining in wild-type cells is most likely in the nucleolus.
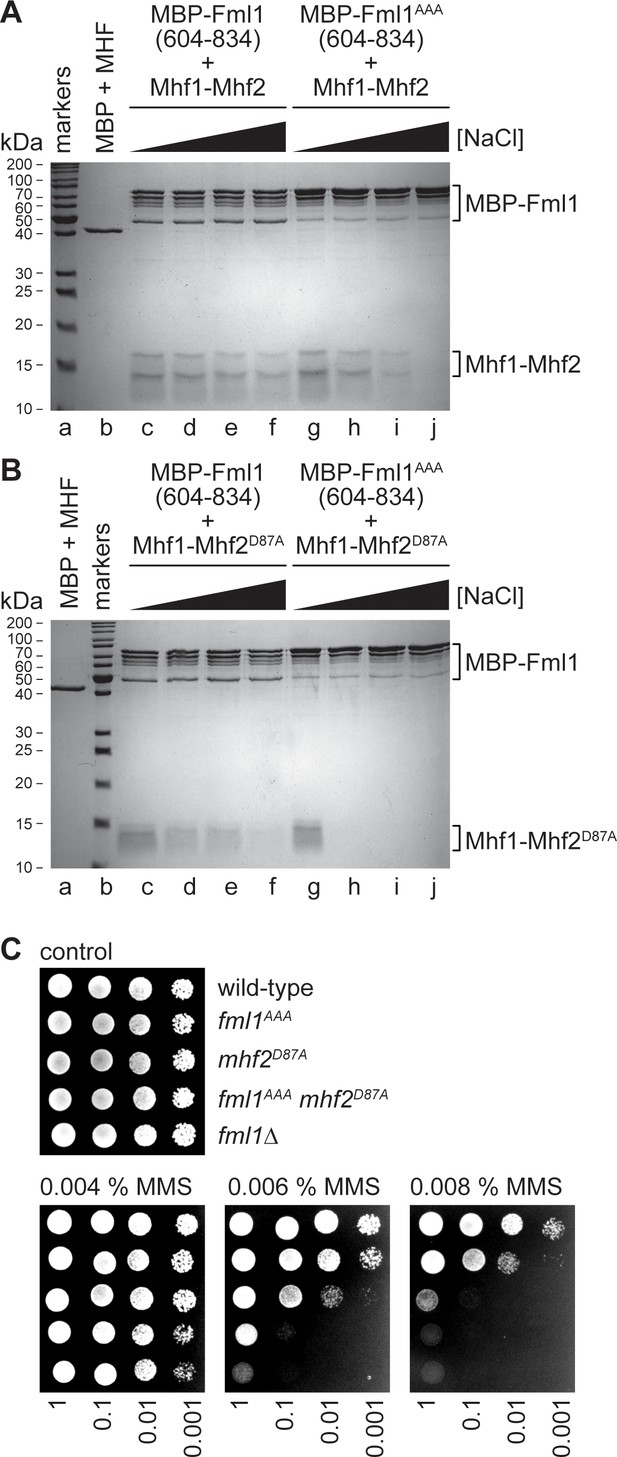
Effect of the AAA mutation in Fml1 and D87A mutation in Mhf2 on the interaction between Fml1 and MHF.
(A) Coomassie blue stained SDS-PAGE gel showing the results of pull-down experiments (see Figure 2A) using MBP-Fml1604-834/MBP-Fml1604-834AAA and Mhf1-Mhf2 performed with increasing NaCl concentration washes (50 mM, 200 mM, 300 mM and 600 mM). The result of a control pull-down experiment using purified MBP and Mhf1-Mhf2 at 10 mM NaCl is also included. (B) The same as in A) except Mhf1-Mhf2D87A was used instead of Mhf1-Mhf2. (C) Spot assay comparing the MMS sensitivities of strains MCW1221 (wild-type), MCW6976 (fml1AAA), MCW6321 (mhf2D87A), MCW7057 (fml1AAA mhf2D87A) and MCW2080 (fml1∆).
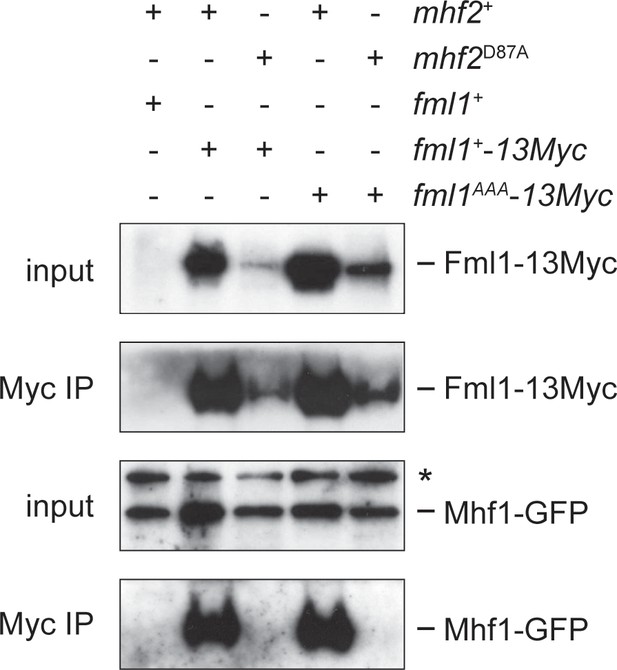
Co-immunoprecipitation of Fml1+−13Myc/Fml1AAA-13Myc and Mhf1-GFP from mhf2+ and mhf2D87A cells.
The strains used for this experiment were: MCW5846 (fml1+ mhf1+-GFP), MCW9616 (fml1+−13Myc mhf1+-GFP), MCW9593 (fml1+−13Myc mhf1+-GFP mhf2D87A), MCW9594 (fml1AAA-13Myc mhf1+-GFP) and MCW9595 (fml1AAA-13Myc mhf1+-GFP mhf2D87A). The asterisk indicates a non-specific band.
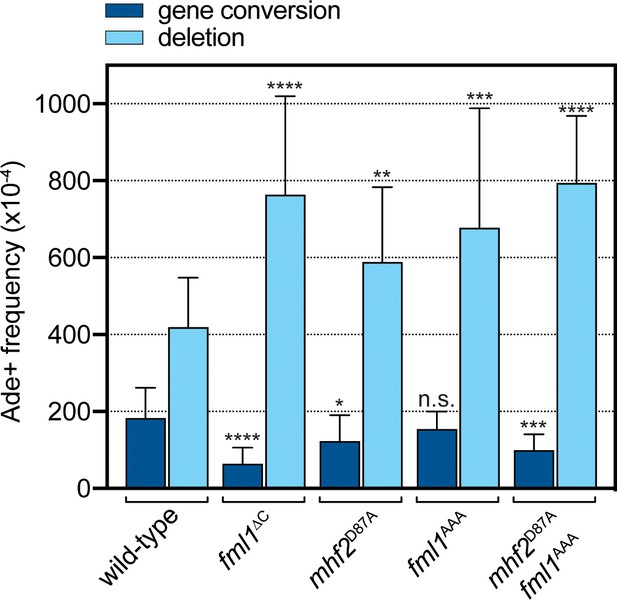
Effect of the AAA mutation in Fml1 and D87A mutation in Mhf2 on RTS1-induced recombination.
Comparison of SDD and gene conversion frequency in strains MCW8020 (wild-type), MCW9266 (fml1∆C), MCW9220 (mhf2D87A), MCW9283 (fml1AAA) and MCW9269 (fml1AAA mhf2D87A). Ade+ recombinant frequencies with statistical analysis are also shown in Supplementary file 1.
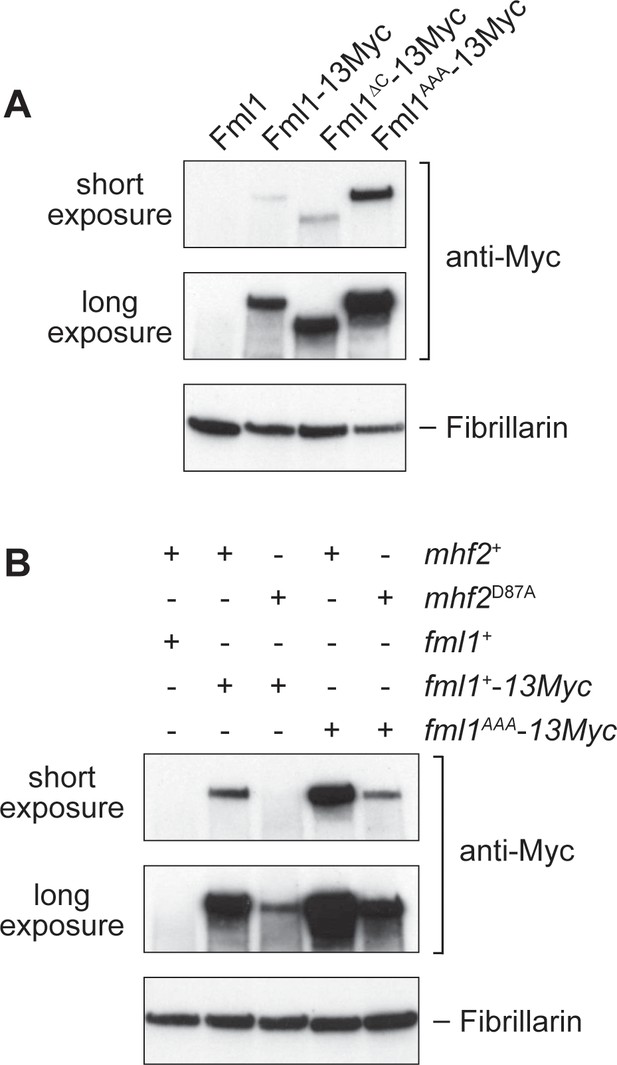
Effect of the AAA mutation in Fml1 and D87A mutation in Mhf2 on Fml1 stability.
(A) Western blot showing the relative amounts of Fml1+−13Myc, Fml1∆C-13Myc and Fml1AAA-13Myc in whole cell extracts from strains MCW1221 (fml1+), MCW4406 (fml1+−13Myc), MCW6977 (fml1∆C-13Myc), and MCW6980 (fml1AAA-13Myc). (B) Western blot showing the relative amounts of Fml1+−13Myc/Fml1AAA-13Myc in whole cell lysates from strains MCW1221 (fml1+ mhf2+), MCW9616 (fml1+−13Myc mhf2+), MCW9593 (fml1+−13Myc mhf2D87A), MCW9594 (fml1AAA-13Myc mhf2+) and MCW9595 (fml1AAA-13Myc mhf2D87A). Fibrillarin serves as a loading control.
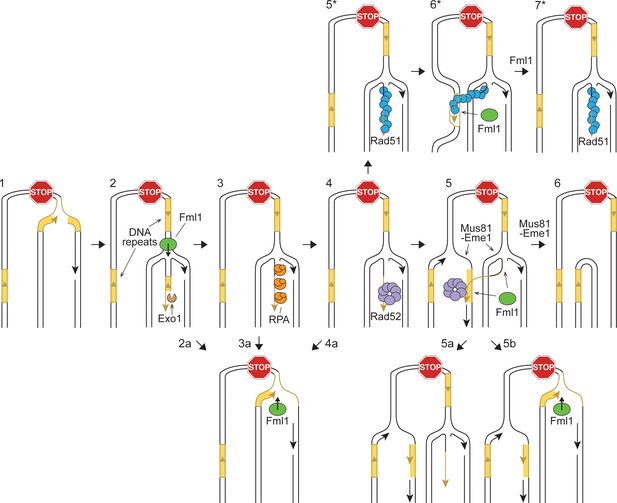
Hypothetical model showing potential ways in which Fml1 could promote gene conversions and suppress SDDs formed by recombination between DNA repeats at a collapsed replication fork.
Step 1: Replication fork collapses at RFB. Step 2: Replication fork regresses (Fml1 may help to catalyse this process) and Exo1 resects the exposed dsDNA end. Step 3: RPA binds to the ssDNA tail at the resected regressed fork. Step 4: Rad52 binds to the ssDNA tail at the resected regressed fork. Step 5: Rad52 anneals the ssDNA at the regressed fork to the lagging strand gap of the oncoming replication fork to form an ‘IFSA junction’. Step 6: Mus81-Eme1 cleaves the IFSA junction generating a SDD. Steps 2a-4a: Fml1 catalyses fork restoration limiting Rad52-mediated IFSA. Step 5a: Fml1 unwinds the IFSA junction. Alternatively, it may catalyse fork restoration like in Steps 2a-4a and in so-doing disrupt the IFSA junction (Step 5b). Step 5*: Rad52 mediates the loading of Rad51 onto the ssDNA tail at the regressed fork. Step 6*: Rad51 catalyses strand invasion of the second DNA repeat leading to the formation of a D-loop at which DNA synthesis is primed. Following limited extension of the invading strand by a DNA polymerase (not shown), the D-loop is unwound by Fml1 (Steps 6* - 7*). If the DNA copied during the strand invasion event is not identical in sequence to the first repeat, then fork restoration will lead to the formation of a heteroduplex DNA with the potential for gene conversion either by mismatch repair or by DNA replication (not shown).
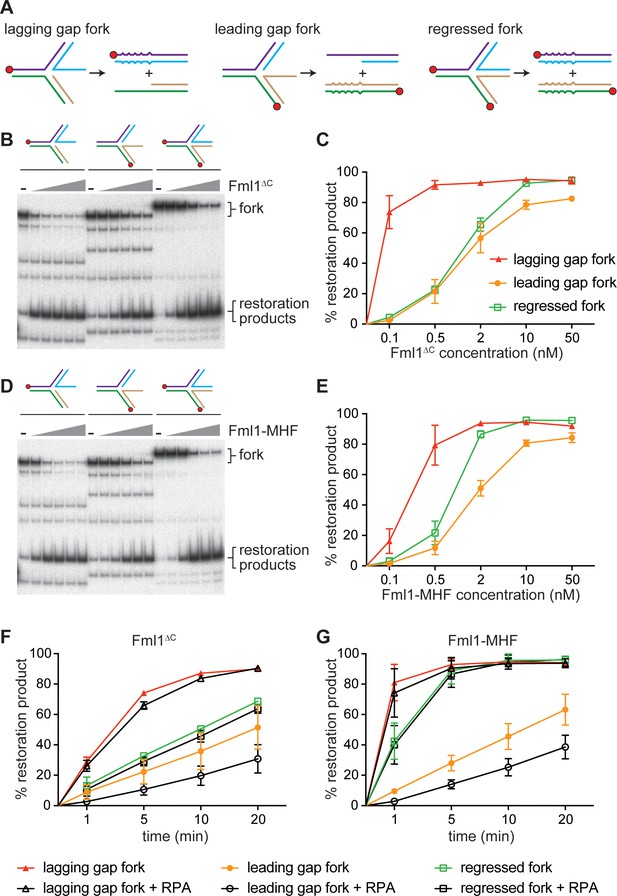
Replication fork restoration by Fml1∆C and Fml1-MHF.
(A) Schematic of the regressed fork substrates and reaction products. Component oligonucleotides are colour coded and red circles indicate 5’ 32P labels. Base pair mismatches in the reaction products are indicated by zigzagged lines. (B) Comparison of the restoration of the different regressed fork substrates shown in (A) by increasing concentrations of Fml1∆C. (C) Quantification of data like in B. (D) The same as (B) but with Fml1-MHF instead of Fml1∆C. (E) Quantification of data like in C. (F – G) Time course reactions for the restoration of the different regressed fork substrates shown in A by Fml1∆C (0.5 nM with lagging gap fork and 2 nM with leading gap fork and regressed fork) and Fml1-MHF (0.5 nM with lagging gap fork and 2 nM with leading gap fork and regressed fork) in the presence and absence of RPA (20 nM). Data in (C) and (E – G) are mean values + /- SD from three independent experiments.
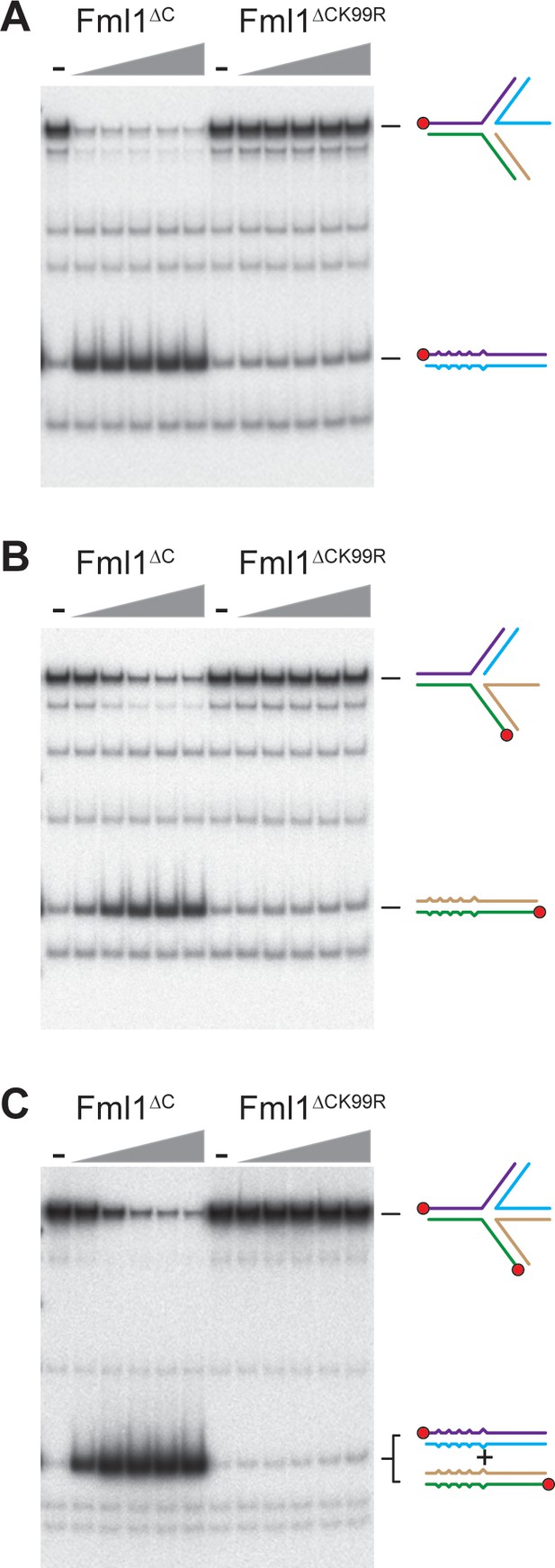
Restoration of regressed fork substrates by Fml1 requires its ATPase activity.
(A – C) Comparison of fork restoration of (A) a lagging gap fork, (B) a leading gap fork and (C) a regressed fork by increasing concentrations (0.1 nM, 0.5 nM, 2 nM, 10 nM and 50 nM) of Fml1∆C and Fml1∆C K99R.
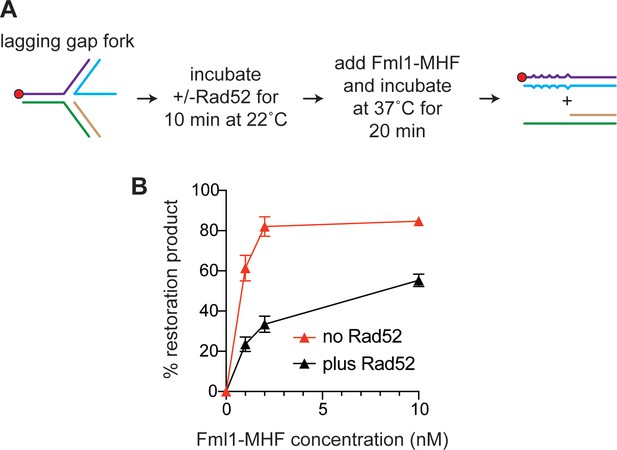
Effect of Rad52 on lagging gap fork restoration by Fml1-MHF.
(A) Schematic outlining the order of addition of Rad52 and Fml1-MHF. (B) Graph showing the inhibitory effect of Rad52 (210 nM) on lagging gap fork restoration by different concentrations of Fml1-MHF. Data are mean values + /- SD from three independent experiments.
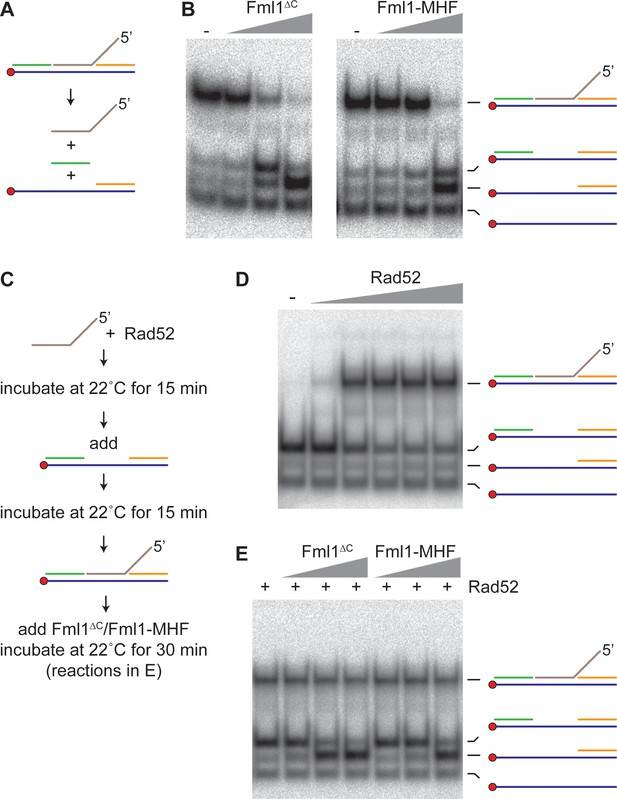
Effect of Rad52 on 5’ flap unwinding by Fml1∆C and Fml1-MHF.
(A) Schematic of the 5’ flap DNA substrate and Fml1∆C/Fml1-MHF reaction products. Component oligonucleotides are colour coded and the red circle indicates the 5’ 32P label. (B) 5’ flap (0.1 nM) unwinding by increasing concentrations (0.7 nM, 7 nM and 70 nM) of Fml1∆C/Fml1-MHF. (C) Reaction scheme for Rad52 annealing reactions with and without Fml1∆C/Fml1-MHF. (D) Annealing of 5’ flap DNA strand (0.1 nM) into a ssDNA gap (0.1 nM) by increasing concentrations (20 nM, 50 nM, 100 nM, 200 nM and 310 nM) of Rad52. (E) Rad52 (150 nM) catalysed annealing of 5’ flap DNA strand (0.1 nM) into a ssDNA gap (0.1 nM) plus addition of Fml1∆C/Fml1-MHF (0.7 nM, 7 nM and 70 nM) following the reaction scheme in C.
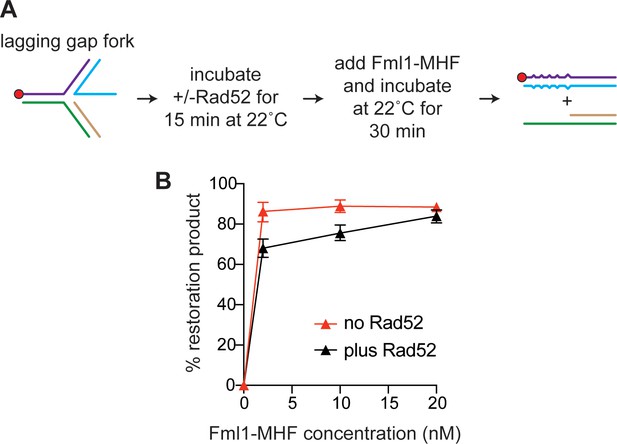
Effect of Rad52 on lagging gap fork restoration by Fml1-MHF under similar reaction conditions as used in the experiment in Figure 9E.
(A) Schematic showing the order of addition of Rad52 and Fml1-MHF. (B) Graph showing the inhibitory effect of Rad52 (210 nM) on lagging gap fork restoration by different concentrations of Fml1-MHF. Data are mean values + /- SE from two independent experiments.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (S. pombe) | MCW8020 | PMID: 28586299 | standard laboratory strain (972) derivatives; seeSupplementary file 2 | |
| Strain, strain background (S. pombe) | MCW8300 | PMID: 28586299 | standard laboratory strain (972) derivatives; seeSupplementary file 2 | |
| Strain, strain background (S. pombe) | MCW2080 | PMID: 18851838 | standard laboratory strain (972) derivatives; seeSupplementary file 2 | |
| Strain, strain background (S. pombe) | MCW5846, MCW5963 and MCW4406 | PMID: 24026537 | standard laboratory strain (972) derivatives; seeSupplementary file 2 | |
| Strain, strain background (S. pombe) | various strains | this paper | standard laboratory strain (972) derivatives; seeSupplementary file 2 | |
| Antibody | anti-GFP (Mouse monoclonal) | Clontech | RRID: AB_2313808 | dilution used (1:1000) |
| Antibody | anti-PCNA (Mouse monoclonal) | Santa Cruz Biotechnology | RRID: AB_628110 | dilution used (1:1000) |
| Antibody | anti-Myc tag (Goat polyclonal) | Abcam | RRID: AB_307033 | dilution used (1:10000) |
| Antibody | anti-Fibrillarin (38F3) (Mouse monoclonal) | Novus Biologicals | RRID: AB_2100980 | dilution used (1:1000) |
| Antibody | Myc-Trap Magnetic Agarose | Chromotek | RRID: AB_2631370 | amount used (20 µl) |
| Recombinant DNA reagent | various plasmids | this paper | plasmid; see Materials and methods | |
| Recombinant DNA reagent | pSN3 | PMID: 18851838 | plasmid; see Materials and methods | |
| Recombinant DNA reagent | pMW891 | PMID: 24026537 | plasmid; see Materials and methods | |
| Recombinant DNA reagent | pET19b-SpSSB | PMID: 8702843 | plasmid; see Materials and methods | |
| Recombinant DNA reagent | pMW601 | PMID: 15486206 | plasmid; see Materials and methods | |
| Sequence-based reagent | various oligonucleotides | this paper | oligonucleotide; see Materials and methods | |
| Sequence-based reagent | regressed fork substrate oligonucleotides | PMID: 23746452 | oligonucleotide; see Materials and methods | |
| Sequence-based reagent | X-12 substrate oligonucleotides | PMID: 9857040 | oligonucleotide; see Materials and methods | |
| Peptide, recombinant protein | Fml1∆C1-603 | this study | recombinant protein; see Materials and methods | |
| Peptide, recombinant protein | Fml1-MHF | this study | recombinant protein; see Materials and methods | |
| Peptide, recombinant protein | Mhf1-Mhf2 | this study | recombinant protein; see Materials and methods | |
| Peptide, recombinant protein | Mhf1-Mhf2D87A | this study | recombinant protein; see Materials and methods | |
| Peptide, recombinant protein | RPA | this study | recombinant protein; see Materials and methods | |
| Peptide, recombinant protein | Rad52 | this study | recombinant protein; see Materials and methods |
Additional files
-
Supplementary file 1
Direct repeat recombinant frequencies.
- https://cdn.elifesciences.org/articles/49784/elife-49784-supp1-v2.docx
-
Supplementary file 2
Schizosaccharomyces pombe strains.
- https://cdn.elifesciences.org/articles/49784/elife-49784-supp2-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/49784/elife-49784-transrepform-v2.docx





