Drosophila PSI controls circadian period and the phase of circadian behavior under temperature cycle via tim splicing
Figures
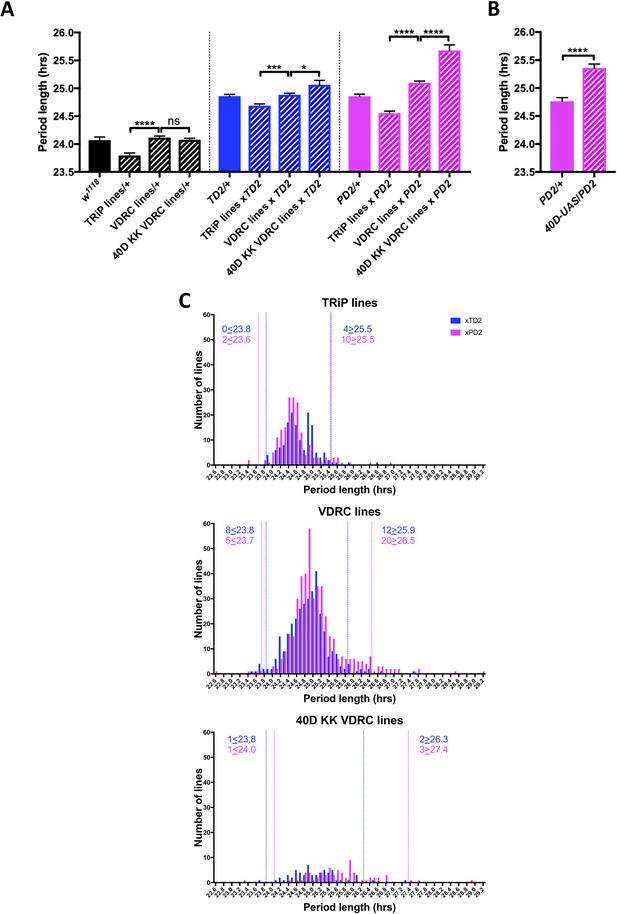
An RNAi screen of RNA associated proteins identifies long and short period hits.
(A–B) Background effect of TRiP and VDRC collections on circadian period length. Circadian period length (hrs) is plotted on the y axis. RNAi collection and genotypes are labeled. Error bars represent SEM. (A) Left group (black bars): Patterned bars are the average of period lengths of a subset of RNAi lines in the screen crossed to w1118 (TRiP/+ N = 17 crosses, VDRC/+ N = 46 crosses, 40D KK VDRC/+ N = 20 crosses). Solid bar is the w1118 control (N = 20 crosses). Middle group (blue bars): Patterned bars are the average of period lengths of all RNAi lines in the screen crossed to tim-GAL4, UAS-Dicer2 (TD2) (TRiP/TD2 N = 151 crosses, VDRC/TD2 N = 340 crosses, 40D KK VDRC/TD2 N = 61 crosses). Solid bar is the TD2/+ control (N = 35 crosses). Right group (magenta bars): Patterned bars are the average of period lengths of all RNAi lines in the screen crossed to Pdf-GAL4, UAS-Dicer2 (PD2) (TRiP/PD2 N = 176 crosses, VDRC/PD2 N = 448 crosses, 40D KK VDRC/PD2 N = 69 crosses). Solid bar is the PD2/+ control (N = 36 crosses). One-way ANOVA followed by Tukey’s multiple comparison test: *p<0.05, ***p<0.001, ****p<0.0001. Note that the overall period lengthening, relative to wild-type (w1118), when RNAi lines are crossed to TD2 or PD2 is a background effect of our drivers (see main text), while the period differences between the TRiP (shorter) and VDRC (longer) collections is most likely a background effect of the RNAi lines themselves. There is also a lengthening effect of the 40D insertion site in the VDRC KK collection that cannot be explained by a background effect, as it is not present in the RNAi controls (Left panel). Instead the lengthening was only observed when these lines were crossed to our drivers. A modest effect was seen with TD2 (middle panel) and a larger effect was seen with PD2 (right panel). (B) The period lengthening effect of the VDRC 40D KK lines is likely due to overexpression of tio, as we observed lengthening when a control line that lacks a RNAi transgene, but still has a UAS insertion in the 40D (40D-UAS) locus was crossed to PD2. N = 32 flies per genotype, ****p<0.0001, Unpaired Student’s t-test. (C) Histogram of period lengths obtained in the initial round of screening. Number of lines per bin is on the y axis. Binned period length (hrs) is on the x axis. Bin size is 0.1 hr. TD2 crosses are in blue and PD2 crosses are in magenta. Dashed lines indicate our cutoff of 2 standard deviations from the mean. Number of crosses that fell above or below the cutoff is indicated. Top panel: TRiP lines. 0 lines crossed to TD2 and 2 lines crossed to PD2 gave rise to short periods and were selected for repeats. four lines crossed to TD2 and 10 lines crossed to PD2 gave rise to long periods and were selected for repeats. Middle panel: VDRC lines. eight lines crossed to TD2 and 5 lines crossed to PD2 gave rise to short periods and were selected for repeats. 12 lines crossed to TD2 and 20 lines crossed to PD2 gave rise to long periods and were selected for repeats. Bottom panel: VDRC 40D KK lines. one line crossed to TD2 and 1 line crossed to PD2 gave rise to short periods and were selected for repeats. two lines crossed to TD2 and 3 lines crossed to PD2 gave rise to long periods and were selected for repeats.
-
Figure 1—source data 1
40D insertion control – behavior data.
- https://cdn.elifesciences.org/articles/50063/elife-50063-fig1-data1-v2.xlsx
-
Figure 1—source data 2
Figure statistics – Figure 1.
- https://cdn.elifesciences.org/articles/50063/elife-50063-fig1-data2-v2.xlsx
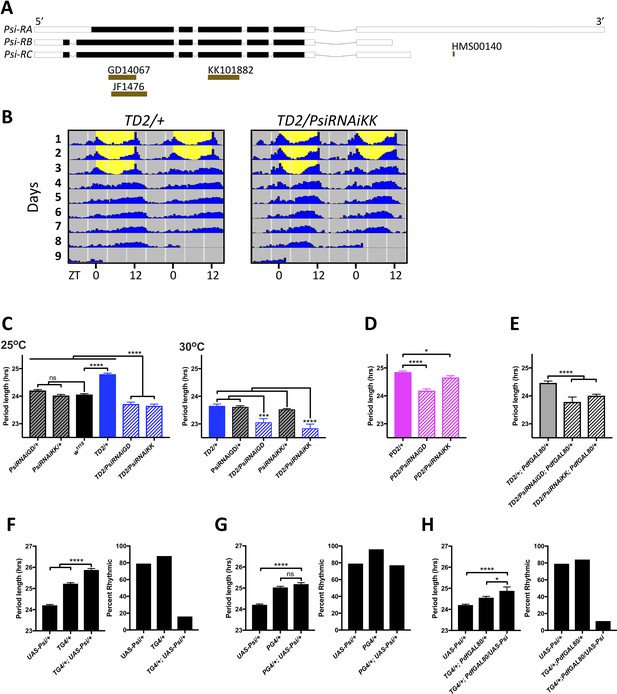
Expression level of Psi affects the circadian behavior period length and circadian rhythmicity.
(A) Schematic of Psi isoforms and position of the long and short hairpins used in this study. Adapted from Ensembl 94 (Zerbino et al., 2018). (B–E) Knockdown of Psi shortens the behavioral period. (B) Double-plotted actograms showing the average activities during 3 days in LD and 5 days in DD. Left panel: TD2/+ (control) flies. Right panel: TD2/PsiRNAi (Psi knockdown) flies. Note the short period of Psi knockdown flies. n = 8 flies/genotype. (C–E) Circadian period length (hrs) is plotted on the y axis. Genotypes are listed on the x axis. Error bars represent SEM. Solid black bar is w1118 (WT) control; solid blue, magenta and gray bars are driver controls; patterned bars are Psi knockdown with two non-overlapping RNAi lines: GD14067 (PsiRNAiGD) and KK101882 (PsiRNAiKK). *p<0.05, ***p<0.001, ****p<0.0001, one-way ANOVA followed by Tukey’s multiple comparison test (C) Dunnett’s multiple comparison test (D and E). (C) Knockdown in all circadian tissues. Left panel 25°C, right panel 30°C. Note that even at 25°C, the experimental flies are shorter than their respective RNAi/+ control, despite the dominant period lengthening caused by TD2 (D) Knockdown in PDF+ circadian pacemaker neurons. (E) Knockdown in PDF- circadian tissues. In D and E, only the driver controls are shown, since they are the controls which the experimental flies need to be compared to because of the dominant period lengthening caused by PD2 and TD2. (F–H) Overexpression of Psi lengthens the behavioral period and decreases rhythmicity. Left panels: Circadian period length (hrs) is plotted on the y axis. Error bars represent SEM. Right panels: Percent of flies that remained rhythmic in DD is plotted on the y axis. Both panels: Genotypes are listed on the x axis. Not significant (ns)p>0.05, *p<0.05, ****p<0.0001, one-way ANOVA followed by Tukey’s multiple comparison test. (F) Overexpression of Psi in all circadian tissues lengthened the circadian period and decreased the percent of rhythmic flies. (G) Overexpression of Psi in PDF+ circadian pacemaker neurons caused a slight but non-significant period lengthening compared to the driver control (PG4/+), which is the relevant comparison because of the dominant period lengthening caused by PG4. Rhythmicity was slightly reduced compared to PG4/+ but not compared to UAS-Psi/+. (H) Overexpression of Psi in PDF- circadian tissues lengthened the circadian period and decreased rhythmicity.
-
Figure 2—source data 1
Psi downregulation and overexpression – behavior data.
- https://cdn.elifesciences.org/articles/50063/elife-50063-fig2-data1-v2.xlsx
-
Figure 2—source data 2
Figure statistics – Figure 2.
- https://cdn.elifesciences.org/articles/50063/elife-50063-fig2-data2-v2.xlsx
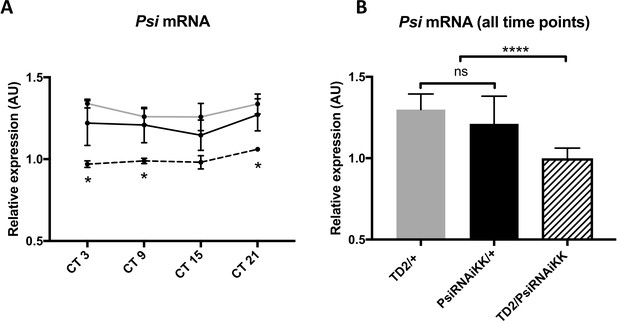
Psi mRNA expression does not cycle and its level is reduced in heads of Psi knockdown flies.
(A) Psi mRNA expression does not cycle in DD. Relative expression of Psi mRNA (normalized to the average of all Psi knockdown time points) in heads on the y axis measured by qPCR. Circadian time (CT) on the x axis. Error bars represent SEM. Gray line: driver control. Black line: RNAi control. Dashed line: Psi knockdown. Controls, N = 3. Psi knockdown, N = 5. Both driver and RNAi control relative to Psi knockdown, two-way ANOVA followed by Tukey’s multiple comparison test: *p<0.05. (B) Knockdown of Psi with RNAiKK causes a significant reduction in Psi mRNA levels relative to both driver and RNAi controls. Since no cycling of Psi was observed, all time points were pooled to increase statistical strength. Relative expression of Psi mRNA (normalized to the average of all Psi knockdown time points) in heads on the y axis measured by qPCR. Genotypes are on the x axis. Error bars represent SEM. Gray bar: driver control. Black bar: RNAi control. Patterned bar: Psi knockdown. Both driver and RNAi control relative to Psi knockdown, one-way ANOVA followed by Tukey’s multiple comparison test: ****p<0.0001.
-
Figure 2—figure supplement 1—source data 1
Psi qPCR data.
- https://cdn.elifesciences.org/articles/50063/elife-50063-fig2-figsupp1-data1-v2.xlsx
-
Figure 2—figure supplement 1—source data 2
Figure statistics – Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/50063/elife-50063-fig2-figsupp1-data2-v2.xlsx
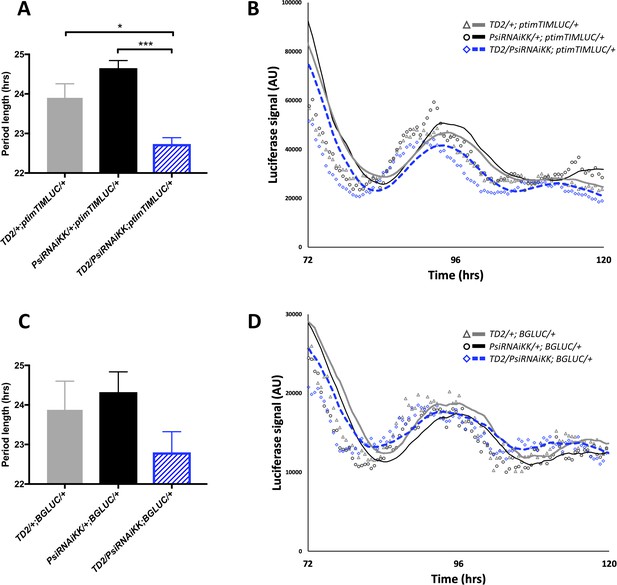
Knockdown of Psi shortens circadian period of PER and TIM rhythms in peripheral tissues.
(A) Period length (hrs) of light output generated from luciferase rhythms of ptim-TIM-LUC in whole flies. 9–24 wells/run (with one exception for control genotype PsiRNAiKK/+; ptimTIMLUC/+), three flies/well. N = 6 runs. *p<0.05, ***p<0.001, one-way ANOVA followed by Tukey’s multiple comparison test. Error bars represent SEM. Gray bar: driver control. Black bar: RNAi control. Blue patterned bar: Psi knockdown. (B) Representative traces from (A) Markers are raw data and lines are 6 hr moving averages. Gray marker (triangle) and gray line: driver control. Black marker (circle) and black line: RNAi control. Blue marker (diamond) and blue dashed line: Psi knockdown. Luciferase signal (arbitrary units, AU) on the y axis and time (hrs) from start of experiment on the x axis. 72 hr = start of DD. (C) Period length (hrs) of average light output generated from luciferase rhythms of BG-LUC in whole flies. 12–30 wells/run, three flies/well. N = 4 runs. Error bars represent SEM. Gray bar: driver control. Black bar: RNAi control. Blue patterned bar: Psi knockdown. (D) Representative traces from (C) Markers are raw data and lines are 6 hr moving averages. Gray marker (triangle) and gray line: driver control. Black marker (circle) and black line: RNAi control. Blue marker (diamond) and blue dashed line: Psi knockdown. Luciferase signal (arbitrary units, AU) on the y axis and time (hrs) from start of experiment on the x axis. 72 hr = start of DD.
-
Figure 2—figure supplement 2—source data 1
TIMLUC signal.
- https://cdn.elifesciences.org/articles/50063/elife-50063-fig2-figsupp2-data1-v2.xlsx
-
Figure 2—figure supplement 2—source data 2
BGLUC signal.
- https://cdn.elifesciences.org/articles/50063/elife-50063-fig2-figsupp2-data2-v2.xlsx
-
Figure 2—figure supplement 2—source data 3
Figure statistics – Figure 2—figure supplement 2.
- https://cdn.elifesciences.org/articles/50063/elife-50063-fig2-figsupp2-data3-v2.xlsx
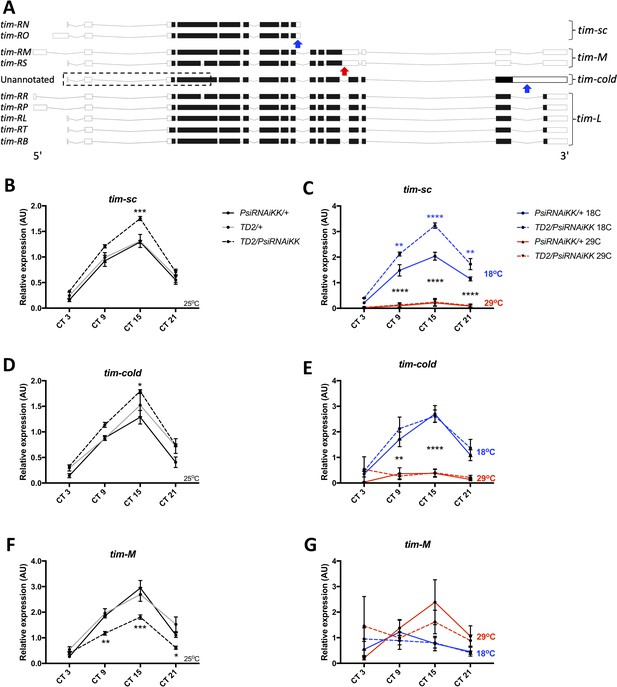
Knockdown of Psi increases the expression of cold induced tim isoforms and decreases the expression of a warm induced tim isoform.
(A) Schematic of tim isoforms. Flybase transcript nomenclature on left, intron retention events studied here on right (tim-L refers to tim transcripts that do not produce C-terminal truncations of TIM via intron retention). Arrows indicate the location of retained introns: blue, upregulated at cold temperature; red, upregulated at warm temperature. The retained intron that gives rise to the tim-cold isoform is not annotated in Flybase (Thurmond et al., 2019). It is possible that multiple tim-cold transcripts may exist due to alternative splicing and alternative transcription/translation start sites in the 5’ region of the gene (dashed box). However, for simplicity, we depict this region of tim-cold using the most common exons. Adapted from Ensembl 94 (Zerbino et al., 2018). (B, D, F) Relative expression of tim mRNA isoforms at 25°C (normalized to the average of all Psi knockdown time points) in heads on the y axis measured by qPCR. Circadian time (CT) on the x axis. Error bars represent SEM. Gray line: driver control. Black line: RNAi control. Dashed line: Psi knockdown. Controls, N = 3. Psi knockdown, N = 5 (3 technical replicates per sample). Both driver and RNAi control compared to Psi knockdown, two-way ANOVA followed by Tukey’s multiple comparison test: *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. (C, E, G) Relative expression of tim mRNA isoforms at 18°C and 29°C (normalized to the average of all Psi knockdown time points). Solid line: RNAi control. Dashed line: Psi RNAi knockdown. Blue indicates flies were transferred to 18°C at CT0 (start of subjective day) on the first day of DD. Red indicates flies were transferred to 29°C. N = 3 (3 technical replicates per sample). 18°C samples compared to 29°C samples, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, two-way ANOVA followed by Tukey’s multiple comparison test. (C) Blue asterisks refer to RNAi control compared to Psi knockdown.
-
Figure 3—source data 1
tim qPCR data.
- https://cdn.elifesciences.org/articles/50063/elife-50063-fig3-data1-v2.xlsx
-
Figure 3—source data 2
Figure statistics – Figure 3.
- https://cdn.elifesciences.org/articles/50063/elife-50063-fig3-data2-v2.xlsx
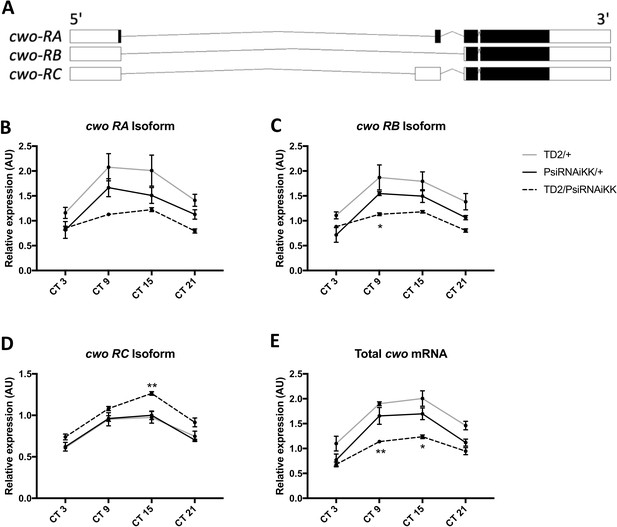
Knockdown of Psi affects the balance of cwo isoform expression.
(A) Schematic of cwo isoforms. Adapted from Ensembl 94 (Zerbino et al., 2018). (B–D) Relative expression of cwo mRNA isoforms (normalized to the average of all Psi knockdown time points) in heads on the y axis measured by qPCR. (E) Relative expression of total cwo mRNA on the y axis. (B–E) Circadian time (CT) on the x axis. Error bars represent SEM. Gray line: driver control. Black line: RNAi control. Dashed line: Psi knockdown. Driver control, N = 3. RNAi control, N = 4. Psi knockdown, N = 6 (3 technical replicates per sample). Both driver and RNAi control compared to Psi knockdown, two-way ANOVA followed by Tukey’s multiple comparison test: *p<0.05, **p<0.01.
-
Figure 3—figure supplement 1—source data 1
cwo qPCR data.
- https://cdn.elifesciences.org/articles/50063/elife-50063-fig3-figsupp1-data1-v2.xlsx
-
Figure 3—figure supplement 1—source data 2
Figure statistics – Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/50063/elife-50063-fig3-figsupp1-data2-v2.xlsx
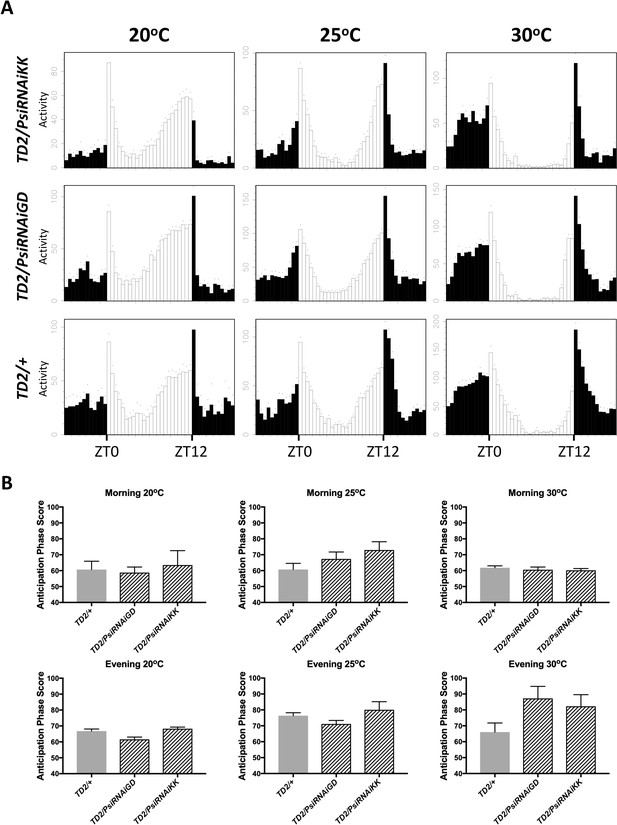
Psi knockdown flies have normal behavioral adaptation to temperature.
(A) Eductions showing the average activity of flies during 3 days of LD (days 2–4). Left panels: flies were entrained at 20°C. Center panels: flies were entrained at 25°C. Right panels: flies were entrained at 30°C. Top panels: TD2/PsiRNAiKK (Psi knockdown) flies. Middle panels: TD2/PsiRNAiGD (Psi knockdown) flies. Bottom panels: TD2/+ (control) flies. Note that, similar to the TD2/+ control, Psi knockdown flies advance the phase of their evening activity at 20°C and delay the phase of their evening activity at 30°C. Psi knockdown flies also show reduced morning activity and increased evening activity at 20°C, and increased morning activity and decreased evening activity at 30°C, similar to the TD2/+ control. (B) Quantification of the morning and evening anticipation phase score indicates that the phase of behavior in LD (day 2–3) is not affected by knockdown of Psi. Genotypes are on the x axis. Error bars represent SEM. Gray bar: driver control. Patterned bars: Psi knockdown. One-way ANOVA followed by Tukey’s multiple comparison test: p>0.05 for all comparisons. N = 3–5 runs (6–16 flies per genotype in each run).
-
Figure 3—figure supplement 2—source data 1
Psi downregulation – anticipation phase.
- https://cdn.elifesciences.org/articles/50063/elife-50063-fig3-figsupp2-data1-v2.xlsx
-
Figure 3—figure supplement 2—source data 2
Figure statistics – Figure 3—figure supplement 2.
- https://cdn.elifesciences.org/articles/50063/elife-50063-fig3-figsupp2-data2-v2.xlsx
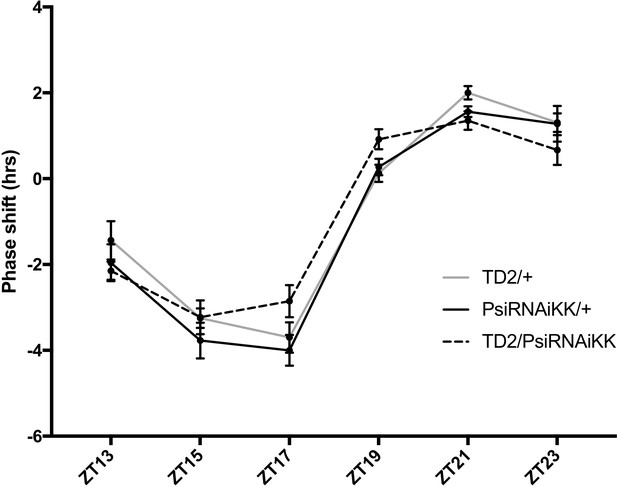
Psi knockdown flies have a normal photic phase response.
Behavioral phase response curve to brief 5 min 1500 lux light pulses. Behavioral phase shifts are on the y-axis. The time of the light pulse administration is on the x-axis. N = 4 for all time points except ZT23 where N = 3. For each genotype, 16 flies per timepoint were tested in each run. No significant effect of genotype was detected, two-way ANOVA. Note that the phase of the Psi knockdown curve is slightly shifted to the left, which probably reflects the short period of Psi knockdown flies.
-
Figure 3—figure supplement 3—source data 1
Psi downregulation – PRC.
- https://cdn.elifesciences.org/articles/50063/elife-50063-fig3-figsupp3-data1-v2.xlsx
-
Figure 3—figure supplement 3—source data 2
Figure statistics – Figure 3—figure supplement 3.
- https://cdn.elifesciences.org/articles/50063/elife-50063-fig3-figsupp3-data2-v2.xlsx
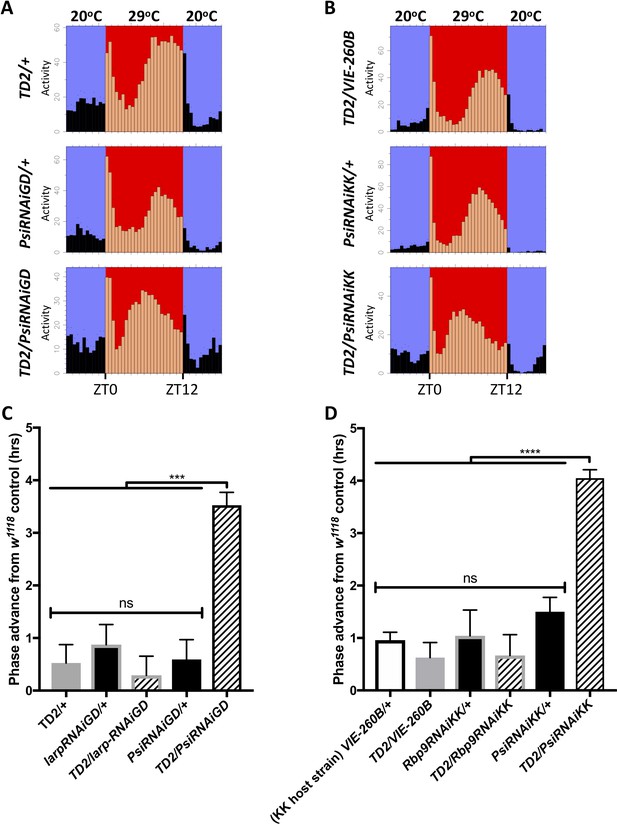
Knockdown of Psi advances the phase of circadian behavior under temperature cycle.
(A) Eductions showing the average activity of flies during 4 days of 12:12 29°C(red)/20°C(blue) temperature entrainment (days 7–10) in DD. Top panels: (driver controls) TD2/+ (left), TD2/VIE-260B (right). Middle panels: (RNAi controls) PsiRNAiGD/+ (left), PsiRNAiKK/+ (right). Bottom panels: (Psi knockdown) TD2/PsiRNAiGD (left), TD2/PsiRNAiKK (right). Note that, Psi knockdown flies advance the phase of their evening activity by about 2.5 hr relative to controls. (C–D) Evening peak phase relative to an internal control in each run (w1118) (hrs) is plotted on the y axis. Genotypes are listed on the x axis. Error bars represent SEM. ***p<0.001, ****p<0.0001, one-way ANOVA followed by Tukey’s multiple comparison test. N = 3–5 runs (C) Quantification of PsiRNAiGD knockdown and controls. Note additional RNAi controls: larpRNAiGD/+ (black bar, gray border) and TD2/larpRNAiGD (patterned bar, gray border). larpRNAiGD (GD8214) is an RNAi line from the GD collection that targets a RAP from our screen that was not a hit. (D) Quantification of PsiRNAiKK knockdown and controls. Note additional RNAi controls: VIE260B/+ (white bar, black border), TD2/VIE260B (gray bar), Rbp9RNAiKK/+ (black bar, gray border) and TD2/Rbp9RNAiKK (patterned bar, gray border). VIE260B is a KK collection host strain control containing the 30B transgene insertion site. Rbp9RNAiKK (KK109093) is an RNAi line from the KK collection targeting a RAP from our screen that was not a hit.
-
Figure 4—source data 1
Psi downregulation – temperature cycle phase.
- https://cdn.elifesciences.org/articles/50063/elife-50063-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Figure statistics – Figure 4.
- https://cdn.elifesciences.org/articles/50063/elife-50063-fig4-data2-v2.xlsx
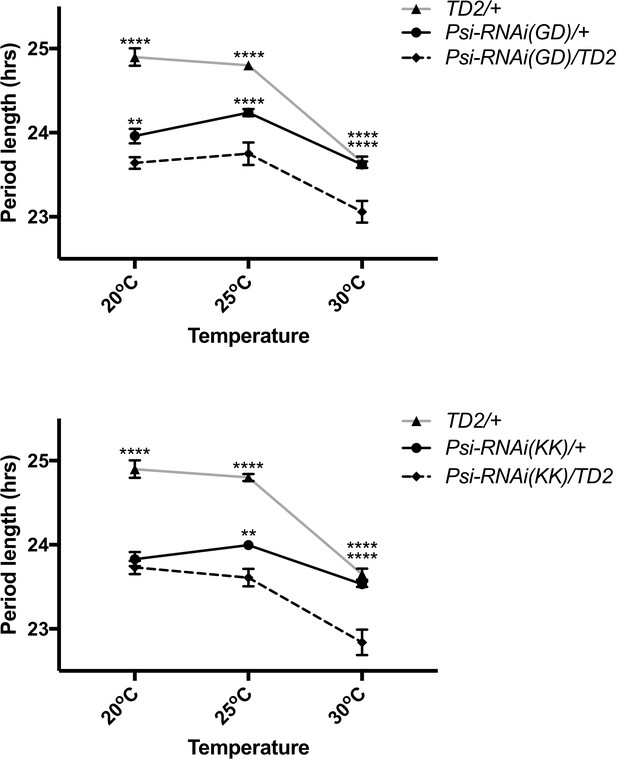
Free-running circadian behavior of Psi knockdown flies and controls at different temperatures in DD.
Circadian period length (hrs) is plotted on the y axis. The temperature at which the experiment was conducted is listed on the x axis. Error bars represent SEM. Gray line and triangle marker is the driver control. Black lines and circle markers are the RNAi controls (top, PsiRNAiGD/+; bottom, PsiRNAiKK/+). Dashed lines and diamond markers are Psi knockdown (top, PsiRNAiGD/TD2; bottom, PsiRNAiKK/TD2). **p<0.01, ****p<0.0001 two-way ANOVA followed by Tukey’s multiple comparison test.
-
Figure 4—figure supplement 1—source data 1
Psi downregulation – temperature compensation.
- https://cdn.elifesciences.org/articles/50063/elife-50063-fig4-figsupp1-data1-v2.xlsx
-
Figure 4—figure supplement 1—source data 2
Figure statistics – Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/50063/elife-50063-fig4-figsupp1-data2-v2.xlsx
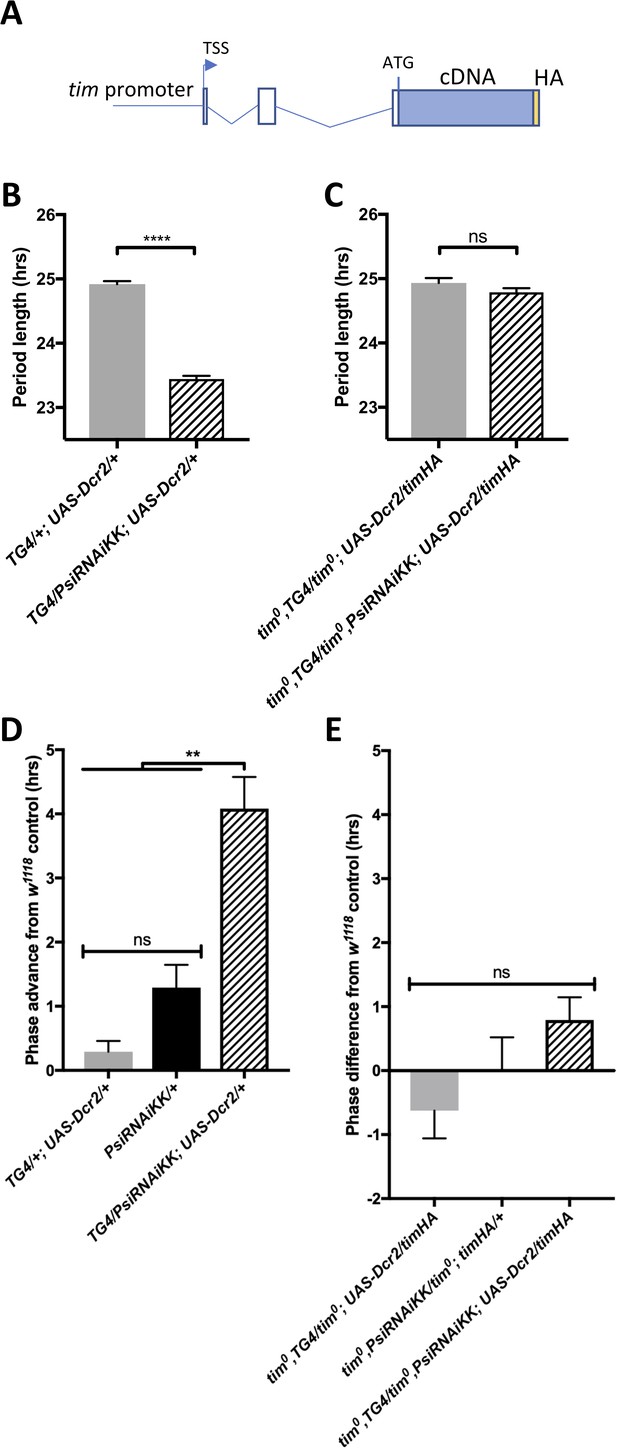
The short period and temperature cycle phase advance effects of Psi knockdown are dependent on tim introns.
(A) Schematic of timHA transgene. The tim promoter is fused upstream of the transcription start site (TSS). Two introns remain in the 5’UTR, upstream of the start codon; however, they are not, to our knowledge, temperature sensitive. A C-terminal HA tag is fused to full length tim cDNA, which lacks any of the introns that are known to be retained at high or low temperatures. (B) Knockdown of Psi with tim-GAL4 and a UAS-dcr2 transgene inserted on the 3rd chromosome also causes period shortening. We used this insertion to more easily generate stocks in a tim0 background, since the tim gene is on the second chromosome, instead of the TD2 combination that has both the tim-GAL4 and UAS-dcr2 transgenes on the 2nd chromosome. ****p<0.0001, Student’s t-test. (C) Period shortening in response to Psi knockdown with tim-GAL4 and UAS-dcr2 is abolished in tim0, ptim-timHA flies that can only produce the full length tim isoform. ns, p=0.1531, Student’s t-test. (B, C) Circadian period length (hrs) is plotted on the y axis. Genotypes are listed on the x axis. Error bars represent SEM. (D) Knockdown of Psi with tim-GAL4 and a UAS-dcr2 3rd chromosome transgene also causes a phase advance in a 12:12 29°C/20°C temperature cycle. (E) The phase advance is abolished in tim0, ptim-timHA flies that can only produce the full length tim isoform. (D, E) Evening peak phase relative to an internal control in each run (w1118) (hrs) is plotted on the y axis. Genotypes are listed on the x axis. Error bars represent SEM. **p<0.01, one-way ANOVA followed by Tukey’s multiple comparison test. N = 3 runs.
-
Figure 5—source data 1
Psi downregulation in a tim0; timHA background – behavioral period length in DD and temperature cycle phase.
- https://cdn.elifesciences.org/articles/50063/elife-50063-fig5-data1-v2.xlsx
-
Figure 5—source data 2
Figure statistics – Figure 5.
- https://cdn.elifesciences.org/articles/50063/elife-50063-fig5-data2-v2.xlsx
Tables
Circadian behavior in DD of screen candidates
| Gene | RNAi Line | Driver | n | % of Rhythmic Flies | Period Average ±SEM | Power Average ±SEM |
|---|---|---|---|---|---|---|
| Atx-1 | GD11345 | TD2 | 24 | 75 | 26 ± 0.1 | 61.5 ± 4.1 |
| PD2 | 17 | 76 | 26.4 ± 0.1 | 50.7 ± 5.6 | ||
| KK108861 | TD2 | 24 | 79 | 25.7 ± 0.1 | 49.1 ± 4.7 | |
| PD2 | 23 | 74 | 26.2 ± 0.1 | 61.8 ± 4.5 | ||
| barc | GD9921 | PD2 | 20 | 75 | 26.5 ± 0.2 | 46.9 ± 5.6 |
| KK101606** | TD2 | 6 | 83 | 25.3 ± 0.5 | 55.4 ± 12.7 | |
| PD2 | 16 | 75 | 27 ± 0.4 | 43.9 ± 5.1 | ||
| bsf | JF01529 | TD2 | 24 | 88 | 25.8 ± 0.1 | 68.4 ± 4.6 |
| PD2 | 24 | 67 | 25.7 ± 0.1 | 47.6 ± 4.1 | ||
| CG16941 | GD9241 | PD2 | 8 | 0 | ||
| HMS00157 | PD2 | 24 | 4 | 23.4 | 28.3 | |
| KK102272 | PD2 | 8 | 0 | |||
| CG32364 | HMS03012 | PD2 | 24 | 88 | 25.7 ± 0.1 | 58.9 ± 3 |
| CG42458 | KK106121 | TD2 | 23 | 35 | 26.5 ± 0.2 | 38.3 ± 4.9 |
| PD2 | 22 | 82 | 26.2 ± 0.1 | 71 ± 4.1 | ||
| CG4849 | KK101580 | TD2 | 1 | 0 | ||
| PD2 | 24 | 63 | 27.3 ± 0.2 | 48.8 ± 4.1 | ||
| CG5808 | KK102720* | TD2 | 23 | 70 | 27.4 ± 0.1 | 45.3 ± 5.1 |
| PD2 | 24 | 54 | 28.5 ± 0.6 | 34.8 ± 2.7 | ||
| CG6227 | GD11867 | TD2 | 1 | 0 | ||
| PD2 | 16 | 63 | 26.7 ± 0.2 | 51.4 ± 7 | ||
| KK108174 | TD2 | 4 | 0 | |||
| PD2 | 20 | 30 | 24.2 ± 0.4 | 30.9 ± 3.5 | ||
| CG7903 | KK103182* | TD2 | 24 | 8 | 23.6 | 26.3 |
| PD2 | 24 | 75 | 26.4 ± 0.2 | 49.1 ± 3.7 | ||
| CG8273 | GD13870 | TD2 | 24 | 83 | 25.9 ± 0.1 | 47.3 ± 4.6 |
| PD2 | 14 | 100 | 25.4 ± 0.1 | 51.2 ± 4.8 | ||
| KK102147 | TD2 | 24 | 58 | 25.5 ± 0.1 | 41.1 ± 5 | |
| PD2 | 23 | 100 | 25.7 ± 0.1 | 64.3 ± 3.9 | ||
| CG8636 | GD13992 | PD2 | 12 | 50 | 26.9 ± 0.2 | 36 ± 6.4 |
| KK110954 | TD2 | 1 | 0 | |||
| PD2 | 19 | 63 | 26.3 ± 0.3 | 51.4 ± 5.6 | ||
| CG9609 | HMS01000 | PD2 | 24 | 46 | 26.3 ± 0.2 | 46.1 ± 6.5 |
| KK109846 | TD2 | 23 | 78 | 25.3 ± 0.1 | 48.5 ± 4.2 | |
| PD2 | 23 | 91 | 26.3 ± 0.1 | 56.4 ± 3.9 | ||
| Cnot4 | JF03203 | TD2 | 23 | 26 | 23.7 ± 0.1 | 39.8 ± 6 |
| PD2 | 31 | 77 | 23.9 ± 0.1 | 51.1 ± 3.2 | ||
| KK101997 | TD2 | 32 | 47 | 23.9 ± 0.1 | 37.3 ± 2.9 | |
| PD2 | 27 | 93 | 25 ± 0.1 | 48 ± 4.1 | ||
| Dcp2 | KK101790 | TD2 | 22 | 64 | 26 ± 0.1 | 49.7 ± 5.3 |
| PD2 | 24 | 92 | 25.9 ± 0.1 | 62.5 ± 4.1 | ||
| eIF1 | KK109232* | PD2 | 24 | 4 | 23.2 | 68.9 |
| eIF3l | KK102071 | TD2 | 24 | 21 | 26 ± 0.2 | 28.9 ± 2.4 |
| PD2 | 23 | 100 | 25.7 ± 0.1 | 62.5 ± 3.9 | ||
| Hrb98DE | HMS00342 | PD2 | 22 | 91 | 25.8 ± 0.1 | 60.2 ± 4.1 |
| l(1)G0007 | GD8110 | PD2 | 24 | 63 | 26.3 ± 0.2 | 42.4 ± 3.7 |
| KK102874 | TD2 | 24 | 17 | 26.9 ± 0.4 | 32.6 ± 5.5 | |
| PD2 | 23 | 48 | 26.7 ± 0.2 | 48 ± 6.1 | ||
| LSm7 | GD7971 | PD2 | 22 | 36 | 28 ± 0.4 | 43.5 ± 5.6 |
| ncm | GD7819 | PD2 | 8 | 0 | ||
| KK100829* | PD2 | 19 | 32 | 23.3 ± 0.1 | 34.4 ± 5.6 | |
| Nelf-A | KK101005 | TD2 | 24 | 63 | 26.4 ± 0.1 | 52.9 ± 4.4 |
| PD2 | 23 | 74 | 24.8 ± 0.1 | 59.4 ± 4.5 | ||
| Not1 | GD9640 | PD2 | 23 | 4 | 22.6 | 43.6 |
| KK100090 | PD2 | 10 | 30 | 23.8 ± 0.3 | 39.4 ± 4.7 | |
| Not3 | GD4068 | PD2 | 8 | 0 | ||
| KK102144 | PD2 | 21 | 14 | 23.6 ± 0.1 | 30.8 ± 2.1 | |
| Patr-1 | KK104961* | TD2 | 23 | 30 | 26.3 ± 0.2 | 33.6 ± 3 |
| PD2 | 24 | 63 | 27.1 ± 0.2 | 38.3 ± 3.6 | ||
| Pcf11 | HMS00406 | PD2 | 8 | 13 | 24 | 20.1 |
| KK100722 | PD2 | 24 | 21 | 23.3 ± 0.1 | 35.4 ± 5 | |
| pcm | GD10926 | TD2 | 16 | 63 | 25.7 ± 0.1 | 36.6 ± 4.1 |
| PD2 | 20 | 55 | 26.3 ± 0.2 | 40.4 ± 3.8 | ||
| KK108511 | TD2 | 24 | 21 | 25.7 ± 0.2 | 40.7 ± 7.8 | |
| PD2 | 24 | 17 | 27.7 ± 0.6 | 32.9 ± 6.1 | ||
| Psi | GD14067 | TD2 | 48 | 79 | 23.7 ± 0.07 | 49.6 ± 3.0 |
| PD2 | 32 | 84 | 24.2 ± 0.1 | 53.3 ± 4.1 | ||
| HMS00140 | TD2 | 24 | 100 | 24 ± 0.1 | 61.8 ± 4.2 | |
| PD2 | 20 | 85 | 24.5 ± 0.1 | 52.9 ± 5.6 | ||
| JF01476 | TD2 | 24 | 92 | 24 ± 0.1 | 64.7 ± 4.9 | |
| PD2 | 24 | 92 | 24.3 ± 0.1 | 53.2 ± 4 | ||
| KK101882 | TD2 | 35 | 77 | 23.6 ± 0.06 | 61.9 ± 3.7 | |
| PD2 | 47 | 89 | 24.7 ± 0.06 | 56.3 ± 3.4 | ||
| Rga | GD9741 | TD2 | 24 | 21 | 26.2 ± 0.1 | 32.8 ± 3.2 |
| PD2 | 22 | 36 | 25.4 ± 0.2 | 36.1 ± 4.7 | ||
| RpS3 | GD4577 | PD2 | 14 | 57 | 26.4 ± 0.2 | 48.9 ± 5.9 |
| JF01410 | PD2 | 24 | 50 | 25.6 ± 0.2 | 34.9 ± 2.3 | |
| KK109080 | PD2 | 8 | 38 | 26 ± 1.3 | 34.5 ± 6.3 | |
| Rrp6 | GD12195 | PD2 | 10 | 10 | 24.5 | 27.2 |
| KK100590 | PD2 | 21 | 10 | 23.6 | 43.2 | |
| sbr | HMS02414 | TD2 | 13 | 85 | 26.8 ± 0.2 | 48.7 ± 5.3 |
| PD2 | 21 | 100 | 24.9 ± 0.1 | 57.2 ± 4.6 | ||
| Set1 | GD4398 | TD2 | 20 | 90 | 25.8 ± 0.1 | 52.1 ± 4.2 |
| PD2 | 13 | 77 | 25.3 ± 0.1 | 42.1 ± 5.5 | ||
| HMS01837 | TD2 | 23 | 78 | 25.6 ± 0.1 | 47.9 ± 3.6 | |
| PD2 | 24 | 92 | 24.8 ± 0.1 | 50 ± 3.8 | ||
| SmB | GD11620 | PD2 | 13 | 69 | 26.2 ± 0.1 | 52.1 ± 8 |
| HM05097 | PD2 | 24 | 58 | 25.6 ± 0.1 | 45.2 ± 4.4 | |
| KK102021 | PD2 | 2 | 100 | 25.6 | 67.1 | |
| SmE | GD13663 | PD2 | 24 | 58 | 25.7 ± 0.3 | 37.3 ± 3.3 |
| HMS00074 | PD2 | 8 | 100 | 24.5 ± 0.1 | 55.1 ± 7.4 | |
| KK101450 | PD2 | 15 | 67 | 26.5± | 51.3 ± 7.8 | |
| SmF | JF02276 | PD2 | 24 | 75 | 25.8 ± 0.1 | 46.3 ± 3.9 |
| KK107814 | PD2 | 21 | 57 | 27.3 ± 0.3 | 45.4 ± 4.2 | |
| smg | GD15460 | PD2 | 24 | 58 | 26.5 ± 0.2 | 39 ± 3.5 |
| Smg5 | KK102117 | TD2 | 23 | 52 | 23.7 ± 0.1 | 38.9 ± 3.7 |
| PD2 | 24 | 79 | 23.9 ± 0.1 | 58.5 ± 4.3 | ||
| Smn | JF02057 | TD2 | 3 | 67 | 24.2 | 25.9 |
| PD2 | 24 | 54 | 25.7 ± 0.1 | 47.2 ± 3.6 | ||
| KK106152 | TD2 | 24 | 67 | 25.3 ± 0.1 | 39.7 ± 3.5 | |
| PD2 | 24 | 96 | 26.3 ± 0.2 | 48.7 ± 2.7 | ||
| snRNP-U1-C | GD11660 | PD2 | 11 | 82 | 25.7 ± 0.1 | 56.5 ± 6.1 |
| HMS00137 | PD2 | 24 | 92 | 25.8 ± 0.1 | 55.9 ± 4.1 | |
| Spx | GD11072 | PD2 | 14 | 64 | 26.5 ± 0.2 | 56.1 ± 7.4 |
| KK108243 | TD2 | 4 | 100 | 24 ± 0.2 | 47.5 ± 10.2 | |
| PD2 | 19 | 79 | 26.9 ± 0.3 | 56.4 ± 5 | ||
| Srp54k | GD1542 | PD2 | 5 | 0 | ||
| KK100462 | PD2 | 24 | 17 | 23.7 ± 0.4 | 31.3 ± 6 | |
| Zn72D | GD11579 | TD2 | 28 | 89 | 26.3 ± 0.1 | 46.1 ± 4.6 |
| PD2 | 22 | 82 | 26.4 ± 0.1 | 59.4 ± 6.9 | ||
| KK100696 | TD2 | 26 | 73 | 26.8 ± 0.1 | 57 ± 3.6 | |
| PD2 | 24 | 83 | 26 ± 0.1 | 57 ± 4.5 |
-
*Line contains insertion at 40D.
** Unknown if line contains insertion at 40D.
Predicted or known functions of screen candidates
| Gene | Molecular function (based on information from Flybase) (Thurmond et al., 2019) |
|---|---|
| Atx-1 | RNA binding |
| barc | mRNA splicing; mRNA binding; U2 snRNP binding |
| bsf | mitochondrial mRNA polyadenylation, stability, transcription, translation; polycistronic mRNA processing; mRNA 3'-UTR binding |
| CG16941/Sf3a1 | alternative mRNA splicing; RNA binding |
| CG32364/tut | translation; RNA binding |
| CG42458 | mRNA binding |
| CG4849 | mRNA splicing; translational elongation |
| CG5808 | mRNA splicing; protein peptidyl-prolyl isomerization; regulation of phosphorylation of RNA polymerase II C-terminal domain; mRNA binding |
| CG6227 | alternative mRNA splicing; ATP-dependent RNA helicase activity |
| CG7903 | mRNA binding |
| CG8273/Son | mRNA processing; mRNA splicing; RNA binding |
| CG8636/eIF3g1 | translational initiation; mRNA binding |
| CG9609 | transcription; proximal promoter sequence-specific DNA binding |
| Cnot4 | CCR4-NOT complex |
| Dcp2 | deadenylation-dependent decapping of mRNA; cytoplasmic mRNA P-body assembly; RNA binding |
| eIF1 | ribosomal small subunit binding; RNA binding; translation initiation |
| eIF3l | translational initiation |
| Hrb98DE | translation; alternative mRNA splicing; mRNA binding |
| l(1)G0007 | alternative mRNA splicing; 3'−5' RNA helicase activity |
| LSm7 | mRNA splicing; mRNA catabolic process; RNA binding |
| ncm | mRNA splicing; RNA binding |
| Nelf-A | transcription elongation; RNA binding |
| Not1 | translation; poly(A)-specific ribonuclease activity; CCR4-NOT complex |
| Not3 | translation; transcription; poly(A)-specific ribonuclease activity; CCR4-NOT complex |
| Patr-1 | cytoplasmic mRNA P-body assembly; deadenylation-dependent decapping of mRNA; RNA binding |
| Pcf11 | mRNA polyadenylation; transcription termination; mRNA binding |
| pcm | cytoplasmic mRNA P-body assembly; 5'−3' exonuclease activity |
| Psi | alternative mRNA splicing; transcription; mRNA binding |
| Rga | translation; transcription; poly(A)-specific ribonuclease activity; CCR4-NOT complex |
| RpS3 | DNA repair; translation; RNA binding; structural constituent of ribosome |
| Rrp6 | chromosome segregation; mRNA polyadenylation; nuclear RNA surveillance; 3'−5' exonuclease activity |
| sbr | mRNA export from nucleus; mRNA polyadenylation; RNA binding |
| Set1 | histone methyltransferase activity; nucleic acid binding; contains an RNA Recognition Motif |
| SmB | mRNA splicing; RNA binding |
| SmE | mRNA splicing; spliceosomal snRNP assembly |
| SmF | mRNA splicing; spliceosomal snRNP assembly; RNA binding |
| smg | RNA localization; translation; mRNA poly(A) tail shortening; transcription; mRNA binding |
| Smg5 | nonsense-mediated decay; ribonuclease activity |
| Smn | spliceosomal snRNP assembly; RNA binding |
| snRNP-U1-C | mRNA 5'-splice site recognition; mRNA splicing, alternative mRNA splicing |
| Spx | mRNA splicing; mRNA binding |
| Srp54k | SRP-dependent cotranslational protein targeting to membrane; 7S RNA binding |
| Zn72D | mRNA splicing; RNA binding |
PSI affects circadian behavior
| Genotype | Period ±SEM | Power ±SEM | n | % of Rhythmic Flies |
|---|---|---|---|---|
| Psi downregulation and overexpression at 25°C | ||||
| TD2/+ | 24.8 ± 0.04 | 48.2 ± 2.3 | 71 | 82 |
| TD2/PsiRNAiGD | 23.7 ± 0.07 | 49.6 ± 3.0 | 48 | 79 |
| TD2/PsiRNAiKK | 23.6 ± 0.06 | 61.9 ± 3.7 | 35 | 77 |
| PD2/+ | 24.9 ± 0.04 | 50.4 ± 2.1 | 77 | 83 |
| PD2/PsiRNAiGD | 24.2 ± 0.06 | 53.3 ± 4.1 | 32 | 84 |
| PD2/PsiRNAiKK | 24.7 ± 0.06 | 56.3 ± 3.4 | 47 | 89 |
| TD2/+; PdfGAL80/+ | 24.5 ± 0.07 | 49.4 ± 2.8 | 40 | 75 |
| TD2/PsiRNAiGD; PdfGAL80/+ | 23.8 ± 0.17 | 45.8 ± 5.5 | 24 | 50 |
| TD2/PsiRNAiKK; PdfGAL80/+ | 24.0 ± 0.05 | 71.9 ± 4.0 | 39 | 95 |
| w1118 | 24.1 ± 0.03 | 84.8 ± 2.5 | 70 | 99 |
| PsiRNAiGD/+ | 24.2 ± 0.04 | 58.9 ± 2.9 | 63 | 94 |
| PsiRNAiKK/+ | 24.0 ± 0.04 | 67.1 ± 3.7 | 55 | 96 |
| TG4/+ | 25.2 ± 0.05 | 52.5 ± 2.2 | 68 | 88 |
| TG4/+; UAS-Psi/+ | 25.9 ± 0.07 | 31.3 ± 1.2 | 302 | 16 |
| PG4/+ | 25.0 ± 0.05 | 66.0 ± 3.5 | 26 | 96 |
| PG4/+; UAS-Psi/+ | 25.2 ± 0.07 | 44.0 ± 2.7 | 48 | 77 |
| TG4/+; PdfGAL80/+ | 24.6 ± 0.06 | 42.8 ± 2.8 | 37 | 84 |
| TG4/+; PdfGAL80/UAS-Psi | 24.9 ± 0.19 | 31.3 ± 2.8 | 116 | 11 |
| UAS-Psi/+ | 24.2 ± 0.04 | 46.4 ± 1.8 | 80 | 79 |
| Psi downregulation at 20°C | ||||
| TD2/+ | 24.9 ± 0.10 | 42.0 ± 3.1 | 39 | 59 |
| TD2/PsiRNAiGD | 23.6 ± 0.07 | 52.2 ± 4.7 | 44 | 66 |
| TD2/PsiRNAiKK | 23.7 ± 0.08 | 43.8 ± 5.5 | 44 | 36 |
| PsiRNAiGD/+ | 24.0 ± 0.09 | 46.0 ± 3.7 | 32 | 72 |
| PsiRNAiKK/+ | 23.8 ± 0.08 | 39.1 ± 4.9 | 32 | 38 |
| Psi downregulation at 30°C | ||||
| TD2/+ | 23.7 ± 0.07 | 48.2 ± 2.9 | 39 | 87 |
| TD2/PsiRNAiGD | 23.1 ± 0.13 | 38.3 ± 3.8 | 42 | 40 |
| TD2/PsiRNAiKK | 22.8 ± 0.15 | 43.1 ± 4.2 | 41 | 41 |
| PsiRNAiGD/+ | 23.6 ± 0.04 | 43.2 ± 3.4 | 32 | 75 |
| PsiRNAiKK/+ | 23.5 ± 0.03 | 63.0 ± 3.7 | 31 | 90 |
| TIM-HA suppression of PSI's effect on circadian behavior | ||||
| TG4/PsiRNAiKK; UAS-Dcr2/+ | 23.4 ± 0.04 | 59.5 ± 4.3 | 57 | 75 |
| TG4/+; UAS-Dcr2/+ | 24.9 ± 0.04 | 59.4 ± 3.1 | 36 | 92 |
| tim0,TG4/tim0; UAS-Dcr2/timHA | 24.9 ± 0.07 | 44.3 ± 4.0 | 28 | 75 |
| tim0,TG4/tim0,PsiRNAiKK; UAS-Dcr2/timHA | 24.8 ± 0.06 | 50.0 ± 2.9 | 38 | 79 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Drosophila melanogaster) | Psi | FLYB:FBgn0014870 | Flybase name:P-element somatic inhibitor | |
| Gene (Drosophila melanogaster) | tim | FLYB:FBgn0014396 | Flybase name: timeless | |
| Gene (Drosophila melanogaster) | tio | FLYB:FBgn0028979 | Flybase name: tiptop | |
| Gene (Drosophila melanogaster) | per | FLYB:FBgn0003068 | Flybase name: period | |
| Gene (Drosophila melanogaster) | cwo | FLYB:FBgn0259938 | Flybase name:clockwork orange | |
| Gene (Drosophila melanogaster) | RpL32 | FLYB:FBgn0002626 | qPCR control Flybase name:Ribosomal protein L32 | |
| Gene (Drosophila melanogaster) | larp | FLYB:FBgn0261618 | Flybase name: La related protein | |
| Gene (Drosophila melanogaster) | Rbp9 | FLYB:FBgn0010263 | Flybase name:RNA-binding protein 9 | |
| Gene (Drosophila melanogaster) | Dcr-2 | FBgn0034246 | Flybase name: Dicer-2 | |
| Genetic reagent (D. melanogaster) | tim-GAL4 | Kaneko et al., 2000 | FLYB:FBtp0010385 | |
| Genetic reagent (D. melanogaster) | Pdf-GAL4 | Renn et al., 1999 | FLYB:FBtp0011844 | |
| Genetic reagent (D. melanogaster) | Pdf-GAL80, Pdf-GAL80 | Stoleru et al., 2004 | ||
| Genetic reagent (D. melanogaster) | UAS-Dcr2 | Dietzl et al., 2007 | FLYB:FBti0100275 RRID:BDSC_24650 | Chromosome 2 |
| Genetic reagent (D. melanogaster) | UAS-Dcr2 | Dietzl et al., 2007 | FLYB:FBti0100276 | Chromosome 3 |
| Genetic reagent (D. melanogaster) | PsiRNAi KK101882 | FLYB:FBal0231542 | ||
| Genetic reagent (D. melanogaster) | PsiRNAi GD14067 | Dietzl et al., 2007 | FLYB:FBst0457756 | |
| Genetic reagent (D. melanogaster) | UAS-Psi | Labourier et al., 2001 | ||
| Genetic reagent (D. melanogaster) | BG-LUC | Stanewsky et al., 1997 | ||
| Genetic reagent (D. melanogaster) | ptim-TIMLUC | Lamba et al., 2018 | ||
| Genetic reagent (D. melanogaster) | timHA | Rutila et al., 1998 | FLYB:FBal0143160 | |
| Genetic reagent (D. melanogaster) | tim0 | Sehgal et al., 1994 | FLYB:FBal0035778 | |
| Genetic reagent (D. melanogaster) | VIE260B | VDRC_ID: 60100 | ||
| genetic reagent (D. melanogaster) | larpRNAi GD8214 | Dietzl et al., 2007 | VDRC_ID: 17366 | |
| Genetic reagent (D. melanogaster) | Rbp9RNAi KK109093 | VDRC_ID: 101412 | ||
| Genetic reagent (D. melanogaster) | w1118 | VDRC_ID: 60000 | ||
| Genetic reagent (D. melanogaster) | 40D-UAS | VDRC_ID: 60101 | ||
| Sequence-based reagent | RpL32-forward | Dubruille et al., 2009 | PCR primers | ATGCTAAGCTGTCGCACAAA |
| Sequence-based reagent | RpL32-reverse | Dubruille et al., 2009 | PCR primers | GTTCGATCCGTAACCGATGT |
| Sequence-based reagent | psi-forward | This paper | PCR primers | GGTGCCTTGAATGGGTGAT |
| Sequence-based reagent | psi-reverse | This paper | PCR primers | CGATTTATCCGGGTCCTCG |
| Sequence-based reagent | tim-M-forward | This paper | PCR primers | TGGGAATCTCGCCCGAAAC |
| Sequence-based reagent | tim-M-reverse | This paper | PCR primers | AGAAGGAGGAGAAGGAGAGAGG |
| Sequence-based reagent | tim-sc-forward | This paper | PCR primers | ACTGTGCGATGACTGGTCTG |
| Sequence-based reagent | tim-sc-reverse | This paper | PCR primers | TGCTTCAAGGAAATCTTCTG |
| Sequence- based reagent | tim-cold-forward | This paper | PCR primers | CCTCCATGAAGTCCTCGTTCG |
| Sequence-based reagent | tim-cold-reverse | This paper | PCR primers | ATTGAGCTGGGACACCAGG |
| Sequence-based reagent | cwo-foward | This paper | PCR primers | TTCCGCTGTCCACCAACTC |
| Sequence-based reagent | cwo-reverse | This paper | PCR primers | CGATTGCTTTGCTTTACCAGCTC |
| Sequence-based reagent | cwoRA-forward | This paper | PCR primers | TCAAGTATGAGAGCGAAGCAGC |
| Sequence-based reagent | cwoRA-reverse | This paper | PCR primers | TGTCTTATTACGTCTTCCGGTGG |
| Sequence-based reagent | cwoRB-forward | This paper | PCR primers | GTATGAGAGCAAGATCCACTTTCC |
| Sequence-based reagent | cwoRB-reverse | This paper | PCR primers | GATGATCTCCGTCTTCTCGATAC |
| Sequence-based reagent | cwoRC-forward | This paper | PCR primers | GTATGAGAGCCAAGCGACCAC |
| Sequence-based reagent | cwoRC-reverse | This paper | PCR primers | CCAAATCCATCTGTCTGCCTC |
| Commercial assay or kit | Direct-zol RNA MiniPrep kit | Zymo Research | Zymo Research: R2050 | |
| Commercial assay or kit | iSCRIPT cDNA synthesis kit | Bio-RAD | Bio-RAD: 1708891 | |
| Commercial assay or kit | iTaq Universal SYBR Green Supermix | Bio-RAD | Bio-RAD: 1725121 | |
| Chemical compound, drug | D-Luciferin, Potassium Salt | Goldbio | Goldbio: LUCK-1G | |
| Chemical compound, drug | TRIzol Reagent | Invitrogen | ThermoFisher Scientific:15596026 | |
| Software, algorithm | FaasX software | Grima et al., 2002 | http://neuro-psi.cnrs.fr/spip.php?article298&lang=en | |
| Software, algorithm | MATLAB (MathWorks) signal-processing toolbox | Levine et al., 2002 | MATLAB RRID: SCR_001622 | |
| Software, algorithm | MS Excel | RRID: SCR_016137 | ||
| Software, algorithm | GraphPad Prism version 7.0 c for Mac OS X | GraphPad Software, La Jolla, CA USA | RRID: SCR_002798 | www.graphpad.com |
Additional files
-
Supplementary file 1
RAP Screen Dataset.
Circadian behavior analysis for all RNAi lines included in our screen. Period, Power (i. e. rhythm amplitude), and percentage of rhythmic flies are indicated. SD: Standard Deviation. Each lines is crossed to TD2 or PD2, or in some cases to w1118.
- https://cdn.elifesciences.org/articles/50063/elife-50063-supp1-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/50063/elife-50063-transrepform-v2.pdf





