A terminal selector prevents a Hox transcriptional switch to safeguard motor neuron identity throughout life
Figures
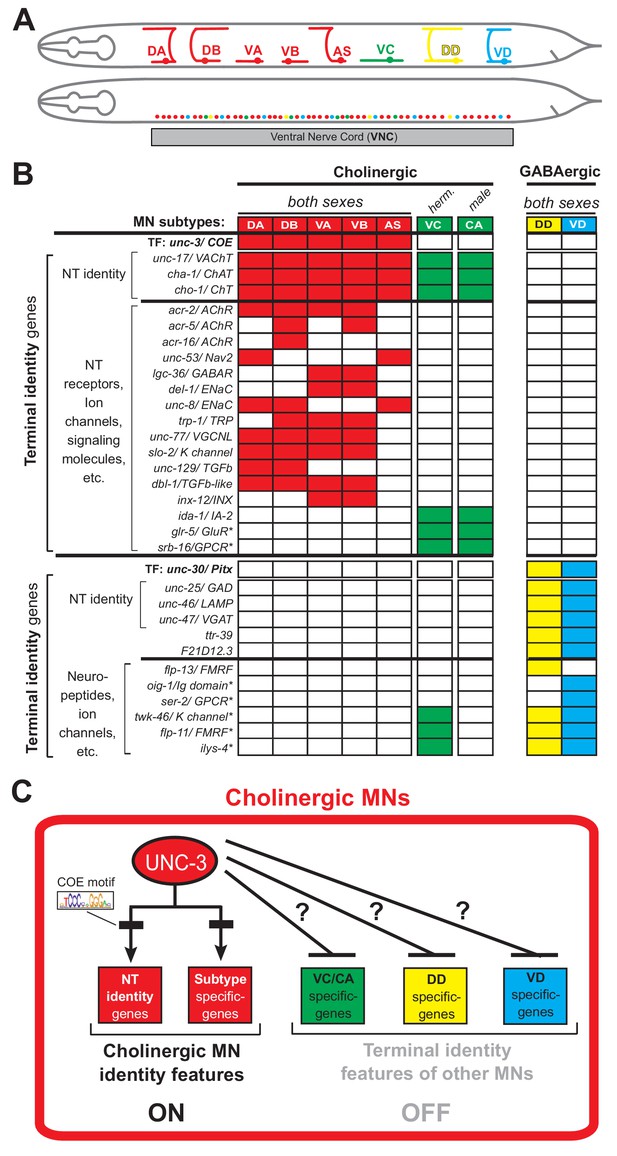
An extensive collection of terminal identity markers for distinct motor neuron subtypes of the C.
elegans ventral nerve cord. (A) Schematic showing distinct morphology for each motor neuron subtype in the C. elegans hermaphrodite. Below, colored dots represent the invariant cell body position of all MNs of the ventral nerve cord (VNC). Red: 39 sex-shared cholinergic MNs (DA2−7 = 6 neurons, DB3−7 = 5, VA2−11 = 10, VB3−11 = 9, AS2−10 = 9); Green: six hermaphrodite-specific VC MNs; Yellow: four sex-shared GABAergic DD neurons (DD2−5 = 4); Blue: nine sex-shared GABAergic VD neurons (VD3−11 = 9). With the exception of VC, all other subtypes have 1–3 extra neurons located at the flanking ganglia (retrovesicular and pre-anal) of the VNC (not shown). Individual neurons of each subtype intermingle along the VNC. (B) Table summarizing expression of terminal identity markers for VNC MNs. The sex-shared GABAergic MNs (DD, VD) and the sex-specific MNs (VC, CA) do not express UNC-3. Conversely, the sex-shared cholinergic MNs (DA, DB, VA, VB, AS) and the sex-specific MNs (VC, CA) do not express UNC-30/Pitx. For the genes indicated with an asterisk (*), a detailed expression pattern is provided in Figure 1—figure supplement 1. Of note, the male-specific MNs of the CP subtype are also not shown. (C) Schematic that summarizes the known function of UNC-3 (activator of cholinergic MN identity genes) and the question under investigation: does UNC-3 prevent expression of terminal identity features reserved for other MN subtypes?.
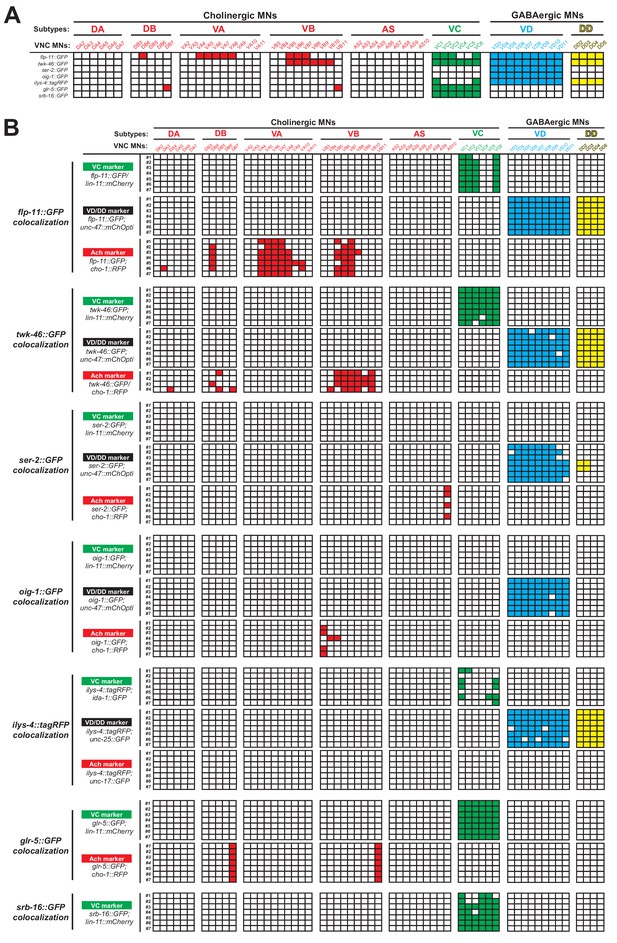
Detailed characterization of the expression pattern of VC and VD terminal identity markers.
(A) An expression map of VC and VD terminal identity genes with single-cell resolution at larval stage 4 (L4). Each column represents an individual motor neuron (MN) in the ventral nerve cord (VNC) of a hermaphrodite worm. MN subtypes are color-coded. This table represents a summary of the co-localization analysis described in panel B. Apart from the seven genes listed, we also examined five additional terminal identity genes (bra-1, dhc-1, rgs-4, vhp-1, vps-25) but found no evidence for expression in VD or VC neurons. (B) Co-localization analysis of VD and VC terminal identity genes. Each column represents an individual MN in the VNC of a hermaphrodite worm. Each row represents a randomly-selected worm co-expressing the respective VD or VC marker and one of the known identity markers: unc-47::mChOpti or unc-25::gfp for GABAergic (VD and DD) MN identity, cho-1::rfp or unc-17::gfp for cholinergic MN identity and lin-11::mCherry or ida-1::gfp for VC MN identity. A color-filled lattice indicates co-expression of the known identity marker and the selected marker in the individual MN.
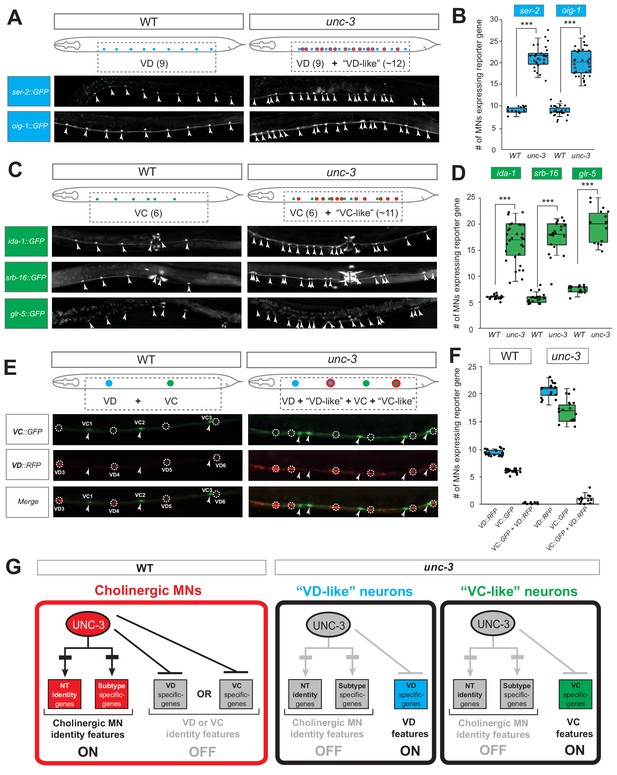
UNC-3 has a dual role in cholinergic ventral cord motor neurons.
(A) Terminal identity markers of VD neurons (ser-2, oig-1) are ectopically expressed in unc-3-depleted MNs. Representative images of larval stage 4 (L4) hermaphrodites are shown. Similar results were obtained in adult animals. Arrowheads point to MN cell bodies with gfp marker expression. Green fluorescence signal is shown in white for better contrast. Dotted black box indicates imaged area. (B) Quantification of VD markers (ser-2, oig-1) in WT and unc-3 (n3435) at L4. N > 15. ***p<0.001. For details on box plots, see Materials and methods. (C) Terminal identity markers of VC neurons (ida-1, srb-16, glr-5) are ectopically expressed in unc-3-depleted MNs. Representative images of larval stage 4 (L4) hermaphrodites are shown. Similar results were obtained in adult animals. Arrowheads point to MN cell bodies with gfp marker expression. Green fluorescence signal is shown in white for better contrast. Dotted black box indicates imaged area. (D): Quantification of VC markers (ida-1, srb-16, glr-5) in WT and unc-3 (n3435) at L4. Individual data points are dot-plotted. N > 15. ***p<0.001. (E) Distinct MNs acquire VC-like or VD-like terminal identity features in unc-3 (n3435) mutants. The VC marker in green (ida-1::gfp) and the VD marker in red (ser-2::rfp) do not co-localize in WT or unc-3 (n3435) mutants. Representative images are shown. Individual VC/VC-like and VD/VD-like neurons are pointed and circled, respectively,(VD: dotted circles; VC: arrowheads) to highlight that an individual MN never expresses both markers. (F) Quantification of data shown in E. N > 16. (G) Schematic that summarizes the dual role of unc-3. Apart from activating cholinergic MN terminal identity genes, UNC-3 prevents expression of VD and VC terminal features in distinct cells (‘VD-like’ versus ‘VC-like’).
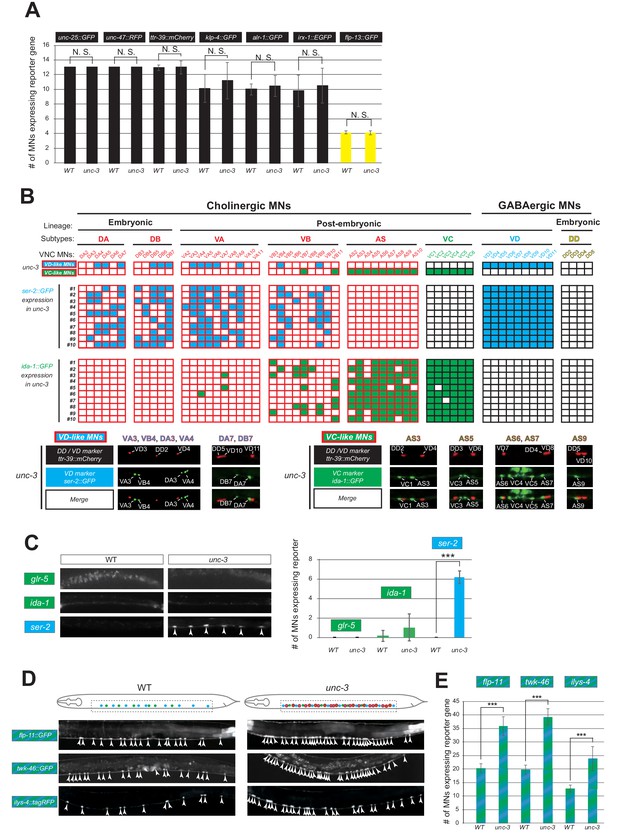
UNC-3 selectively prevents expression of VD and VC terminal identity features in distinct cholinergic MNs.
(A) Quantification of six VD/DD (unc-25::gfp, unc-47::rfp, ttr-39::mCherry, klp-4::gfp, alr-1::gfp and irx-1::egfp) and one DD (flp-13::gfp) markers in WT and unc-3 (n3435) animals at L4. No statistically significant effects were observed in unc-3 mutants. N > 13. N. S: not significant. (B) A table showing that VD and VC identity genes are ectopically expressed in distinct cholinergic MNs upon unc-3 depletion. Top two rows: A summary map of VD and VC gene expression with single-cell resolution in unc-3 mutants, which is based on the co-localization analysis shown below. Each column represents an individual MN in the VNC of an unc-3-depleted hermaphrodite worm. Ten randomly selected worms that co-express VD/DD marker ttr-39::mCherry and VD marker ser-2::gfp or VC marker ida-1::gfp are analyzed. The expression of ttr-39::mCherry is not affected by unc-3 depletion, which serves as a positional landmark for reference. This landmark together with the invariant MN cell body position along the VNC enable the identification of distinct unc-3-depleted MNs that lose cholinergic features and concomitantly gain either ‘VD-like’ or ‘VC-like’ features. Below this table, examples of the co-localization analysis are provided. Representative magnified images are shown. The identity of unc-3-depleted MNs that acquire VC or VD terminal features is shown. Note that VC-like terminal features are mainly acquired by MNs of the AS cholinergic subtype, whereas VD-like terminal features are acquired by select members of four cholinergic MN subtypes (DA, DB, VA, VB). (C) The DA and DB cholinergic MNs are born embryonically and therefore present in the VNC at L1. Corroborating our results from panel B, only the VD terminal identity marker (ser-2) shows ectopic expression in DA and DB neurons of unc-3 mutants at L1. This is not the case for the VC terminal identity markers (glr-5, ida-1). Representative images are shown on the left. Arrowheads point to MN cell bodies with ectopic gfp marker expression. Green fluorescence signal is shown in white for better contrast. Quantification is on the right. N > 22. ***p<0.001. (D) Terminal identity markers of VD/VC MNs (flp-11, twk-46 and ilys-4) are ectopically expressed in unc-3-depleted cholinergic MNs. Representative images of larval stage 4 (L4) hermaphrodites are shown. Similar results were obtained in adult animals. Arrowheads point to MN cell bodies with marker expression. Fluorescence signal is shown in white for better contrast. Dotted black box indicates imaged area. (E) Quantification of data shown in panel D.. N > 18. ***p<0.001.
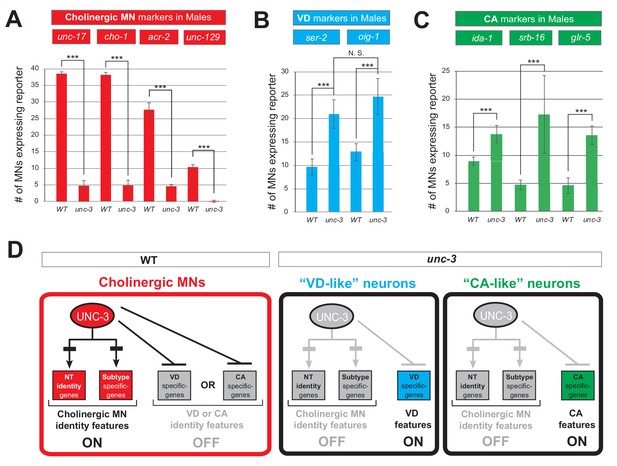
The dual role of UNC-3 in cholinergic MNs extends to both C. elegans sexes.
(A) Quantification of four sex-shared cholinergic MN markers (unc-17, cho-1, acr-2, unc-129) expression in WT and unc-3 (n3435) male animals at L4. N > 13. ***p<0.001. (B) Quantification of VD marker (ser-2 and oig-1) expression in L4 stage WT and unc-3 (n3435) male animals. No significant difference was found between the number of MNs ectopically expressing each marker. N > 11. ***p<0.001. N. S: not significant. (C) Quantification of the male-specific CA markers (ida-1, srb-16 and glr-5) in young adult stage WT or unc-3 (n3435) male animals. N > 14. ***p<0.001. (D) Schematic summarizing the dual role of UNC-3 in C. elegans males.
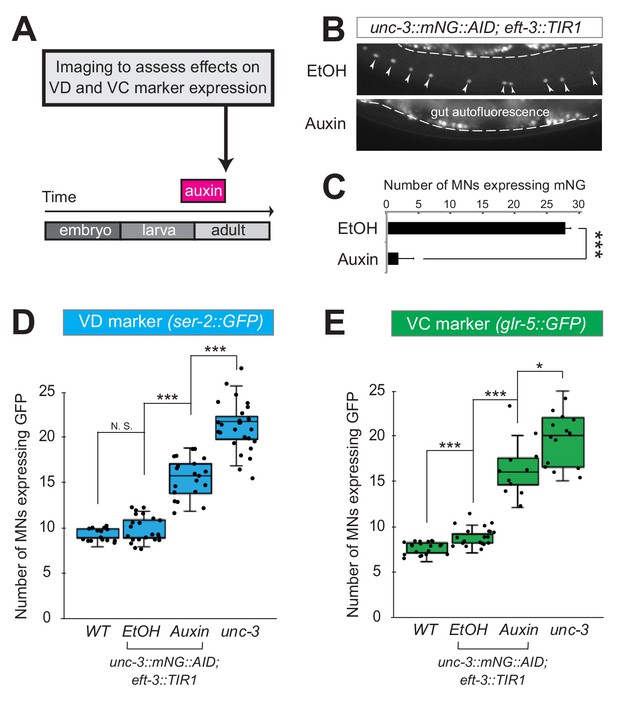
UNC-3 is continuously required to prevent expression of VD and VC terminal identity features.
(A) Schematic showing time window of auxin administration. (B) Animals of the unc-3::mNG::AID; eft-3::TIR1 genotype were either administered ethanol (EtOH) or auxin at the L4 stage. Twenty four hours later, expression of endogenous unc-3 reporter (unc-3::mNG::AID) is severely reduced in the nuclei of VNC MNs (arrowheads) at the young adult stage (day 1). The same exact region was imaged in EtOH- and auxin-treated worms. mNG green fluorescent signal is shown in white for better contrast. White dotted line indicates the boundary of intestinal tissue (gut), which tends to be autofluorescent in the green channel. (C) Quantification of number of MNs expressing the unc-3::mNG::AID reporter after EtOH (control) and auxin treatment. N > 12. ***p<0.001. (D) Auxin or ethanol (control) were administered at larval stage 3 (L3) on unc-3::mNG::AID; eft-3::TIR1 animals carrying the VD marker ser-2::gfp. Images were taken at the young adult stage (day 1.5). A significant increase in the number of MNs expressing the VD marker was evident in the auxin-treated animals compared to EtOH-treated controls. For comparison, quantification is provided for ser-2::gfp expressing MNs of wild-type animals and unc-3(n3435) mutants. Similar results were obtained when auxin was applied at L4 or day 1 adult animals. N > 20. ***p<0.001. (E) Auxin or ethanol (control) were administered at larval stage 4 (L4) on unc-3::mNG::AID; eft-3::TIR1 animals carrying the VC marker glr-5::gfp. Images were taken at the young adult stage (day 2). A significant increase in the number of MNs expressing the VC marker was evident in the auxin-treated animals compared to EtOH-treated controls. N > 11. *p<0.05; ***p<0.001.
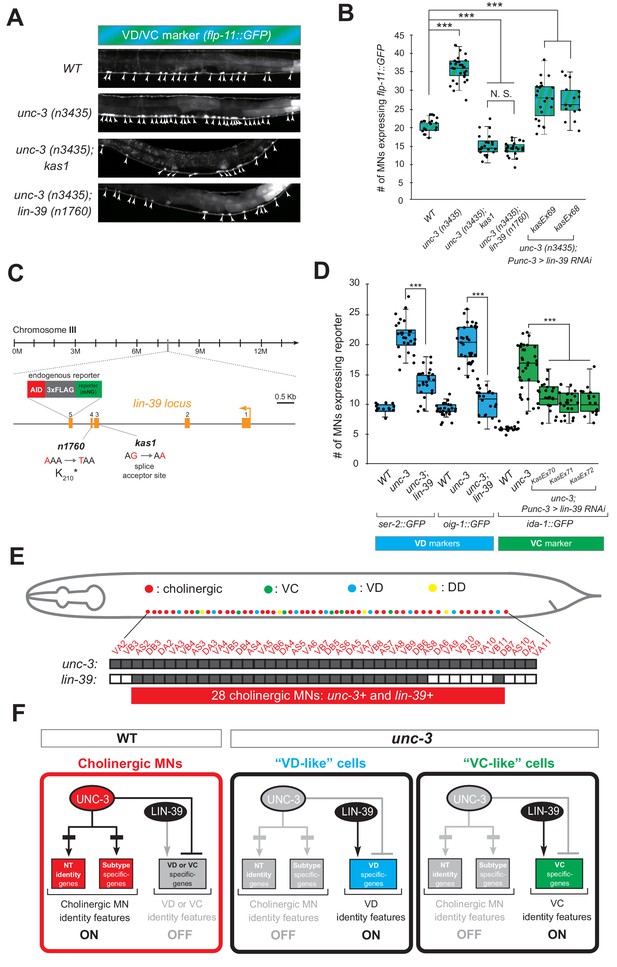
A genetic screen identifies the mid-body Hox protein LIN-39 (Scr/Dfd/Hox4-5) as necessary for ectopic expression of VD and VC terminal features.
(A) Representative images of L4-stage WT, unc-3(n3435), unc-3(n3435); kas1, and unc-3(n3435); lin-39(n1760) animals carrying flp-11::gfp (VD/VC marker). Arrowheads point to MN cell bodies with gfp marker expression. (B) Quantification graph summarizing results from panel A. The two right-most bars show quantification of two independent transgenic lines driving lin-39 RNAi specifically in cholinergic MNs (Punc-3 >lin-39 RNAi) of unc-3 (n3435) mutants. N > 15. ***p<0.001. N.S: not significant. (C) Genetic locus of lin-39. Molecular lesions for kas1 and n1760 alleles are shown, as well as the AID::3xFLAG::mNG cassette inserted at the C-terminus (endogenous reporter). (D) Quantification of two VD (ser-2::gfp, oig-1::gfp) and one VC (ida-1::gfp) markers in WT, unc-3 (n3435), unc-3(n3435); lin-39(n1760) animals at L4. The three right-most bars show quantification of three independent transgenic lines driving lin-39 RNAi specifically in cholinergic MNs (Punc-3 >lin-39 RNAi) of unc-3 (n3435) mutants. N > 15. ***p<0.001. (E) Summary of unc-3 and lin-39 expression in cholinergic MNs. See Figure 4—figure supplement 2 for raw data. (F) Schematic that summarizes our findings. In the wild type (Faumont et al., 2011) panel on the left, lin-39 is normally expressed in cholinergic MNs but unable to induce expression of VD or VC genes. In the unc-3 mutant, lin-39 is now able to induce expression of alternative identity features (VD or VC) in distinct MN populations.
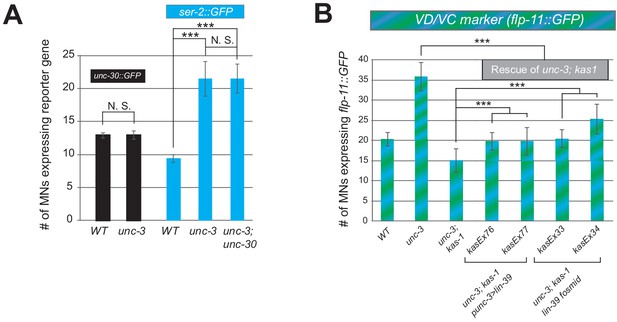
Ectopic expression of VD terminal identity markers in unc-3 mutants requires LIN-39 but not UNC-30.
(A) Quantification of unc-30::gfp and VD marker ser-2::gfp in WT and unc-3 (n3435) animals at L4, with ser-2::gfp further examined in unc-3 (n3435); unc-30 (e191) double mutants. Expression of ser-2::gfp is equally affected in unc-3 (n3435) single and unc-3 (n3435); unc-30 (e191) double mutants. N > 15. ***p<0.001. N. S: not significant. (B) Quantification of the VD/VC marker flp-11::gfp expression in L4 stage WT, unc-3 (n3435) mutant, unc-3 (n3435); kas1, and transgenic animals that rescue unc-3 (n3435); kas1 background with expression of lin-39 in cholinergic MNs (Punc-3 >lin-39) or expression of GFP-tagged lin-39 fosmid. Two independent transgenic lines were used for Punc-3 >lin-39 OE and lin-39 fosmid. Of note, the partial rescue observed with the Punc-3 >lin-39 OE lines likely arises because the fragment of unc-3 promoter used to drive lin-39 is not expressed in all 39 cholinergic MNs.
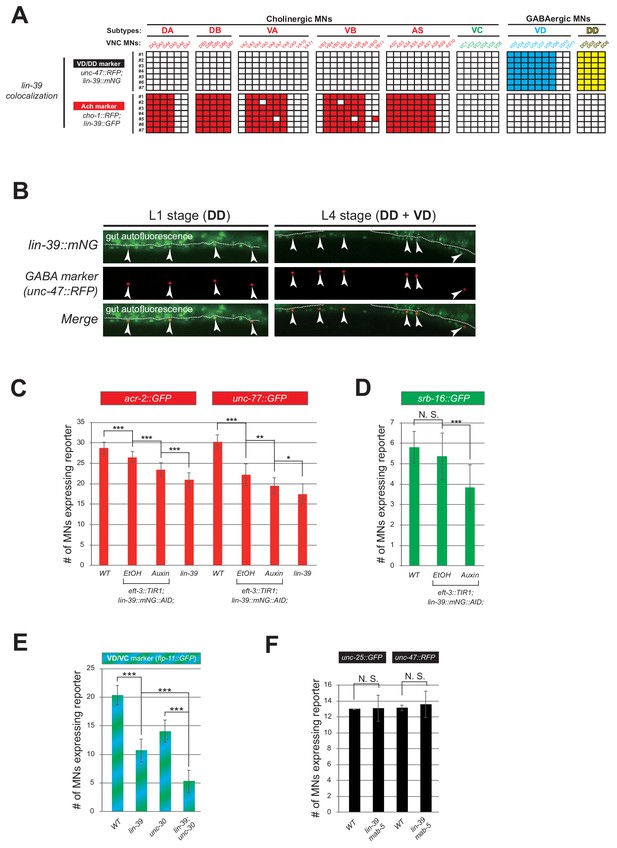
LIN-39 is continuously required to activate distinct terminal identity genes in sex-shared and sex-specific cholinergic MNs.
(A) Co-localization analysis to determine the lin-39 expression pattern with single-cell resolution. Each column represents an individual MN in the VNC of a hermaphrodite worm. Top seven rows represent seven randomly-selected worms carrying a GABA marker (unc-47::RFP) and the lin-39::mNG::3xFLAG::AID endogenous reporter allele. Bottom seven rows represent seven randomly-selected worms carrying a cholinergic marker (cho-1::RFP and the lin-39 fosmid-based reporter wgIs18 [lin-39fosmid::GFP]. A color-filled lattice indicates co-expression of the known identity marker and lin-39 in the individual MN. (B) Representative images of L1- and L4-stage animals co-expressing unc-47::rfp (labels only DD at L1, labels both DD and VD at L4) and endogenous lin-39 marker (lin-39::mNG). Arrowheads point to cell bodies of DD and VD MNs co-expressing both markers. White dotted line indicates the boundary of intestinal tissue (gut), which tends to be autofluorescent in the green channel. (C) Auxin or ethanol (control) were administered at larval stage 4 (L4) on lin-39::mNG::3xFLAG::AID; eft-3::TIR1 animals carrying the sex-shared cholinergic MN markers acr-2::gfp and unc-77::gfp. Images were taken at the young adult stage (day 1 for acr-2::gfp and day 2.4 for unc-77::gfp) and the number of MNs expressing these markers was quantified. A statistically significant decrease is evident in the auxin-treated animals compared to EtOH-treated controls. For comparison, quantification of marker expression is also provided in WT and lin-39 (n1760) animals. N > 12. *p<0.05, **p<0.01, ***p<0.001. (D) Auxin or ethanol (control) were administered at larval stage 3 (L3) on lin-39::mNG::3xFLAG::AID; eft-3::TIR1 animals carrying the VC marker srb-16::gfp. Quantification was performed at the young adult stage (day 1.5). A significant decrease in the number of MNs expressing the VC marker was evident in the auxin-treated animals compared to EtOH-treated controls. For comparison, quantification of marker expression is also provided in WT animals. N > 15. **p<0.01, ***p<0.001. N. S: not significant. (E) Quantification of VD/VC marker flp-11::gfp in WT, lin-39 (n1760), unc-30 (e191) and lin-39 (n1760); unc-30 (e191) double mutant animals at L4 stage. Double mutants showed a more severe reduction in flp-11::gfp expression compared to each single mutant. N > 20. ***p<0.001. (F) Expression of GABA biosynthesis markers (unc-25, unc-47) is not affected in lin-39 (n1760); mab-5 (e1239) double mutants at the L4 stage. N > 15; N. S: not significant.
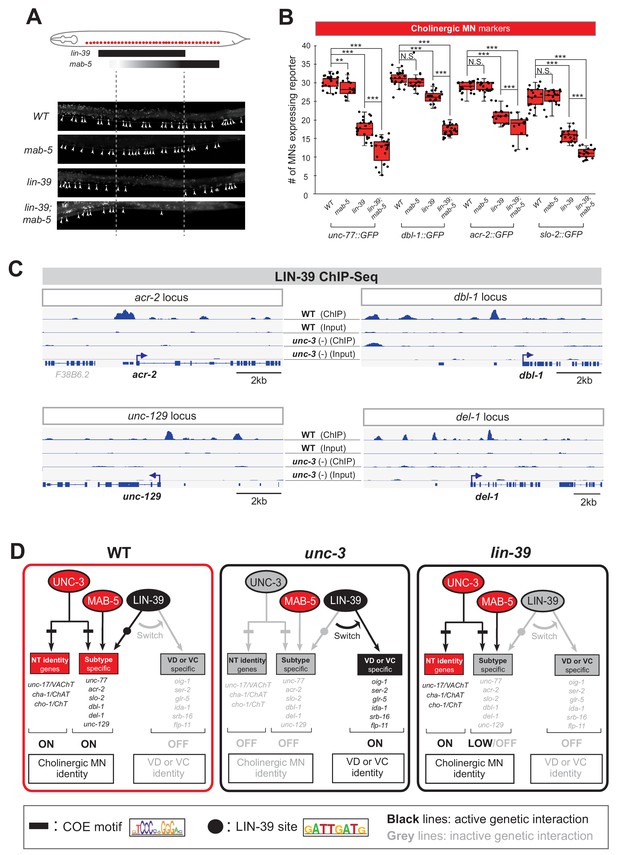
UNC-3 prevents a switch in the transcriptional targets of LIN-39 in cholinergic motor neurons.
(A) Schematic summarizing the expression pattern of lin-39 and mab-5 in VNC cholinergic MNs. Below, representative images are shown of unc-77::gfp in WT, lin-39 (n1760), mab-5 (1239) and lin-39 (n1760); mab-5 (1239) animals at L4 stage. Arrowheads point to MN cell bodies with gfp marker expression. Green fluorescence signal is shown in white for better contrast. Dotted black box indicates imaged area. (B) Quantification of cholinergic MN terminal identity markers (unc-77, dbl-1, acr-2, slo-2) in WT, lin-39 (n1760), mab-5 (1239) and lin-39 (n1760); mab-5 (1239) animals at L4. N > 15. **p<0.01; ***p<0.001. (C) ChIP-Seq tracks are shown for LIN-39 on four cholinergic MN terminal identity genes (acr-2, unc-129, dbl-1, del-1). The WT data come from the modENCODE project (Boyle et al., 2014), whereas the unc-3 (-) data were obtained by performing ChIP-Seq for LIN-39 on unc-3 (n3435); lin-39 (kas9 [lin-39::mNG::3xFLAG::AID] animals. (D) Schematic showing the transcriptional switch in LIN-39 targets. In WT animals, UNC-3, MAB-5 and LIN-39 co-activate subtype-specific genes in cholinergic MNs (e.g., unc-77, dbl-1, unc-129, acr-2). In unc-3 mutants, LIN-39 is no longer able to activate these genes, and instead switches to VD- or VC-specific terminal identity genes. Black font: gene expressed. Gray font: gene not expressed. Gray arrows indicate inactive genetic interactions. COE motif taken from Kratsios et al. (2012) and LIN-39 site taken from Weirauch et al. (2014) are represented with black rectangles and dots, respectively.
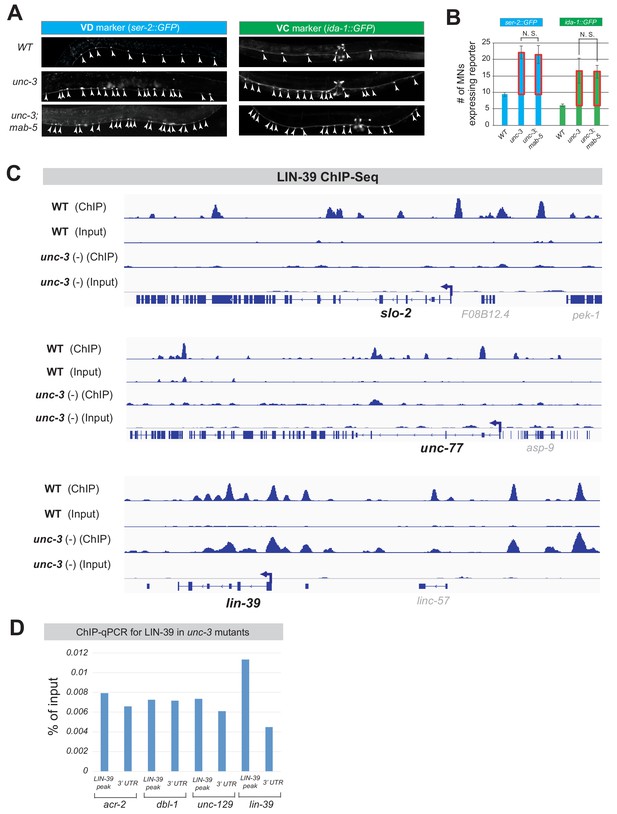
MAB-5 is not required for ectopic VD or VC marker expression and LIN-39 binding on cholinergic MN genes is affected in unc-3 mutants.
(A) The ectopic expression of VD (ser-2) or VC (ida-1) terminal identity markers in unc-3 mutants is not affected by loss of mab-5. Strong loss-of-function (null) alleles for unc-3 (n3435) and mab-5 (e1239) were used. Representative images of L4 stage animals are shown. Arrowheads point to MN cell bodies with gfp marker expression. Green fluorescence signal is shown in white for better contrast. (B) Quantification of data shown in panel D. N > 15. N. S: not significant. (C) ChIP-Seq tracks are shown for LIN-39 on two cholinergic MN terminal identity genes (slo-2, unc-77) and lin-39 locus (positive control). The WT data come from the modENCODE project (Boyle et al., 2014), whereas the unc-3 (-) data were obtained by performing ChIP-Seq for LIN-39 on unc-3 (n3435); lin-39 (kas9 [lin-39::mNG::3xFLAG::AID] animals. (D) ChIP-qPCR was performed on unc-3 (n3435); wgIs18 (lin-39 fosmid::GFP) animals to examine LIN-39 binding at four target genes (acr-2, dbl-1, unc-129, lin-39).
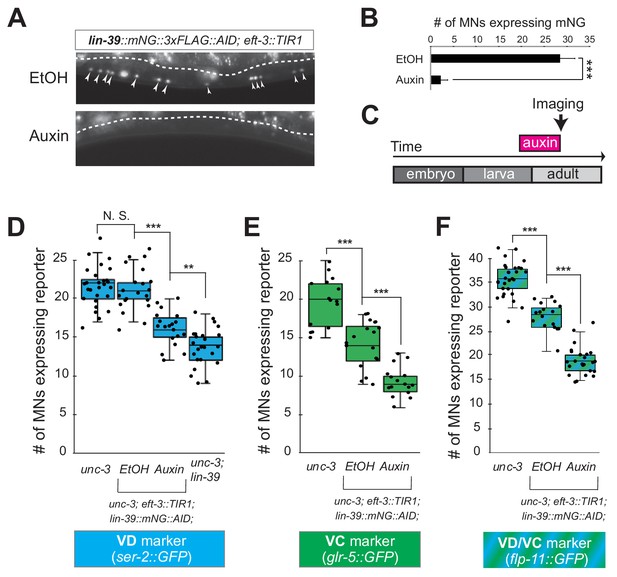
LIN-39 is continuously required to control expression of terminal identity genes.
(A) Animals of the lin-39::mNG::3xFLAG::AID; eft-3::TIR1 genotype were either administered ethanol (EtOH) or auxin at the L3 stage. Twenty four hours later, expression of endogenous lin-39 reporter (lin-39::mNG::3xFLAG::AID) is severely reduced in the nuclei of VNC MNs (arrowheads) at the young adult stage (day 1). mNG green fluorescent signal is shown in white for better contrast. White dotted line indicates the boundary of intestinal tissue (gut), which tends to be autofluorescent in the green channel. (B) Quantification of number of MNs expressing the lin-39::mNG::3xFLAG::AID reporter after EtOH (control) and auxin treatment. N > 14. ***p<0.001. (C) Schematic showing time window of auxin administration. (D–F) Auxin or ethanol (control) were administered at larval stage 4 (L4) on unc-3 (n3435); lin-39::mNG::3xFLAG::AID; eft-3::TIR1 animals carrying either the VD marker ser-2::gfp, the VC marker glr-5::gfp, or the VD/VC marker flp-11::gfp. Images were taken at the young adult stage (day 1.6 for ser-2, day 1.8 for glr-5 and day two for flp-11). A significant decrease in the number of MNs expressing the VD marker was evident in the auxin-treated animals compared to EtOH-treated controls. For comparison, quantification of marker expression is also provided in unc-3 (n3435) mutants. We note that hypomorphic effects in the ethanol treated group have been previously reported for other AID-tagged TFs in C. elegans (Kerk et al., 2017). Such effects appear to be target gene-specific, as they were observed for glr-5 and flp-11, but not ser-2 (Figure 6E–F). N > 15. **p<0.01, ***p<0.001, N. S: not significant.
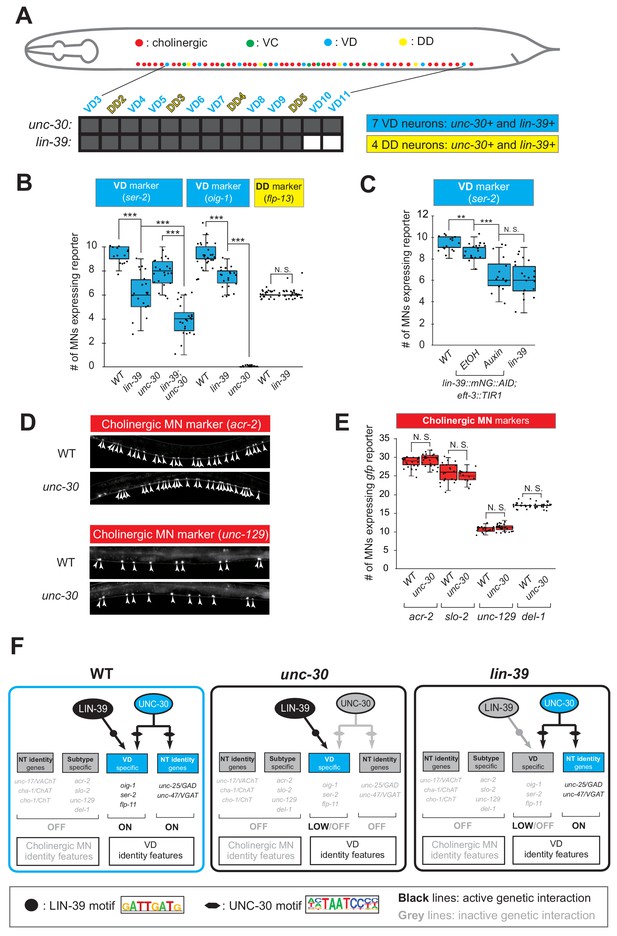
LIN-39 is an activator of VD terminal identity genes.
(A) Schematic summarizing unc-30 and lin-39 expression in VD and DD neurons populating the VNC. In addition, 4 VD and 2 DD neurons are located in ganglia flanking the VNC (not shown because they were excluded from our analysis). Raw data on lin-39 expression described in Figure 4—figure supplement 2. (B) Quantification of two VD (ser-2::gfp, oig-1::gfp) and one DD (flp-13::gfp) markers in WT and lin-39 (n1760) animals at L4. Both VD markers were also tested in unc-30 (e191) mutants. Double lin-39 (n1760); unc-30 (e191) mutants showed a more severe reduction in expression of the VD marker ser-2::gfp compared to each single mutant. N > 15. ***p<0.001. N. S: not significant. (C) Auxin or ethanol (control) were administered at larval stage 3 (L3) on lin-39::mNG::3xFLAG::AID; eft-3::TIR1 animals carrying the VD marker ser-2::gfp. Images were taken at the young adult stage (day 1.5). A significant decrease in the number of MNs expressing the VD marker was evident in the auxin-treated animals compared to EtOH-treated controls. Similar results were obtained when auxin administration occurred at L4 or day one adult animals. For comparison, quantification of marker expression is also provided in WT and lin-39 (n1760) animals. N > 15. **p<0.01, ***p<0.001. N. S: not significant. (D) Several terminal identity markers of cholinergic neurons (acr-2, slo-2, unc-129, del-1) are not ectopically expressed in unc-30-depleted GABAergic MNs. A strong loss-of-function allele e191 for unc-30 was used (Brenner, 1974; Eastman et al., 1999). Arrowheads point to MN cell bodies with gfp marker expression. Green fluorescence signal is shown in white for better contrast. (E) Quantification of data presented in panel D. N. S: not significant. (F) Schematic summarizing the function of LIN-39 and UNC-30 in GABAergic VD neurons. LIN-39 site is taken from Weirauch et al. (2014). UNC-30 site is taken from Yu et al. (2017).
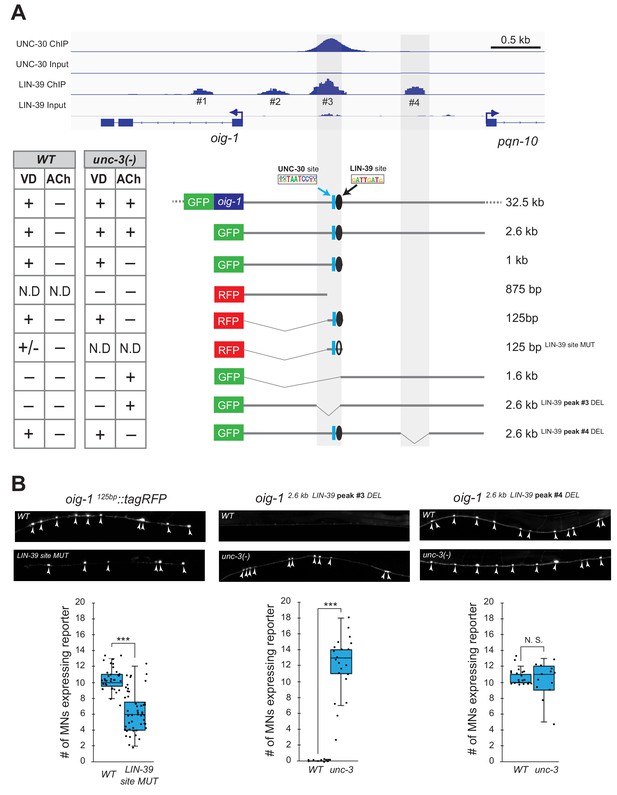
LIN-39 acts through distinct cis-regulatory elements to activate oig-1 expression in VD and VD-like neurons.
(A) ChIP-Seq tracks are shown for UNC-30 (the top two) and LIN-39 (the bottom two) on VD gene oig-1 locus. The UNC-30 data were obtained from Yu et al. (2017). The LIN-39 data come from the modENCODE project (Boyle et al., 2014). Four LIN-39 peaks are annotated with peak#3 largely overlapping with UNC-30 peak. The results of cis-regulatory analysis in both WT and unc-3 mutants are shown in the lower panel (aligned to the ChIP-seq tracks). Expression patterns of at least two transgenic lines were analyzed for each construct. ‘+' indicates consistent and bright expression in ventral nerve cord (VNC) MNs (either VD or cholinergic). ‘+/−' indicates consistent and bright expression in noticeable less number of VNC MNs. ‘−' indicates no or extremely dim expression in VNC MNs. ‘N.D.': Not determined. In the schematic of the transgenes, a known UNC-30 site is shown as a blue box and a bioinformatically predicted LIN-39 site is represented as a black circle (filled circle indicates the presence of the site while unfilled one indicates deletion of the site). MUT indicates deletion of the LIN-39 site and DEL indicates deletion of the respective LIN-39 peak region. (B) Images (top part) and quantifications (bottom part) of selected constructs in the cis-regulatory analysis shown in (A). Animals carrying the oig-1125bp::tagRFP (left panel) with the LIN-39 site deleted show reduced tagRFP reporter expression in VD neurons; animals carrying the oig-12.6kb LIN-39 peak #3 DEL (middle panel) ectopically express the reporter in cholinergic MNs of unc-3 mutants, but not in WT animals; animals carrying the oig-12.6kb LIN-39 peak #4 DEL (right panel) do not show ectopic reporter expression in unc-3-depleted MNs, but do show VD expression in both wild-type and unc-3 mutants. N > 12. ***p<0.001. N. S: not significant.
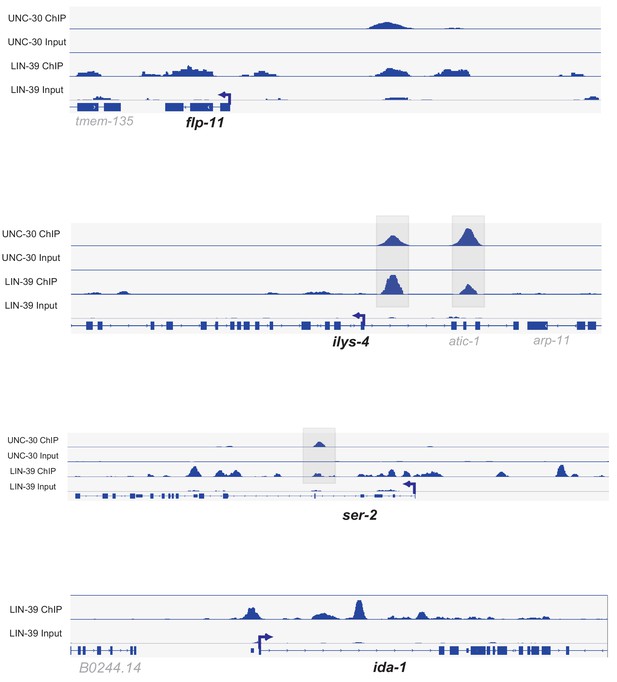
LIN-39 binds directly to the cis-regulatory region of VD and VC terminal identity genes.
ChIP-Seq tracks for UNC-30 and LIN-39 are shown for the VD-expressed genes flp-11, ilys-4, and ser-2. ChIP-Seq tracks for LIN-39 are shown for the VC-specific terminal identity gene ida-1. ChIP-Seq data for LIN-39 were generated through the modENCODE project (Boyle et al., 2014) and ChIP-Seq data for UNC-30 were previously generated (Yu et al., 2017). ChIP-seq data were visualized using Integrative Genomics Viewer (Siponen et al., 2010) (Thorvaldsdóttir et al., 2013) and snapshots of ChIP-Seq tracks were generated.
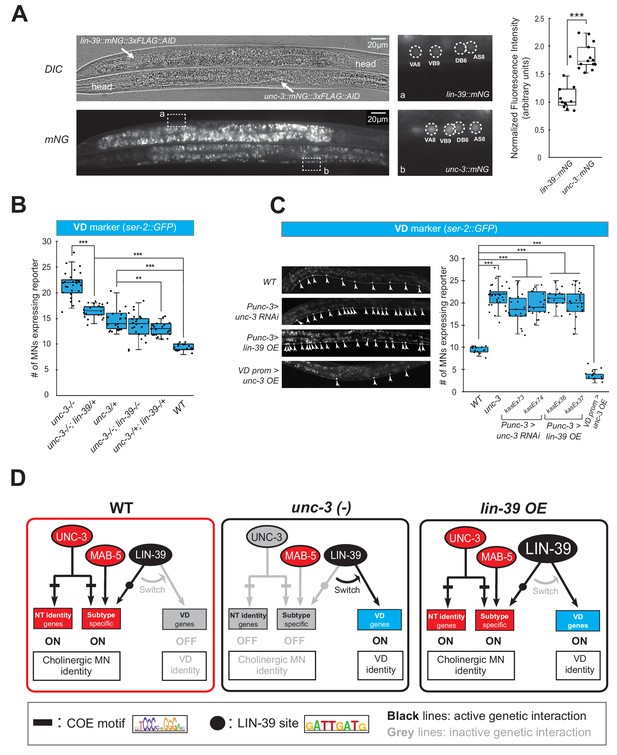
Gene dosage experiments suggest that LIN-39 is the rate-limiting factor.
(A) The endogenous expression levels of UNC-3 are higher than LIN-39. The DIC and mNG channels of two worms next to each other on the same slide with the genotype of lin-39::mNG::3xFLAG::AID (top, anterior right, ventral up) and unc-3::mNG::3xFLAG::AID (bottom, anterior left, ventral down) respectively are shown on the left. VNC regions indicated by dashed frame (a, b) are zoomed in at the middle panel with dashed circles around MN nuclei. The identities of these MNs are shown (e.g., VA8, VB9). The same cholinergic MNs have stronger expression levels of endogenous unc-3::mNG than lin-39::mNG. Quantification of the fluorescence intensities is shown on the right panel. For details on the quantification, see Materials and Materials and methods. N = 12. ***p<0.001. (B) Quantification of the VD marker (ser-2::gfp) in unc-3 (n3435), unc-3 (n3435); lin-39 (n1760)/+, unc-3 (n3435)/+, unc-3 (n3435); lin-39 (n1760), unc-3 (n3435)/+; lin-39 (n1760)/+, and WT animals at L4. N > 15. **p<0.01, ***p<0.001. (C) Representative images of the VD marker (ser-2::gfp) expression on the left in L4 stage transgenic animals that either down-regulate unc-3 in cholinergic MNs (Punc-3 >unc-3 RNAi), over-express lin-39 in cholinergic MNs (Punc-3 >lin-39 OE), or over-express unc-3 in VD neurons (VD prom [unc-47 prom]>unc-3 OE). Arrowheads point to MN cell bodies with gfp marker expression. Green fluorescence signal is shown in white for better contrast. Quantification is provided on the right. Two independent transgenic lines were used for Punc-3 >unc-3 RNAi and Punc-3 >lin-39 OE. N > 13. ***p<0.001. (D) Schematic summarizing the gene dosage experiments.
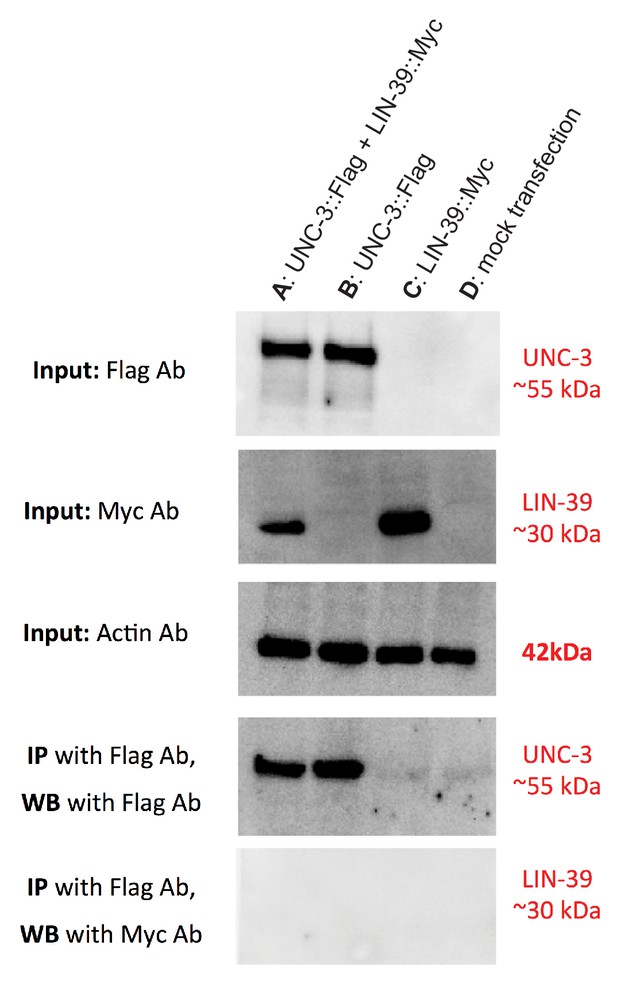
UNC-3 does not physically interact with LIN-39 in a heterologous system.
(A) HEK293 cells were transfected with two mammalian expression plasmids encoding Flag-tagged UNC-3 and a plasmid encoding Myc-tagged LIN-39, or (B) with a single plasmid encoding the Flag-tagged UNC-3, or (C) with a single plasmid encoding the Myc-tagged LIN-39, or (D) were mock transfected. 10 ug of the protein lysate were subjected to western blot analysis using antibodies against Flag, Myc and Actin (upper three panels). 100 ug of these cell lysates were used for immunoprecipitation of the Flag-tagged UNC-3, using anti-Flag antibody coated beads (lower two panels).
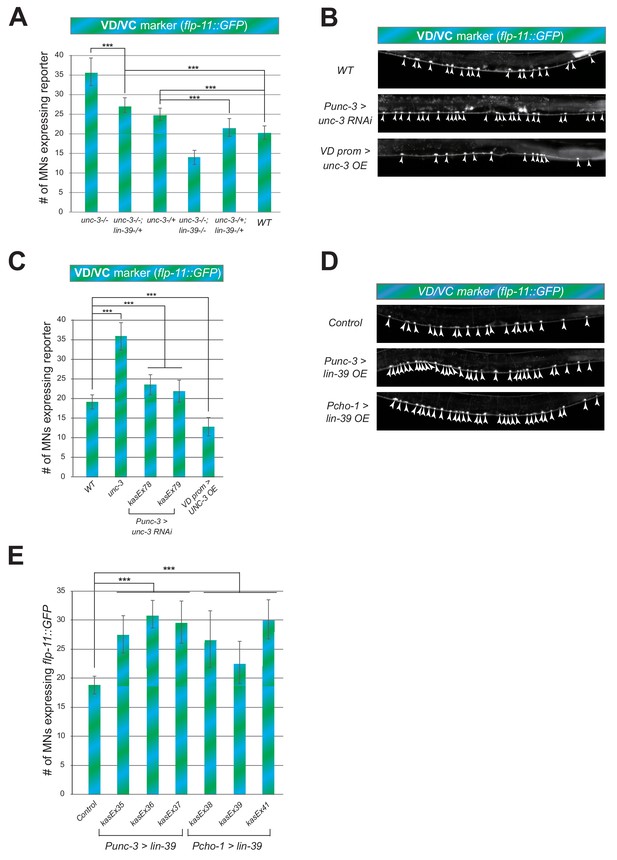
UNC-3 and LIN-39 levels are crucial for ectopic expression of VD/VC terminal identity marker flp-11.
(A) Quantification of the VD/VC marker flp-11::gfp in unc-3 (n3435), unc-3 (n3435); lin-39 (n1760)/+, unc-3 (n3435)/+, unc-3 (n3435); lin-39 (n1760), unc-3 (n3435)/+; lin-39 (n1760)/+, and WT animals at L4. N > 18. ***p<0.001. (B): Representative images of the VD/VC marker flp-11::gfp expression in L4 stage WT or transgenic animals that either down-regulate unc-3 in cholinergic MNs (Punc-3 >unc-3 RNAi) or over-express unc-3 in GABAergic (including VD) neurons (Punc-47 >unc-3 OE). Arrowheads point to MN cell bodies with gfp expression. Green fluorescence signal is shown in white for better contrast. (C) Quantification of data shown in panel B. Two independent transgenic lines were used for Punc-3 >unc-3 RNAi. N > 17. ***p<0.001. Quantification of flp-11::gfp is also shown in WT and unc-3 (n3435) null animals for comparison. (D) Representative images of the VD/VC marker flp-11::gfp expression in L4 stage WT or transgenic animals that over-express lin-39 in cholinergic MNs driven by two different cholinergic MN promoters(Punc-3 >lin-39 OE, Pcho-1 >lin-39 OE). Arrowheads point to MN cell bodies with marker expression. Green fluorescence signal is shown in white for better contrast. (E) Quantification of data shown in panel D. Three independent transgenic lines were used for Punc-3 >lin-39 OE and Pcho-1 >lin-39 OE. N = 19. ***p<0.001.
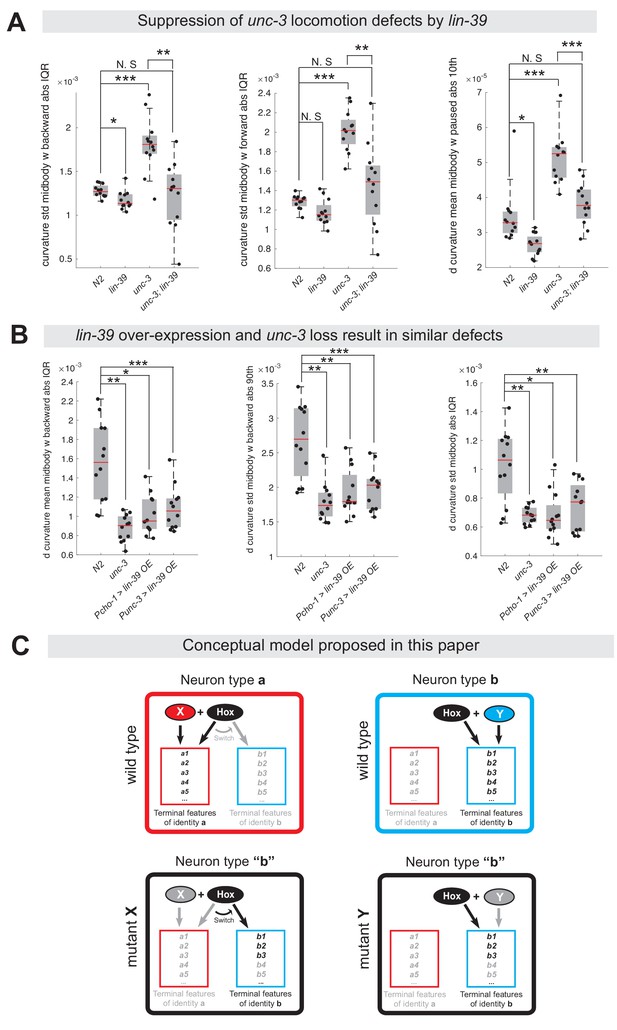
Ectopic expression of VD terminal identity genes in cholinergic motor neurons is associated with locomotion defects.
(A) Examples of three mid-body locomotion features that are significantly affected in unc-3 (n3435) animals, but markedly improved in unc-3 (n3435); lin-39 (n1760) double mutant animals. Each black dot represents a single adult animal. The unit for the first two graphs is 1/microns. The unit for the graph on the right is 1/(microns*seconds). N = 12. Additional mid-body features affected in unc-3 (n3435) animals, but improved in unc-3 (n3435); lin-39 (n1760) mutants are provided in Figure 10—figure supplement 1 and Supplementary file 3. *p<0.01, **p<0.001, ***p<0.0001. (B) Examples of three mid-body locomotion features affected in unc-3 (n3435) mutants and animals over-expressing lin-39 in cholinergic MNs. Each black dot represents a single adult animal. The unit for the Y axis is 1/(microns*seconds). N = 12. Additional mid-body features affected in unc-3 (n3435) and lin-39 over-expressing animals are provided in Figure 10—figure supplement 1 and Supplementary file 3. *p<0.01, **p<0.001, ***p<0.0001. (C) Conceptual model summarizing the findings of this paper. Gray font: not expressed gene. Black font: expressed gene. Gray arrow: inactive genetic interaction. Black arrow: active genetic interaction.
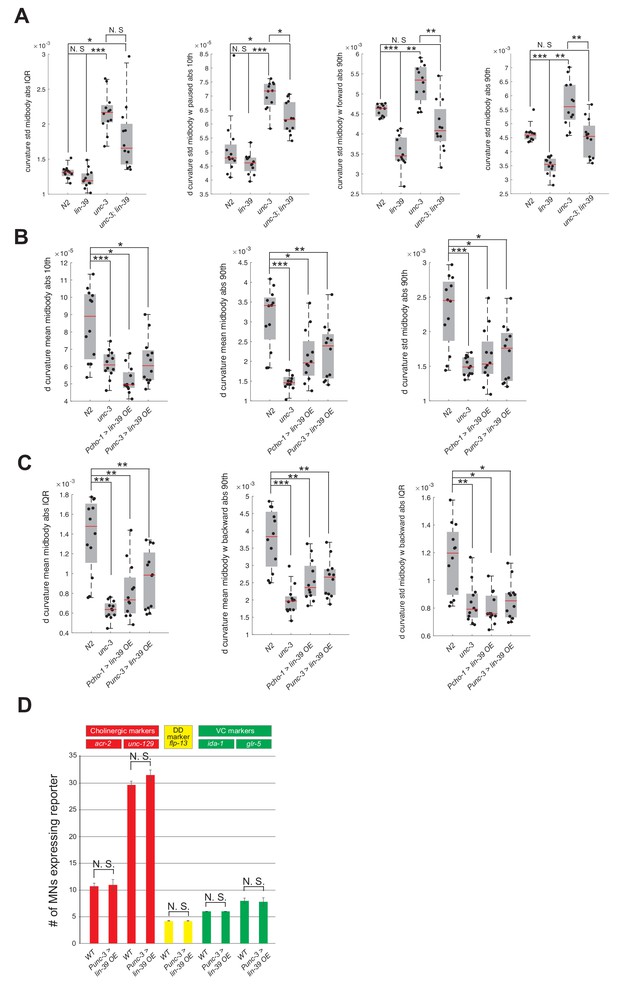
Automated worm tracking analysis on unc-3 and unc-3; lin-39 mutants.
(A) Additional mid-body locomotion features that are significantly affected in unc-3 (n3435) animals, but markedly improved in unc-3 (n3435); lin-39 (n1760) double mutant animals. Each black dot represents a single adult animal. The unit for the first two graphs is 1/microns. The unit for the graph on the right is 1/(microns*seconds). N = 12. For a comprehensive list of mid-body features see Supplementary file 3. *p<0.01, **p<0.001, ***p<0.0001. (B–C) Additional mid-body locomotion features affected in unc-3 (n3435) mutants and animals over-expressing lin-39 in cholinergic MNs. Each black dot represents a single adult animal. The unit for the y axis is 1/(microns*seconds). N = 12. For a comprehensive list of mid-body features see Supplementary file 3. *p<0.01, **p<0.001, ***p<0.0001. (D) Over-expression of LIN-39 in cholinergic MNs (Punc-3 >LIN-39 OE) does not affect expression of terminal identity genes specific to cholinergic (acr-2, unc-129), GABAergic DD (flp-13) or VC (ida-1, glr-5) motor neurons. L4 stage animals were imaged and quantified. N > 15.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Caenorhabditis elegans) | unc-3 | Wormbase | WBGene00006743 | |
| Gene (Caenorhabditis elegans) | unc-30 | Wormbase | WBGene00006766 | |
| Gene (Caenorhabditis elegans) | lin-39 | Wormbase | WBGene00003024 | |
| Gene (Caenorhabditis elegans) | mab-5 | Wormbase | WBGene00003102 | |
| Strain, strain background (Caenorhabditis elegans) | unc-3 (n3435) | Bob Horvitz (MIT, Cambridge MA) | MT10785 | Null Allele: deletion |
| Strain, strain background (Caenorhabditis elegans) | unc-30 (e191) | Caenorhabditis Genetics Center | CB845 | Allele: substitution |
| Strain, strain background (Caenorhabditis elegans) | lin-39(n1760)/dpy-17(e164) unc-32(e189) III. | Caenorhabditis Genetics Center | MT4009 | Null Allele: substitution |
| Strain, strain background (Caenorhabditis elegans) | mab-5 (n1239) III; him-5 (e1490) V | Caenorhabditis Genetics Center | CB3531 | Allele: substitution |
| Strain, strain background (Caenorhabditis elegans) | him-8 (e1489) IV | Caenorhabditis Genetics Center | CB1489 | Allele: substitution |
| Strain, strain background (Caenorhabditis elegans) | ieSi57 II; unc-3 (ot837 [unc-3::mNG::AID]) | Caenorhabditis Genetics Center | OH13988 | CRISPR-generated allele |
| Strain, strain background (Caenorhabditis elegans) | lin-39 (kas9 [lin-39::mNG::AID]) | This paper | KRA110 | See Materials and methods, Section Targeted genome editing |
| Strain, strain background (Caenorhabditis elegans) | ieSi57 [eft-3prom::tir1] | Caenorhabditis Genetics Center | CA1200 | Genotype: ieSi57 II; unc-119(ed3) III. |
| Strain, strain background (Caenorhabditis elegans) | ser-2::gfp | Caenorhabditis Genetics Center | OH2246 | Genotype: otIs107 I |
| Strain, strain background (Caenorhabditis elegans) | oig-1::gfp | Caenorhabditis Genetics Center | OH3955 | Genotype: pha-1(e2123) III; otEx193 |
| Strain, strain background (Caenorhabditis elegans) | ida-1::gfp | Caenorhabditis Genetics Center | BL5717 | Genotype: inIs179 II; him-8(e1489) IV |
| Strain, strain background (Caenorhabditis elegans) | glr-5::gfp | Aixa Alfonso (University of Illinois, Chicago IL) | AL270 | Genotype: icIs270 X |
| Strain, strain background (Caenorhabditis elegans) | srb-16::gfp | Caenorhabditis Genetics Center | BC14820 | Genotype:dpy-5(e907) I; sEx14820 |
| Strain, strain background (Caenorhabditis elegans) | flp-11::gfp | Caenorhabditis Genetics Center | NY2040 | Genotype: ynIs40 V |
| Strain, strain background (Caenorhabditis elegans) | twk-46::gfp | Caenorhabditis Genetics Center | BC13337 | Genotype: dpy-5(e907) I; sIs12928 V |
| Strain, strain background (Caenorhabditis elegans) | ilys-4::tagrfp | This paper | KRA22 | Genotype: pha-1(e2123) III; kasEx22 |
| Strain, strain background (Caenorhabditis elegans) | flp-13::gfp | Caenorhabditis Genetics Center | NY2037 | Genotype: ynIs37 III |
| Strain, strain background (Caenorhabditis elegans) | lin-11::mCherry | Oliver Hobert (Columbia University, New York NY) | OH11954 | Genotype: lin-11::mCherry + myo-2::GFP V |
| Strain, strain background (Caenorhabditis elegans) | klp-4::gfp | Caenorhabditis Genetics Center | BC11799 | Genotype: dpy-5(e907) I; sEx11799 |
| Strain, strain background (Caenorhabditis elegans) | alr-1::egfp | Caenorhabditis Genetics Center | OP200 | Genotype: unc-119(ed3) III; wgIs200 X |
| Strain, strain background (Caenorhabditis elegans) | irx-1::egfp | Caenorhabditis Genetics Center | OP536 | Genotype: unc-119(tm4063) III; wgIs536 I |
| Strain, strain background (Caenorhabditis elegans) | del-1::gfp | Caenorhabditis Genetics Center | NC138 | |
| Strain, strain background (Caenorhabditis elegans) | acr-2::gfp | Caenorhabditis Genetics Center | CZ631 | Genotype: juIs14 IV |
| Strain, strain background (Caenorhabditis elegans) | unc-129::gfp | Caenorhabditis Genetics Center | evIs82b | Genotype: evIs82b IV |
| Strain, strain background (Caenorhabditis elegans) | dbl-1::gfp | Caenorhabditis Genetics Center | BW1935 | Genotype: unc-119(ed3) III; ctIs43 him-5(e1490) V |
| Strain, strain background (Caenorhabditis elegans) | nca-1::gfp | Caenorhabditis Genetics Center | BC15028 | Genotype: dpy-5(e907) I; sEx15028 |
| Strain, strain background (Caenorhabditis elegans) | slo-2::gfp | Caenorhabditis Genetics Center | BC10749 | Genotype: dpy-5(e907) I; sEx10749 |
| Strain, strain background (Caenorhabditis elegans) | ttr-39::mCherry | Caenorhabditis Genetics Center | CZ8332 | Genotype: juIs223 IV |
| Strain, strain background (Caenorhabditis elegans) | cho-1::rfp | Caenorhabditis Genetics Center | OH13646 | Genotype: pha-1(e2123) III; him-5(e1490) otIs544 V |
| Strain, strain background (Caenorhabditis elegans) | unc-17::gfp | Caenorhabditis Genetics Center | LX929 | Genotype: vsIs48 X |
| Strain, strain background (Caenorhabditis elegans) | unc-25::gfp | Caenorhabditis Genetics Center | CZ13799 | Genotype: juIs76 II |
| Strain, strain background (Caenorhabditis elegans) | unc-47::mChOpti | Caenorhabditis Genetics Center | OH13105 | Genotype: him-5(e1490) otIs564 V |
| Strain, strain background (Caenorhabditis elegans) | unc-30::gfp | Caenorhabditis Genetics Center | OP395 | Genotype: unc-119(tm4063) III; wgIs395 |
| Strain, strain background (Caenorhabditis elegans) | ser-2::rfp | Mark Alkema (University of Massachusetts, Worcester MA) | AL270 | Genotype: zfIs8 IV |
| Strain, strain background (Caenorhabditis elegans) | oig-1(fosmid)::GFP | Caenorhabditis Genetics Center | OH11809 | Genotype: otIs450 |
| Strain, strain background (Caenorhabditis elegans) | lin-39::gfp | Caenorhabditis Genetics Center | OP18 | Genotype: unc-119(ed3) III; wgIs18 |
| Genetic reagent (Caenorhabditis elegans) | Poig-1_1 kb::gfp | Oliver Hobert (Columbia University, New York NY) | otEx5993 otEx5994 otEx5995 | |
| Genetic reagent (Caenorhabditis elegans) | Poig-1_1.6 kb::gfp | This paper | kasEx147 kasEx148 | See Materials and methods |
| Genetic reagent (Caenorhabditis elegans) | Poig-1_2.6 kb_LIN-39 site #3 DEL::gfp | This paper | kasEx149 kasEx150 | See Materials and methods |
| Genetic reagent (Caenorhabditis elegans) | Poig-1_2.6 kb_LIN-39 site #4 DEL::gfp | This paper | kasEx151 kasEx152 | See Materials and methods |
| Genetic reagent (Caenorhabditis elegans) | Poig-1_125 bp_::tagrfp | This paper | kasEx80 kasEx81 kasEx82 | See Materials and methods |
| Genetic reagent (Caenorhabditis elegans) | Poig-1_ LIN-39 site mut 125 bp_::tagrfp | This paper | kasEx91 kasEx92 kasEx93 | See Materials and methods |
| Genetic reagent (Caenorhabditis elegans) | Punc-3_558bp > lin-39 RNAi + myo-2::gfp | This paper | kasEx68 kasEx69 kasEx70 kasEx71 kasEx72 | See Materials and methods |
| Genetic reagent (Caenorhabditis elegans) | Punc-3_558bp > unc-3 RNAi + myo-2::gfp | This paper | kasEx73 kasEx74 kasEx78 kasEx79 | See Materials and methods |
| Genetic reagent (Caenorhabditis elegans) | Punc-3_558bp > lin-39 cDNA OE + myo-2::gfp | This paper | kasEx35 kasEx36 kasEx37 kasEx76 kasEx77 | See Materials and methods |
| Genetic reagent (Caenorhabditis elegans) | Punc-47 > unc-3 cDNA + myo-2::gfp | This paper | kasEx75 | See Materials and methods |
| Genetic reagent (Caenorhabditis elegans) | Pcho-1_280bp > lin-39 cDNA OE + myo-2::gfp | This paper | kasEx38 kasEx39 kasEx41 | See Materials and methods |
| Genetic reagent (Caenorhabditis elegans) | lin-39 fosmid WRM0616aE11 + myo-2::gfp | This paper | kasEx33 kasEx34 | See Materials and methods |
| Antibody | anti-Myc (Rabbit polyclonal) | Abcam | #ab9106; RRID:AB_307014 | 1:1000 dilution |
| Antibody | anti-Flag (Mouse monoclonal) | Sigma | #F3165; RRID:AB_259529 | 1:1000 dilution |
| Antibody | anti-Flag (Rabbit polyclonal) | Sigma, | #SAB4301135; RRID: AB_2811010 | 1:1000 dilution |
| Antibody | Clean-Blot IP Detection Reagent (Mouse monoclonal) | Thermo Fisher | #21230; RRID: AB_2576514 | See Materials and methods |
| Antibody | Flag antibody coated beads (Mouse monoclonal) | Sigma, | #A2220; RRID:AB_10063035 | See Materials and methods |
| Antibody | anti-FLAG M2 magnetic beads (Mouse monoclonal) | Sigma-Aldric | M8823; RRID: AB_2637089 | See Materials and methods |
| Recombinant DNA reagent | pcDNA 3.1(+)- C-Flag (Plasmid) | Genscript | pcDNA 3.1(+) | C-terminus Flag-tagged UNC-3 |
| Recombinant DNA reagent | pcDNA 3.1(+)-N-Myc (Plasmid) | Genscript | pcDNA 3.1(+) | N-terminus Myc-tagged LIN-39 |
| Recombinant DNA reagent | Fosmid clone WRM0616aE11 | Source BioScience | WRM0616aE11 | lin-39::GFP fosmid clone |
| Commercial assay or kit | Gibson Assembly Cloning Kit | NEB | #5510S | |
| Commercial assay or kit | QIAquick PCR Purification Kit | QIAGEN | #28104 | |
| Commercial assay or kit | Ampure XP beads | Beckman Coulter Life Sciences | A63881 | |
| Commercial assay or kit | TOPO XL-2 Complete PCR Cloning Kit | Thermo Fisher | K8050 | |
| Chemical compound, drug | Auxin (indole-3-acetic acid) | Alfa Aesar | #10196875 | |
| Software, algorithm | ZEN | ZEISS | Version 2.3.69.1000, Blue edition | RRID:SCR_013672 |
| Software, algorithm | Image J | Image J | Version 1.52i | RRID:SCR_003070 |
| Software, algorithm | RStudio | RStudio | Version 1.2.5001 | |
| Software, algorithm | Adobe Photoshop CS6 | Adobe | Version 13.0 × 64 | |
| Software, algorithm | Adobe Illustrator CS6 | Adobe | Version 16.0.0 × 64 |
Additional files
-
Supplementary file 1
UNC-3 binding sites (COE motifs) are not found in the cis-regulatory region of VD- and VC-expressed terminal identity genes.
- https://cdn.elifesciences.org/articles/50065/elife-50065-supp1-v1.docx
-
Supplementary file 2
LIN-39/Hox targets in cholinergic and GABAergic (VD) motor neurons.
Asterisk (*) highlights novel LIN-39 targets; N. D: Not Determined. The selected cis-regulatory regions are LIN-39 ChIP-seq peaks that fall within the DNA sequence used for our reporter gene constructs (except for del-1). The UNC-3 binding sites (COE motifs 23 bp) have been previously described in Kratsios et al. (2012). The LIN-39 binding sites were predicted by a FIMO search (p<0.005). The UNC-30 binding site on ser-2 locus was predicted by a FIMO search. The UNC-30 site on oig-1 was experimentally validated in Howell et al. (2015).
- https://cdn.elifesciences.org/articles/50065/elife-50065-supp2-v1.docx
-
Supplementary file 3
Locomotion features assessed by automated worm tracking analysis.
- https://cdn.elifesciences.org/articles/50065/elife-50065-supp3-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/50065/elife-50065-transrepform-v1.pdf





