Single-cell analysis uncovers that metabolic reprogramming by ErbB2 signaling is essential for cardiomyocyte proliferation in the regenerating heart
Figures
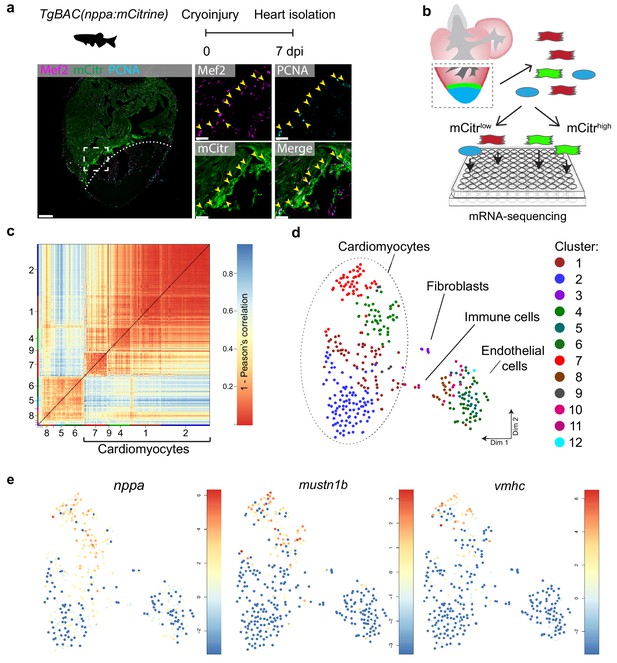
Single-cell mRNA sequencing identifies different cardiomyocyte-populations in the injured zebrafish heart.
(a) Schematic of cryoinjury procedure on adult TgBAC(nppa:mCitrine) fish and immunohistochemistry on section of injured TgBAC(nppa:mCitrine) heart 7 dpi. Overview image on left and zoom-in of boxed region on the right. Mef2 (in magenta) labels cardiomyocytes, nppa:mCitrine (in green) marks the borderr zone, and PCNA (in cyan) marks proliferating cells. Arrows indicate triple-positive cells. Dashed line indicates injury site. Scale bar in overview 50 μm. Scale bar in zoom-ins 20 μm. (b) Experimental outline of the single-cell mRNA-sequencing of injured zebrafish hearts (blue, injury area; green, border zone) (c) Pairwise correlation between individual cells across all genes detected. Color-code indicates cell-to-cell distances measured by [1 – Pearson’s correlation coefficient]. StemID clusters are indicated by color and number on the x- and y-axis. (d) t-distributed stochastic neighbor embedding (tSNE) map representation of transcriptome similarities between individual cells. (e) tSNE maps visualizing log2-transformed read-counts of the border zone marker genes nppa, mustn1b and vmhc.
-
Figure 1—source data 1
Single-cell mRNA sequencing data from cryoinjured TgBAC(nppa:mCitrine) zebrafish hearts at 7 dpi.
Raw reads are represented per gene (rows) and single cell (columns).
- https://cdn.elifesciences.org/articles/50163/elife-50163-fig1-data1-v2.xlsx
-
Figure 1—source data 2
List of the clusters in the adult injured heart (raw data see SD1), as identified by StemID and the associated cells per cluster.
- https://cdn.elifesciences.org/articles/50163/elife-50163-fig1-data2-v2.xlsx
-
Figure 1—source data 3
List of differentially expressed genes between cardiomyocytes clusters 2 and 7 of the adult heart dataset.
Only genes with a p-value<0.05 are listed. Per gene, the log2 fold change, adjusted p-value (padj) and associated gene name are given. GO-terms for genes upregulated between clusters 2 and 7 (p<0.01) are listed in separate excel sheets.
- https://cdn.elifesciences.org/articles/50163/elife-50163-fig1-data3-v2.xlsx
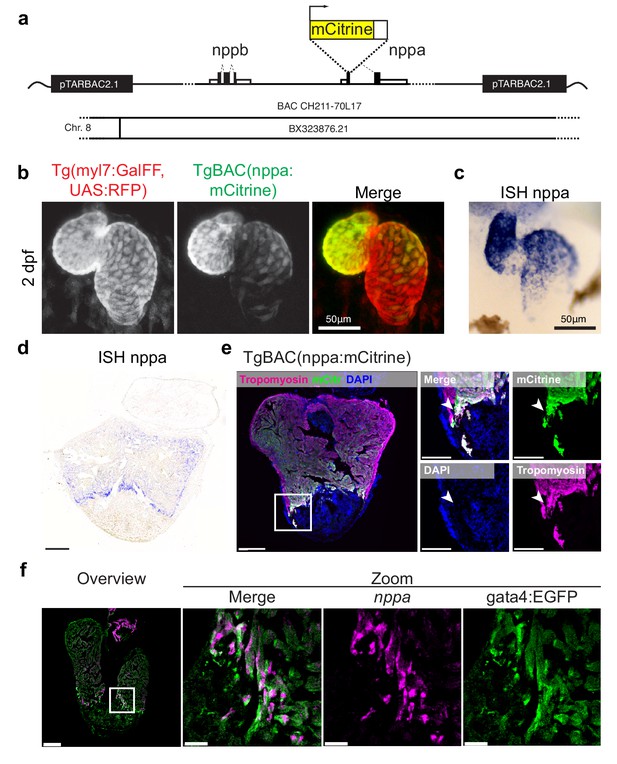
TgBAC(nppa:mCitrine) expression recapitulates endogenous nppa gene expression.
(a) Design of the bacterial artificial chromosome (BAC) used for generation of the transgenic line TgBAC(nppa:mCitrine). (b) Transgenic mCitrine expression in the heart in relation to RFP expression in the whole myocardium of Tg(myl7:GFF, UAS:RFP, nppa:mCitrine) in embryos at 2 days post-fertilization (dpf). (c) Whole mount in situ hybridization for endogenous nppa expression in embryos at two dpf. Note the specific expression in ventricle and atrium and absence of expression in atrioventricular canal of the nppa:mCitrine transgene expression (in panel b) as well as for the endogenous nppa expression (in panel c). (d, e) Endogenous nppa expression (d) and nppa:mCitrine expression (e) in the adult heart at 7 days post-injury (dpi). Note expression of nppa:mCitrine in the cortical borderzone (arrowheads). (f) Fluorescent in situ hybridization for nppa performed on gata4:EGFP hearts 7dpi. Note the overlap of nppa and gata4:EGFP in trabecular borderzone cardiomyocytes.
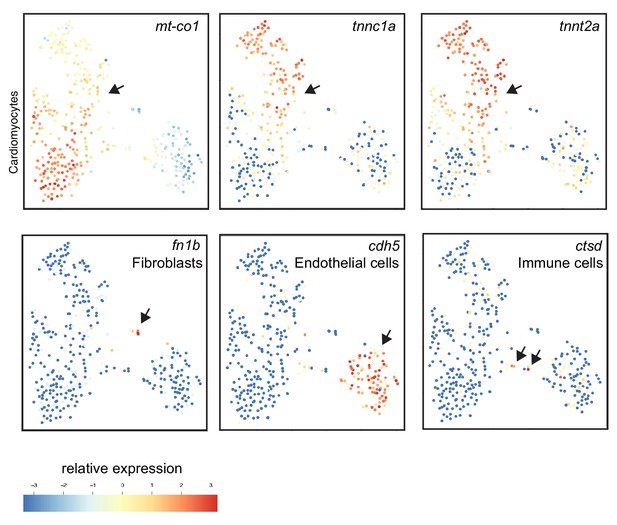
Single-cell mRNA sequencing identifies different cell-populations in the injured zebrafish heart.
tSNE maps visualizing log2-transformed read-counts of genes with high expression in cardiomyocytes (mt-co1, tnnc1a, tnnt2a), in fibroblasts (fn1b), in endothelial cells (cdh5) and immune cells (ctsd). Arrow indicates positive cell populations.
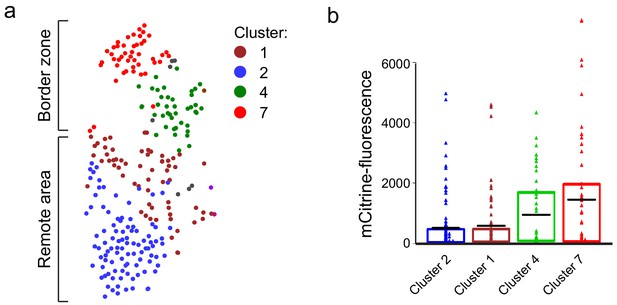
Cluster 7 cells display highest nppa:mCitrine expression.
(a) tSNE map of the cardiomyocyte clusters derived from the single-cell mRNA-sequencing. (b) mCitrine fluorescence levels of the nppa:mCitrine FACS sorted cells from injured hearts that were used for the single-cell RNA-sequencing. Triangles represent individual cells of the four cardiomyocyte clusters. The box indicates the 25–75% quartiles, black lines indicate mean-fluorescence per cluster.
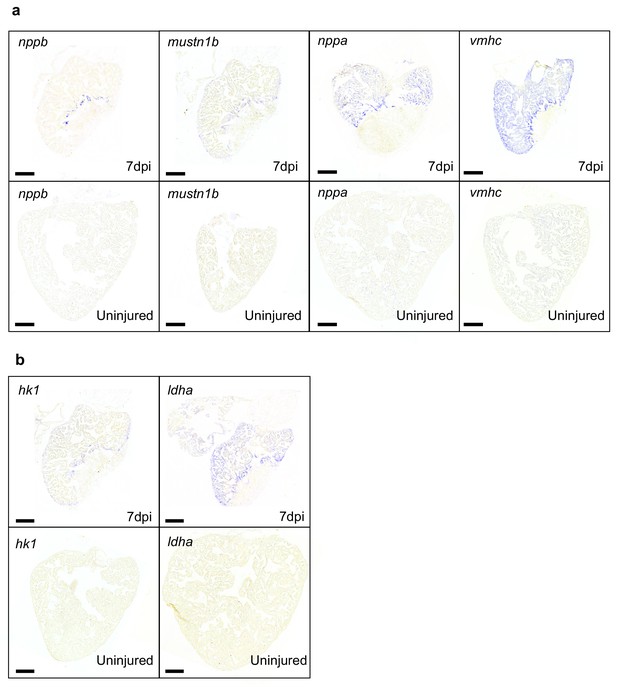
Enhanced expression of genes elevated in cluster 7 versus cluster 2 is injury induced.
(a) In situ hybridizations for border zone genes nppb, mustn1, nppa and vmhc in zebrafish hearts at seven dpi compared to uninjured hearts. (b) In situ hybridizations for glycolysis genes hk1 and ldha in zebrafish hearts at seven dpi compared to uninjured hearts. Staining for injured and uninjured hearts was stopped simultaneously. Scale bars indicate 200 μm. (n = 3 per condition).
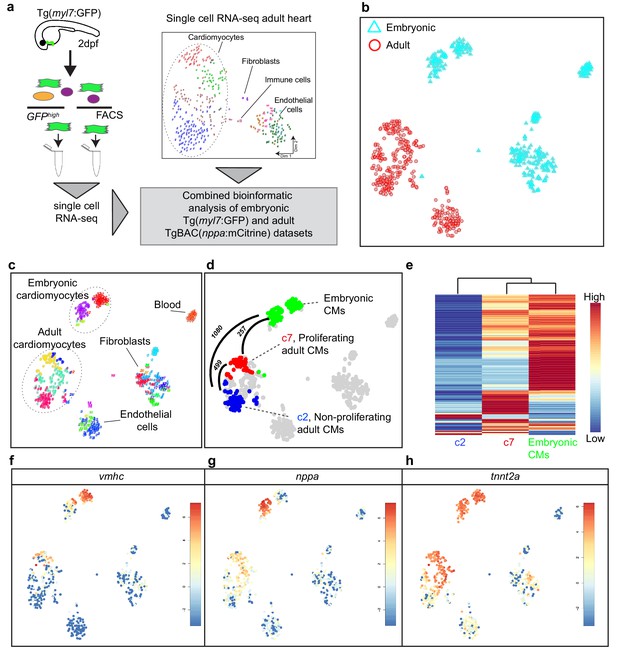
Single-cell transcriptome of border zone cardiomyocytes resembles that of embryonic cardiomyocytes.
(a) Cartoon to illustrate the experimental procedure for single cell analysis of embryonic and adult cardiac cells. (b) tSNE map of combined adult (red) and embryonic datasets (light blue). (c) tSNE map indicating the different cell types based on marker gene expression. (d) tSNE map with the adult cardiomyocytes of the injured heart (cluster 7 in red, cluster 2 in blue and clusters 1 and 4 in gray) and embryonic (2 dpf) cardiomyocytes (in green), with number of pairwise differentially expressed genes (p-value<0.01) indicated between cardiomyocyte clusters. (e) Heatmap with hierarchical clustering based on the 500 most differentially expressed genes between clusters. Red color represents high expression, blue color represents low expression. Rows represent individual genes. (f–h) tSNE maps visualizing log2-transformed read-counts of vmhc (f), nppa (g) and tnnt2a (h).
-
Figure 2—source data 1
Single-cell mRNA sequencing data from Tg(cmlc2:GFP) zebrafish hearts at 2 days post fertilization.
Raw reads are represented per gene (rows) and single cell (columns).
- https://cdn.elifesciences.org/articles/50163/elife-50163-fig2-data1-v2.csv
-
Figure 2—source data 2
List of the clusters from the embryonic heart data (SD5) identified by StemID and the associated cells for each cluster.
- https://cdn.elifesciences.org/articles/50163/elife-50163-fig2-data2-v2.xls
-
Figure 2—source data 3
Combined single-cell mRNA sequencing data from embryonic and adult hearts.
Raw reads represented per gene (rows) and single cell (columns).
- https://cdn.elifesciences.org/articles/50163/elife-50163-fig2-data3-v2.csv
-
Figure 2—source data 4
List of pairwise differentially expressed genes between all cardiomyocyte clusters from the combined embryonic and adult datasets.
Only genes with a p-value<0.01 are listed. Per gene, the log2 fold change, adjusted p-value (padj) and associated gene name are given. GO-terms for genes upregulated between clusters 5 and 6 (p<0.01) are listed in separate excel sheets.
- https://cdn.elifesciences.org/articles/50163/elife-50163-fig2-data4-v2.xls
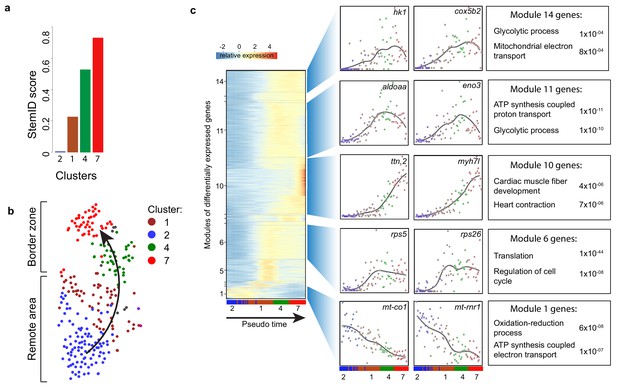
Pseudo time analysis reveals dedifferentiation and metabolic changes in border zone cardiomyocytes.
(a) Bar plot of StemID scores for the cardiomyocyte clusters (clusters #2, 1, 4 and 7) calculated by the formula: number of significant links for each cluster multiplied by the median transcriptome entropy across all cells in a cluster. (b) Cardiomyocyte clusters from adult injured heart. Arrow indicates the dedifferentiation path derived from the StemID scores. (c) Pseudo time analysis. Left; one-dimensional SOM of z-score transformed expression profiles along the differentiation trajectory incurred by StemID analysis. Y-axis represents the fourteen modules with differentially expressed genes. X-axis represents the pseudo time in which the cells were ordered. Middle; expression profiles of representative genes of the major modules. Y-axis shows transcript counts. X-axis represents the pseudo time. Right; Major gene ontology terms derived from all genes expressed in the module with p-values.
-
Figure 3—source data 1
List of genes that are differentially upregulated in the respective modules identified in the pseudo timeline analysis (related to Figure 3c).
- https://cdn.elifesciences.org/articles/50163/elife-50163-fig3-data1-v2.xlsx
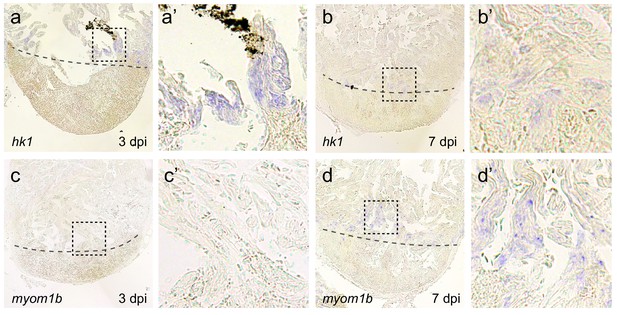
Induction of hexokinase1 expression precedes expression of myomesin1b.
In situ hybridization for the glycolysis gene hexokinase1 (a,b) and the embryonic cardiac gene myomesin1b (c,d) on sections of injured zebrafish hearts at 3 dpi (a,c) and 7 dpi (b,d). While hk1 expression in border zone cardiomyocytes was visible at 3 dpi and 7 dpi, myom1b expression was visible at 7 dpi but undetectable at 3dpi. (a’,b’,c’ and d’) show zoom-in of boxed areas in corresponding panels. Dashed line indicates the injury site.
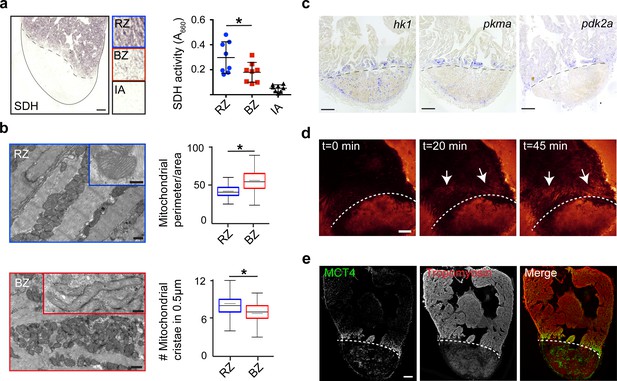
Border zone cardiomyocytes undergo a metabolic switch from mitochondrial OXPHOS to glycolysis.
(a) Succinate dehydrogenase (SDH) enzyme staining on a seven dpi heart section with injury area separated by dashed line. Quantification of SDH activity in remote zone (RZ), border zone (BZ) and injury area (IA). Scale bar indicates 100 μm. Error bars indicate mean and standard deviation. (b) Transmission electron microscopy (TEM) images of mitochondria in cardiomyocytes from the remote zone and the border zone of a 7 dpi injured heart. Note the disorganized and irregular shaped mitochondria in the border zone cardiomyocyte. Scale bar 500 nm (200 nm in inserts). Graphs show quantification of mitochondrial perimeter-to-area as a measurement for roundness and quantification of mitochondrial cristae density. * p-value<0.05. (c) In situ hybridizations for glycolytic genes hk1, pkma and pdk2a on sections of injured zebrafish hearts at 7 dpi. Dashed line indicates injury site. Scale bars indicate 100 μm. (d) Time-lapse multi-photon confocal images of whole heart. The heart was isolated at 7 dpi and incubated with 2-NBDG, a fluorescent glucose analogue, at t = 0. Dotted line indicates injury area. Arrows point to regions of the border zone. Scale bar represents 100 μm. (e) Confocal image of injured zebrafish hearts at 7 dpi stained for the lactate transporter MCT4 (green) and Tropomyosin (red). Dashed line indicates injury site.
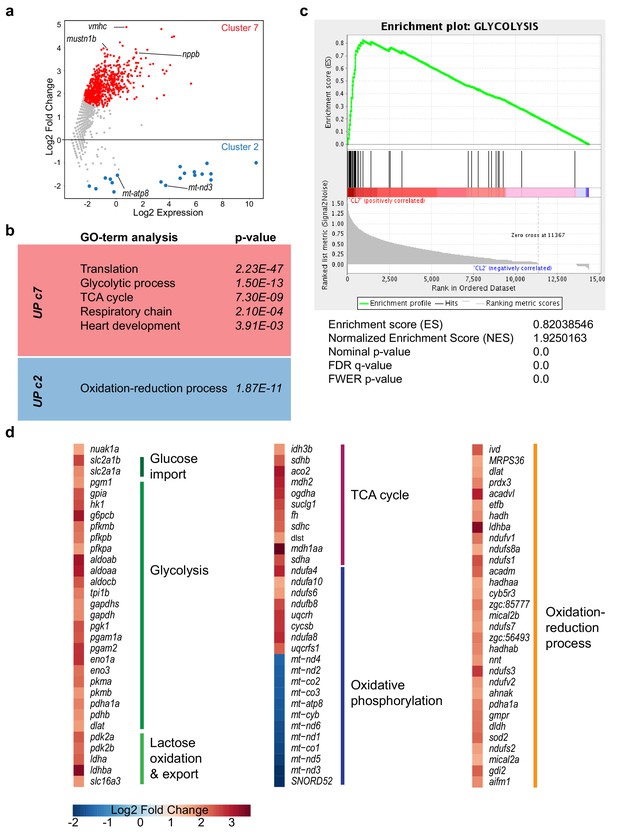
Energy metabolism genes are differentially expressed between cluster 7 and cluster 2 cells.
(a) Plot showing differentially expressed genes between cluster 7 versus cluster 2 cells. Differentially expressed genes (p-value<0.05) are highlighted in red (upregulated in cluster 7 cells) and blue (upregulated in cluster 2 cells). Complete gene list can be found in Figure 1—source data 3. 771 genes were differentially expressed (p<0.05; adjusted p-value after Benjamini-Hochberg correction), of which 752 were specifically upregulated in cluster seven cardiomyocytes, including the border zone genes, nppa, nppb and mustn1b. (b) GO-terms for upregulated genes in cluster 7 and cluster two respectively (p<0.01). (c) Gene set enrichment analysis for glycolysis genes on all genes with differential expression between cluster 7 versus cluster 2 cells (p<0.05). (d) Log2 fold change of differentially expressed genes (cluster 7 vs cluster 2) with a function in energy metabolism. High relative expression in cluster seven depicted in red, high expression in cluster two depicted in blue. In situ hybridization for the glycolysis gene hexokinase1 (a,b) and the embryonic cardiac gene myomesin1b (c,d) on sections of injured zebrafish hearts at three dpi (a,c) and seven dpi (b,d). While hk1 expression in border zone cardiomyocytes was visible at 3 dpi and seven dpi, myom1b expression was visible at seven dpi but undetectable at 3dpi. (a’,b’,c’ and d’) show zoom-in of boxed areas in corresponding panels. Dashed line indicates the injury site.
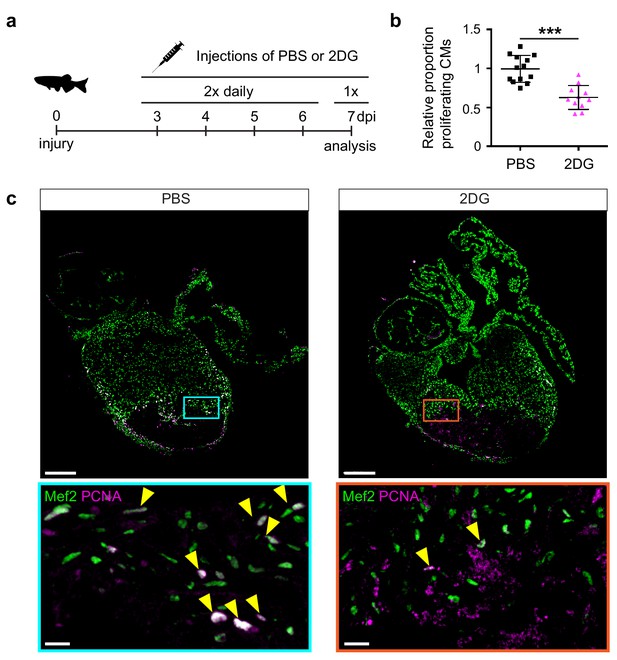
2-Deoxy glucose impairs cardiomyocyte proliferation.
(a) Experimental design for the 2-DG injections to inhibit glycolysis in injured zebrafish hearts. (b) Confocal image of injured zebrafish hearts at seven dpi either injected with PBS or 2-DG stained for Mef2c (green) and PCNA (magenta). Zoom-in images of the borderzone are shown below overview pictures for both PBS (cyan box) and 2-DG (orange box). Arrowheads indicate nuclei positive for Mef2c and PCNA. Scale bar indicates 200 μm (overview) or 20 μm (zoom-in). (c) Quantification of the proliferating cardiomyocytes (double Mef2c/PCNA positive) in the border zone of PBS and 2-DG treated hearts represented as the proportion of proliferating cardiomyocytes compared to the average percentage in the PBS injected group. Each dot represents a single heart (three sections per heart analyzed). Hearts were pooled from two separate experiments. Error bars represent mean ± standard deviation. ***, p<0,0001.
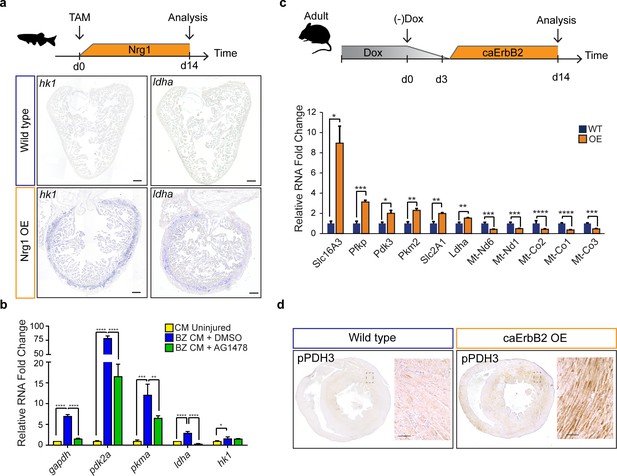
Nrg1/ErbB2 signaling induces glycolysis genes in zebrafish and mouse.
(a) Cartoon showing experimental procedure to induce cardiomyocyte specific Nrg1 expression in zebrafish. Panels show in situ hybridization for hexokinase 1 (hk1) and lactate dehydrogenase a (ldha) expression on sections of control hearts (β-act2:BSNrg1) and Nrg1 OE hearts (cmlc2:CreER; β-act2:BSNrg1). Scale bars represent 100 μm. (b) qPCR results for glycolytic genes showing their relative fold change in DMSO treated (n = 9) (blue) and AG1478 treated (n = 9) (green) nppa:mCitrine high border zone cardiomyocytes at 3dpi compared to uninjured adult cardiomyocytes (n = 4) (yellow). Error bars represent standard deviation. (c) Upper panel: Cartoon showing the experimental procedure to analyse metabolic gene expression after activating ErbB2 signaling in the murine heart. Lower panel: qPCR results for metabolic genes showing their relative fold change in caErbB2 OE (n = 4) heart compared to control WT hearts (n = 4). Error bars represent standard deviation. (d) Immunohistochemistry for phospho-PDH3 on sections of control and caErbB2 OE hearts. Scale bars represent 100 μm. *=p < 0.05, **=p < 0.01, ***=p < 0.001, ****=p < 0.0001.
-
Figure 6—source data 1
Oligonucleotide sequences for real-time PCR analysis.
- https://cdn.elifesciences.org/articles/50163/elife-50163-fig6-data1-v2.xlsx
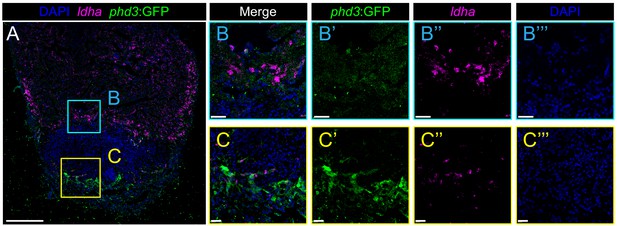
Hypoxia sensor phd3:GFP expression does not correlate with expression of the glycolytic gene ldha.
(a) Overview picture of phd3:GFP heart 7dpi co-stained for expression of ldha by fluorescent ISH. (b) Zoom-ins of trabecular border zone cardiomyocytes. Note high ldha expression and low expression of phd3:GFP. (c) Zoom-ins of the apical region of the injury. Note the abundance of phd3:GFP signal in this region, while ldha expression is low.
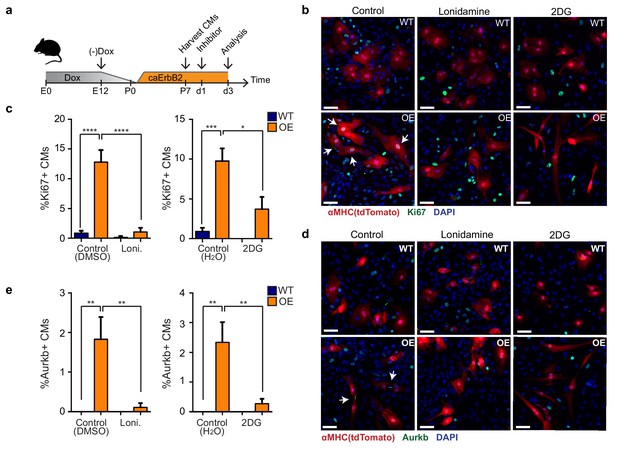
Glycolysis inhibitors impair mitogenic effect of ErbB2 activation in cardiomyocytes.
(a) Cartoon showing the experimental procedure to analyse the effects of glycolysis inhibitors (2-DG and lonidamine) on cardiomyocyte proliferation. (b) Immunofluorescence analysis on P7 cardiac cultures derived from WT and caErbB2 OE hearts that are endogenously fluorescent for tdTomato under the MYH6 promoter, stained for the cell-cycle marker Ki67. Arrows point at Ki67+ CMs. (c) Quantification of % Ki67+ CMs from WT and caErbB2 OE derived from P7 cardiac cultures treated with the glycolysis inhibitors 2-DG (n = 4 for WT and n = 4 for OE), and lonidamine (n = 7 for WT and n = 4 for OE) or their diluents as controls. (d) Immunofluorescence analysis on P7 cardiac cultures derived from WT and caErbB2 OE hearts that are endogenously fluorescent for tdTomato under the MYH6 promoter, stained for the cytokinesis marker Aurora kinase B. Arrows point at Aurkb+ CMs. (e) Quantification of % Aurkb+ CMs from WT and caErbB2 OE derived from P7 cardiac cultures treated with the glycolysis inhibitors 2-DG (n = 4 for WT and n = 4 for OE), and lonidamine (n = 7 for WT and n = 4 for OE) or their diluents as controls. In all panels, bars represent the mean, and error bars represent s.e.m *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, Scale bars represent 50 μm.
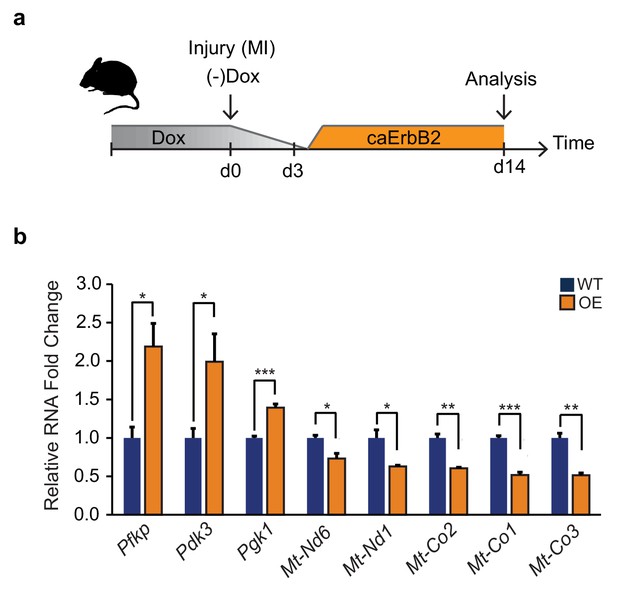
Activation of ErbB2 signaling in murine MI model induces glycolytic gene expression while repressing mitochondrial genes.
(a) Cartoon showing experimental procedure. Myocardial infarction (MI) was induced by left anterior descending coronary artery ligation at the same day when Dox was removed to induce cardiomyocyte specific caErbB2 overexpression (OE). Whole hearts where isolated for mRNA extraction 14 days after the MI and caErbB2 induction. (b) qPCR results with relative mRNA fold changes comparing wild type (WT, blue bars) with caErbB2 OE (OE, orange bars). Note the significant upregulation of glycolytic genes (Pfkp, Pdk3 and Pgk1) and significant downregulation of genes transcribed from the mitochondrial DNA (Mt-Nd1, Mt-Co2, Mt-Co1 and Mt-Co3). Bars represent mean values and error bars represent standard deviation. *p<0,05; **p<0,01; ***p<0001.
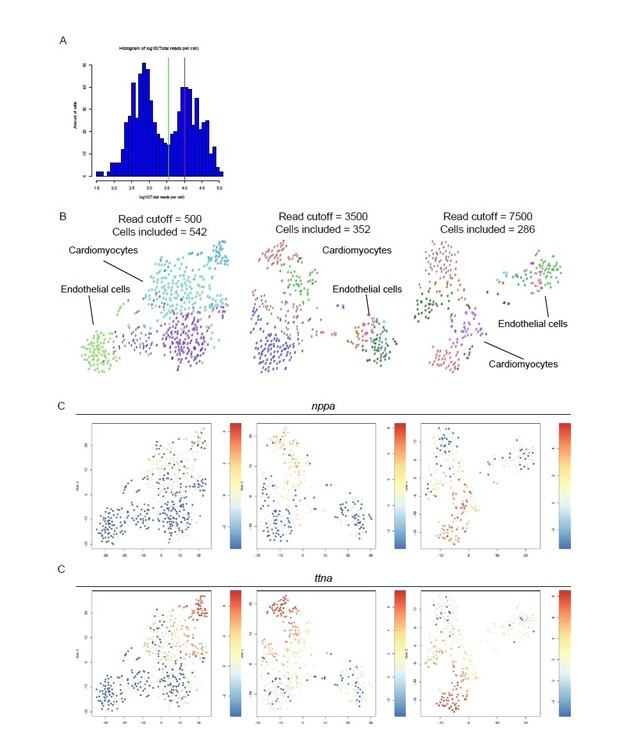
Different read cut-offs do not alter clustering pattern, while altering number of included cells.
(A) Distribution of reads per cell. Green line represents 3,500 reads, red line indicates the mean number of reads per cell (10,443). (B) tSNE maps generated with different read cutoffs. Read cutoffs and included number of cells are indicated on top of tSNE maps. (C) Expression of two marker genes of dedifferentiated cardiomyocytes: nppa and ttna.
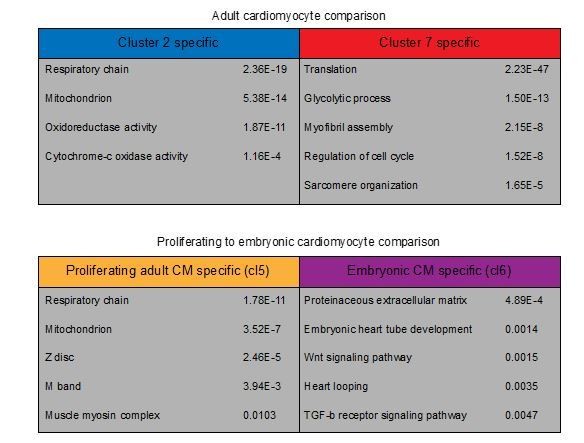
GO-term analysis on differentially expressed genes in cardiomyocyte clusters.
Representative GO-terms for cardiomyocyte clusters. GO-terms in upper table are obtained from differentially expressed genes between adult clusters 2 and 7 (p<0.01). Lower table GO-terms are obtained from differentially expressed genes between cluster 5 (Proliferating adult CM) and cluster 6 (Embryonic CM) of the combined single-cell analysis.
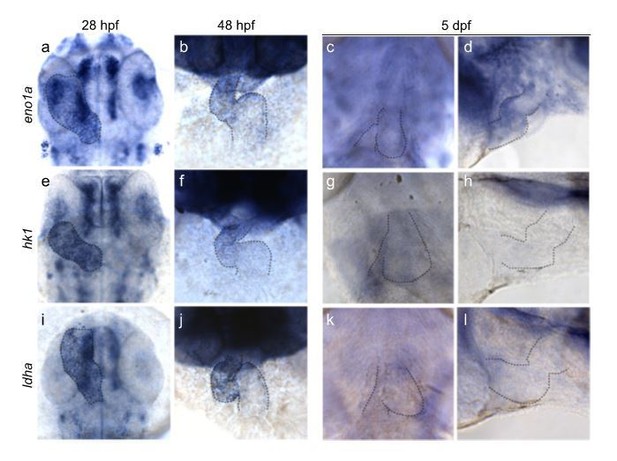
Glycolytic gene expression in the embryonic zebrafish heart.
Expression of eno1a (a, b, c, d), hk1 (e, f, g, h) and ldha (i, j, k, l) during zebrafish development at 28hpf (a, e, i), 48 hpf (b, f, j) and 5dpf (c, d, g, h, k, l) respectively. Outline of the heart is indicated by dashed lines.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Danio rerio) | Tupfel Long Fin (TL) | ZIRC | ZDB-GENO-990623–2 | |
| Genetic reagent (Danio rerio) | Tg(myl7:dsRED)s879Tg | Chi et al., 2008 | ZDB-FISH-150901–3078 | |
| Genetic reagent (Danio rerio) | Tg(myl7:GFP)twu34Tg | Huang et al., 2003 | ZDB-FISH-150901–212 | |
| Genetic reagent (Danio rerio) | Tg(phd3:GFP)sh144 | Santhakumar et al., 2012 | ZDB-FISH-150901–26851 | |
| Genetic reagent (Danio rerio) | Tg(gata4:EGFP)ae1 | Heicklen-Klein and Evans, 2004 | ZDB-FISH-150901–14762 | |
| Genetic reagent (Danio rerio) | TgBAC(nppa:mCitrine) | This paper | More info on generation of this line can be obtained from the Materials and methods section ‘Transgenic zebrafish lines and cryoinjury’. | |
| Cell line (Mus musculus) | TetRE-caErbb2 X MYH6-tTA X MYH6-cre ROSA26-tdTomato | D'Uva et al., 2015 | More info on how to obtain primary cell line can be obtained from Materials and methods sections ‘Transgenic mouse lines and animal procedures’ and ‘Pharmacological inhibition of glycolysis’. | |
| Biological sample (Mus musculus) | TetRE-caErbb2 X MYH6-tTA hearts | D'Uva et al., 2015 from: Xie et al., 1999 and Yu et al., 1996. | The Jackson Laboratory, stock no. 010577; | |
| Biological sample (Danio rerio) | Tg(cmlc2:CreER)pd10 x Tg(β-act2:BSNrg1)pd107hearts | Gemberling et al., 2015 | ZDB-FISH-150901–25249 x ZDB-FISH-150901–25354 | |
| Biological sample (Danio rerio) | Tg(gata4:EGFP)ae1hearts | Lin et al. 2009 | ZDB-FISH-150901–14762 | |
| Biological sample (Danio rerio) | Tg(phd3:GFP)sh144hearts | Santhakumar et al., 2012 | ZDB-FISH-150901–26851 | |
| Antibody | Anti-AuroraB kinase (mouse monoclonal) | BD Transduction Laboratories | #611082, RRID:AB_2227708 | 1:200 |
| Antibody | Anti-Ki67 (Rabbit monoclonal) | Cell Marque | #275R | 1:200 |
| Antibody | Anti-MCT4 (rabbit polyclonal) | Santa Cruz | #SC50329, RRID:AB_2189333 | 1:200 |
| Antibody | Anti-PCNA (mouse monoclonal) | DAKO | #M0879, RRID:AB_2160651 | 1:800 |
| Antibody | Anti-GFP (chicken polyclonal) | Aves | #GFP-1010, RRID:AB_2307313 | 1:1000 |
| Antibody | Anti-Mef2C (rabbit polyclonal) | Santa Cruz/Biorbyt | #SC313, RRID:AB_631920 / #orb256682 | Both 1:1000 |
| Chemical compound, drug | 2-Deoxyglucose | Sigma-Aldrich | #D6134 | 1 mg/g or 3 mM |
| Chemical compound, drug | Lonidamine | Sigma-Aldrich | L4900 | 80 uM |
| Chemical compound, drug | 2NBDG | Caymanchem | #11046 | 400 uM |
| Chemical compound, drug | AG1478 | Selleck Chemical | S2728 | 5M, from 10 mM stock in DMSO |
| Chemical compound, drug | Doxycycline | Harlan Laboratories | TD02503 | |
| Chemical compound, drug | Trizol | Life technologies | #15596026 | |
| Chemical compound, drug | Fast SYBR Green Master Mix | Applied Biosystems | #4385612 | |
| Chemical compound, drug | Collagenase type II | Gibco | 17101015 | 0.1% |
| Chemical compound, drug | TrypLE Express Enzyme (1x), phenol red | Gibco | 12605036 | |
| Software, algorithm | FIJI | Schindelin et al., 2012 | Version 2.0.0, RRID:SCR_002285 | |
| Software, algorithm | RaceID2/StemID | Grün, D. et al. De Novo Prediction of Stem Cell Identity using Single-Cell Transcriptome Data. Cell Stem Cell 19, 266–277 (2016). | RRID:SCR_017242 | |
| Software, algorithm | Rstudio | Rstudio | RRID:SCR_000432 | Version 1.2.1335 |
| Software, algorithm | Imaris | Bitplane | RRID:SCR_0007370 | V9.3.1 |
| Software, algorithm | Gene Set Enrichment Analysis | Genepattern, Broad Institute | RRID:SCR_003199 | # of permutations = 1000 |
| Commercial assay, kit | Superscript III First strand synthesis system | Thermo Fisher Scientific | #18080051 | Input 200 ng RNA |
| Commercial assay, kit | MiRNeasy | Qiagen | 217004 | |
| Commercial assay, kit | High capacity cDNA Reverse Transcription kit | Applied Biosystems | 4374966 | Input 1 ug RNA |
| Commercial assay, kit | Neonatal Dissociation kit | Miltenyi Biotec | 130-098-373 | |
| Commercial assay, kit | TruSeq small RNA primers | Illumina | 20005613 |





