Asymmetrical diversification of the receptor-ligand interaction controlling self-incompatibility in Arabidopsis
Figures
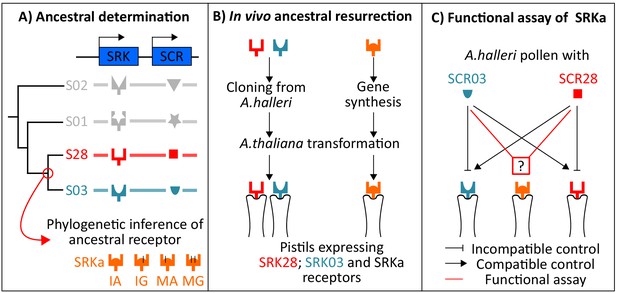
Experimental approach for the ancestral resurrection experiment.
(A) The sequence of the putative last common ancestor of SRK03 and SRK28 was inferred by a phylogenetic approach using codon-based models implemented in PAML. Four different versions of SRKa were defined due to inference uncertainty at two aa positions. (B) SRK03 and SRK28 sequences were cloned from A. halleri DNA BAC clones, whereas SRKa sequences were obtained by gene synthesis. (C) Representation of the controlled cross program to decipher the specificity of SRKa.
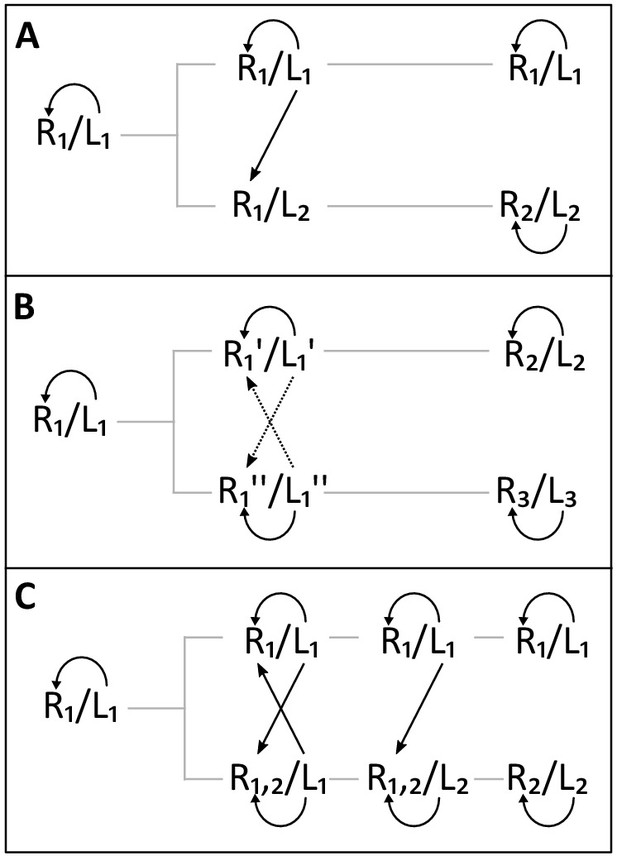
Three models for the emergence of new self-incompatibility specificities.
Docking interaction between ligands and receptors are represented by oriented arrows. (A) In the compensatory mutation model, a non-functional self-compatible intermediate (R1/L2) segregates transiently and is rapidly compensated, creating a novel specificity (R2/L2), while the ancestral one (R1/L1) remains unchanged over time. (B) In the turnover model, slight functional variants with different affinity between them (R1’/L1’ and R1’’/L1’’) segregate in the population and give rise to new specificities (R2/L2 and R3/L3). Dotted lines correspond to weaker affinity interactions. (C) In the promiscuous model, the intermediate receptor (R1,2) has widened up its specificity spectrum enabling it to recognize another potential ligand (L2) while maintaining its capacity to recognize its original ligand (L1). Emergence of the new ligand then favours narrowing of specificity the dual-receptor (R1,2 → R2).
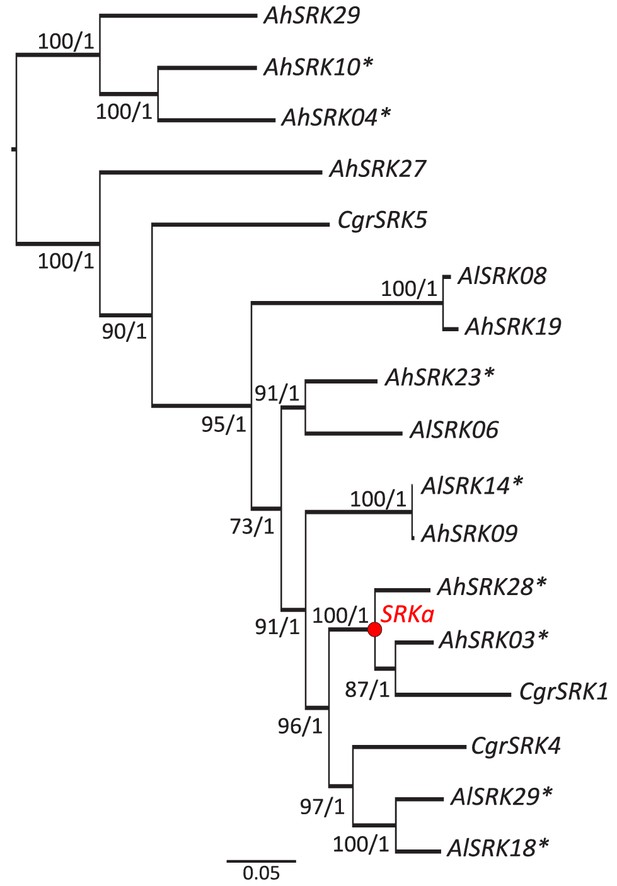
Maximum likelihood phylogenetic tree based on the 17 SRK alleles used for SRKa construction.
The Bayesian reconstruction displays an identical topology. Values at the branches represent the bootstrap percentage/posterior probabilities. The node representing the reconstructed ancestral SRK allele (SRKa) is shown in red. Asterisks highlight alleles with a complete sequence available for ancestral reconstruction.
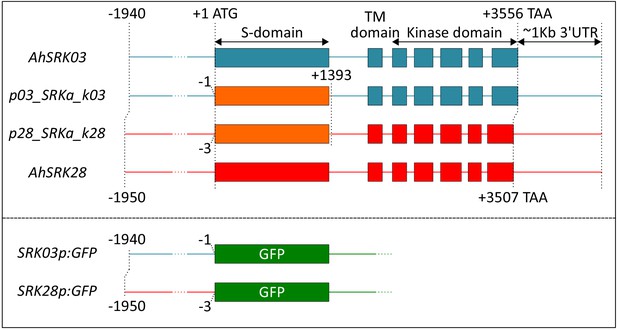
Schematic representation of the molecular constructs used for A. thaliana transformation.
AhSRK03 and AhSRK28 constructs were cloned using a unique genomic fragment starting around −2 Kb and ending 1 Kb after the stop codon. SRKa constructs were each cloned using 3 DNA fragments (amplified promoter and kinase domain of AhSRK03 or AhSRK28 and the synthesized ancestral S-domain). The different parts of the construct were concatenated using the Multisite Gateway Technology at the positions indicated on the figure. SRK03p:GFP and SRK28p:GFP constructs were generated using the same promoter sequence as in the SRKa constructs, but were introduced into the pKGWFS7.0 destination vector.
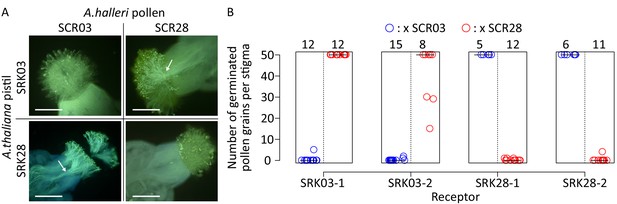
SRK03 and SRK28 are successfully expressed in A. thaliana and represent phenotypically distinct female recognition specificities.
(A) Fluorescence microscopic observation of pistils from SRK-transformed A. thaliana plants pollinated with S03 and S28 pollen from A. halleri. A robust self-incompatibility reaction is observed between cognate alleles, whereas a compatible reaction is observed between non-cognate alleles. White arrows indicate pollen tubes that germinated in the stigma, and are specific to compatible reactions. Bar = 0.3 mm. (B) Number of germinated pollen grains per stigma after pollination. Two SRK03 and SRK28 lines were pollinated with A. halleri pollen expressing either S03 (blue) or S28 (red) specificities. The number of pollinated pistils for each pollination assay is indicated on the top of the figure. The median value for each cross is represented by a horizontal bar.
-
Figure 2—source data 1
Number of germinated pollen grains per stigma after pollination.
Two SRK03 and SRK28 lines were pollinated with A. halleri pollen expressing either S03 or S28 specificities.
- https://cdn.elifesciences.org/articles/50253/elife-50253-fig2-data1-v2.xlsx
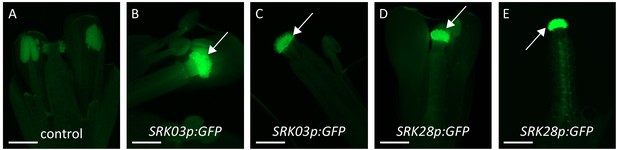
Localization of SRK expression by GFP florescence microscopy.
(A) Flower of an untransformed plant (no SRK expression). (B,C) Flowers of two SRK03p:GFP transformed lines. (D,E) Flowers of two SRK28p:GFP transformed lines. Promoters of both SRK03 and SRK28 drive GFP expression specifically in stigmatic papillae cells (white arrows). Bar = 1 mm.
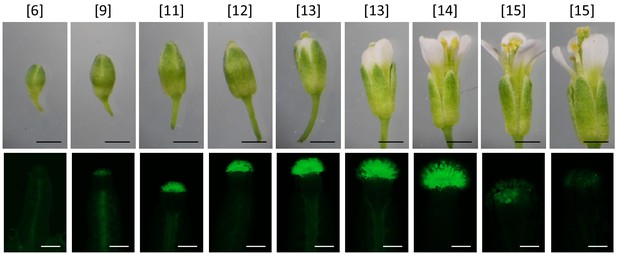
Pattern of proSRK03 activity along development stages of floral buds.
Floral buds and pistils of SRK03p:GFP-transformed plants are observed under optical (above) and UV light (below), respectively. Developmental stages (Smyth et al., 1990) are indicated between brackets. SRK expression is located between stages 11 and 14, with a strong decrease of GFP fluorescence in mature flowers (old stage 15). Similar results were observed for the SRK28p:GFP lines (data not shown). No GFP signal was observed in any other part of the plant (inflorescence stems, leaves and non-reproductive floral organs, data not shown). Black bar = 1 mm, white bar = 0.5 mm.
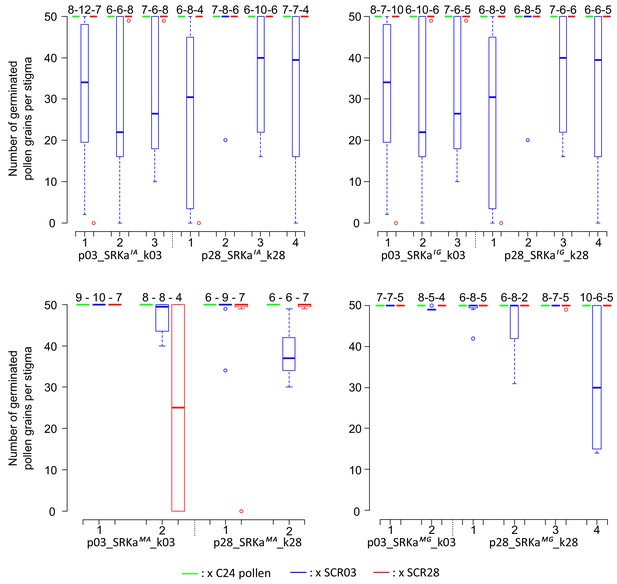
Incompatibility response of SRKa lines.
Each SRKa line was pollinated with A. thaliana C24 (green), A. halleri S03 (blue) and S28 (red) pollen. The number of replicate pollinated pistils is indicated above each boxplot. The horizontal bar represents the median, the box delimitates the 25% and 75% percentiles, the bottom and top whiskers both represent 25% extreme percentiles and outliers are represented by individual dots.
-
Figure 3—source data 1
Number of germinated pollen grains per stigma after pollination.
Each SRKa line was pollinated with A. thaliana C24, A. halleri S03 and S28 pollen.
- https://cdn.elifesciences.org/articles/50253/elife-50253-fig3-data1-v2.xls

Comparison between the S-domain of AhSRK03 and AhSRK28 and their common ancestor.
(A) Arrangement of domains in the full-length SRK protein as assigned by National Center for Biotechnology Information (upper) and protein schematic representation of the S-domain sequence of AhSRK03, AhSRK28 and their common ancestor SRKa (lower). Positions of identical amino acids between the three S-domains are not represented, positions of SRKa amino acids specific to SRK03 and SRK28 are represented by a vertical blue and red line respectively. Positions of hypervariable regions are represented by yellow rectangles. The position of the two amino acid gap presents in both AhSRKa and AhSRK03 is represented by black rectangles. Positions 208 and 305 for which the reconstruction was uncertain and positions 155 and 344 for which the aa identity is different in the two other receptors are represented in black. (B) Amino acid identity and codon sequence (in parentheses) for these four positions in the different SRK variants.
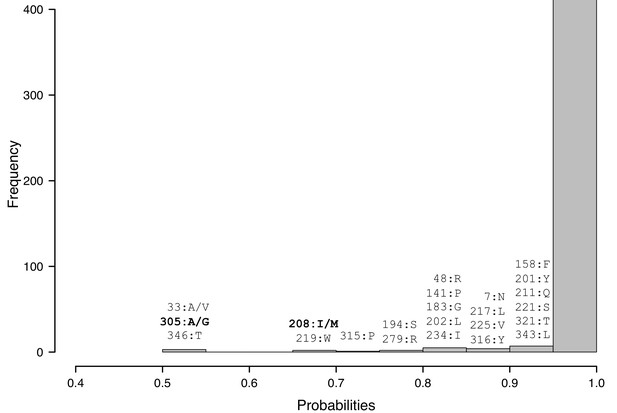
Histogram presenting the distribution of amino acid (aa) probability values obtained with the best fitting-model M3FMutSel.
For each range of values smaller than 0.95, the nature and the position of the aa in the sequence are given. At three sites (33, 208, 305) the models used for the inferences led to different results. The variable sites between the four ancestral sequences used for A. thaliana transformation are in bold.
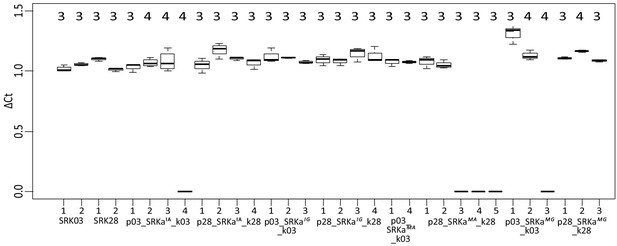
Transgene expression analysis of the different SRK-transformed lines.
ΔCt (Cycle threshold) was calculated as the ratio of Ct-SRK on Ct-Ubiquitin. The number of biological replicates is indicated above each boxplot. For each biological replicate, measurements were realized in technical triplicates.
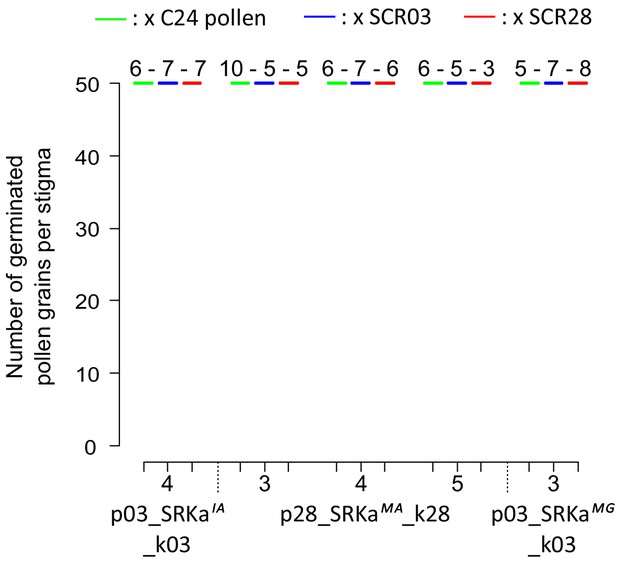
Quantification of incompatibility response of A. thaliana lines with SRKa transcript levels below detection threshold.
Each SRKa line in S03 and S28 context were pollinated with A. thaliana C24 (green), A. halleri S03 (blue) and S28 (red) pollen. The number of pollinated pistils is indicated above each boxplot. These five lines with an undetectable level of transcripts appear compatible with the three pollen types, which is in agreement with the fact that they don’t express any SI receptor at their membrane surface.
-
Figure 3—figure supplement 4—source data 1
Number of germinated pollen grains per stigma after pollination of A. thaliana lines with SRKa transcript levels below detection threshold.
Each SRKa line in S03 and S28 context were pollinated with A. thaliana C24, A. halleri S03 and S28 pollen.
- https://cdn.elifesciences.org/articles/50253/elife-50253-fig3-figsupp4-data1-v2.xls
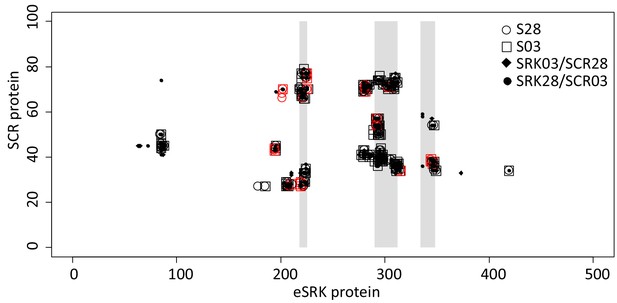
Map of atomic contacts from structural modelling of S03 and S28 complexes.
The X and Y axis represent the eSRK and SCR protein respectively. Open circles and squares represent amino acid contacts between eSRK and SCR proteins in cognate S03 and S28 complexes respectively. Red symbols represent SRK amino acids that differ between SRK03 and SRK28. Full diamonds and dots correspond to amino acid contacts in non-cognate SRK03/SCR28 and SRK28/SCR03 complexes respectively. Hypervariable regions 1, 2 and 3 of eSRK are represented by vertical grey bars at position 219–225, 290–312 and 334–348 respectively (Ma et al., 2016).
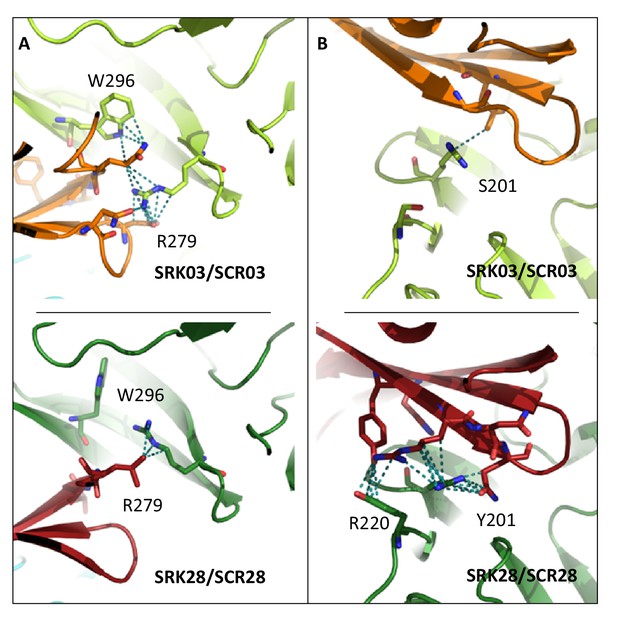
Four amino acid residues establish contrasted patterns of atomic contacts between SCR and SRK in the two cognate complexes.
In (A), residues R279 and W296 of eSRK establish a large number of atomic contacts with SCR in the S03 complex (upper panel), but a very low number of contacts in the S28 complex (lower panel). The situation is reversed in (B), where residues S201 and R220 of eSRK establish a large number of atomic contacts with SCR in the S28 complex (lower panel), but a very low number of contacts in the S03 complex (upper panel). SRK chains are coloured in light and dark green for SRK03 and SRK28, respectively; SCR chains are coloured in orange and red for SCR03 and SCR28, respectively. Amino acid residues are shown in stick representation, with dotted lines indicating atom pair contacts below 4 Å, excluding hydrogen atoms. Note that for clarity a more stringent threshold was used to define atomic contacts here (4 Å) than in Figure 5—figure supplement 1 and Figure 6—figure supplement 1 (where a 5 Å threshold was used for a more comprehensive analysis), but the results are qualitatively similar.
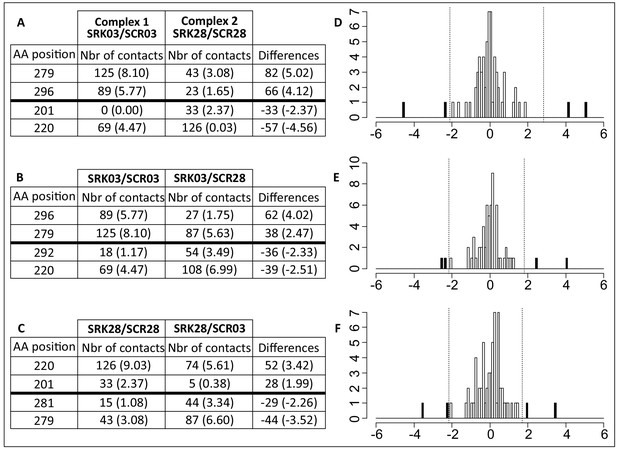
Comparison of amino acid contacts between the different SCR/SRK complexes points to amino acids of SRK potentially involved in specificity of ligand recognition.
The number of atomic contacts between amino acids of the receptor and the ligand was compared between (A) the two cognate complexes (SCR03/SRK03 vs SCR28/SRK28); (B) the cognate vs non-cognate complex established by SRK03 (SCR03/SRK03 vs SCR28/SRK03) and (C) the cognate vs non-cognate complex established by SRK28 (SCR28/SRK28 vs SCR03/SRK28). The three tables list amino acid positions who establish the most extreme differences in atomic interactions in each comparison, values in parentheses correspond to the proportion of contacts realized by each aa independently ((number of contact in the complex/number of contact for one aa) x 100). The full distributions are shown in panels (D), (E) and (F) as the difference of proportion of contacts realized by each aa in both complexes. The vertical lines represent the 5% extreme values. Positive differences correspond to aa involved in numerous contacts in complex one compare to complex two whereas negative differences correspond to aa involved in numerous contact in complex two compare to complex 1.
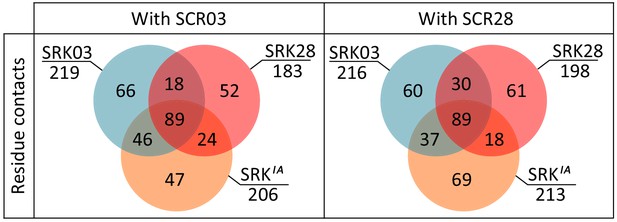
Comparison of predicted binding properties of SRK03, SRK28 and SRKaIA receptors when forming a complex with either the SCR03 or the SCR28 ligand.
Residues in contact between SRKaIA and SCR are more often also in contact between SRK03 and SCR than between SRK28 and SCR. The Venn diagram indicates the number of contacts between amino acid residues that are shared or specific across variants of the SRK receptor and SCR.
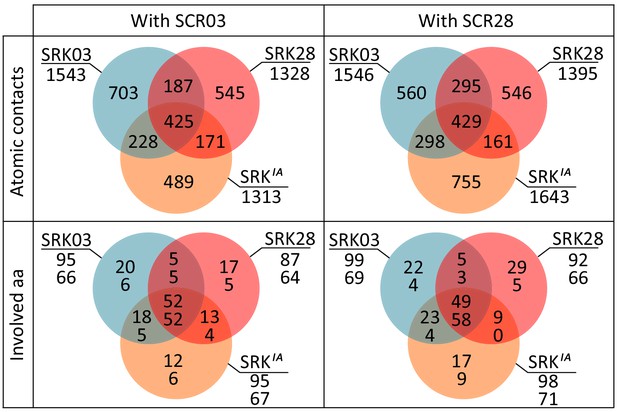
Venn diagram representations of binding features for SRK03, SRK28 and SRKaIA receptors coupled with either SCR03 or SCR28 ligand.
Two features were compared: (i) Atomic contacts are defined by the nature of the amino acid in both proteins, the chain they belong to, their position and the respective atoms involved in the contact (C, N, O, CA main chain atoms or the side chain atoms Cα, Cβ, Cε, …) and (ii) The amino acid involved are defined by the nature of the amino acid, the chain they belong to and their position, independently for SRK (numbers on top) and SCR (bottom numbers). For instance, SRKaIA shared 228 atomic contacts with SRK03 and only 171 contacts with SRK28, when complexed with SCR03. This tendency is also apparent when SRKaIA was complexed with SCR28 (298 identical contact with SRK03 but only 161 with SCR28).
Additional files
-
Supplementary file 1
Detail of the different SRK/SCR interaction protein modelling.
Following the eSRK:SCR template structure where two SCR molecules interact with two SRK proteins to form a heterotetramer (Ma et al., 2016), we indicate the two SRK molecules with their chain identifier A and B and the two SCR molecules with G and H. For each complex, the number of amino acids involved and the number of atomic contacts are defined for each protein chain interaction (AG, AH, BG and BH). Underlined numbers in the column ‘involved aa’ correspond to the number of amino acids involved in both cognate and non-cognate interactions.
- https://cdn.elifesciences.org/articles/50253/elife-50253-supp1-v2.doc
-
Supplementary file 2
Accession numbers for the sequences used in the phylogenetic reconstruction.
- https://cdn.elifesciences.org/articles/50253/elife-50253-supp2-v2.doc
-
Supplementary file 3
PAML ancestral analyses of the SRK protein and model comparison.
np is the number of parameters in the model; lnL is the log likelihood score; AIC (Akaike Information criterion = −2*lnL+2*np) is a measure of the goodness of fit of an estimated statistical model; ω is the nonsynonymous/synonymous substitution ratio; LR is the likelihood ratio: df is the degree of freedom in LRT (Likelihood Ratio Test); *** Highly significant (p-value<0.0001).
- https://cdn.elifesciences.org/articles/50253/elife-50253-supp3-v2.doc
-
Supplementary file 4
Gateway primers used for molecular constructs.
- https://cdn.elifesciences.org/articles/50253/elife-50253-supp4-v2.doc
-
Supplementary file 5
Key resources Table.
- https://cdn.elifesciences.org/articles/50253/elife-50253-supp5-v2.doc
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/50253/elife-50253-transrepform-v2.docx





