Efa6 protects axons and regulates their growth and branching by inhibiting microtubule polymerisation at the cortex
Figures
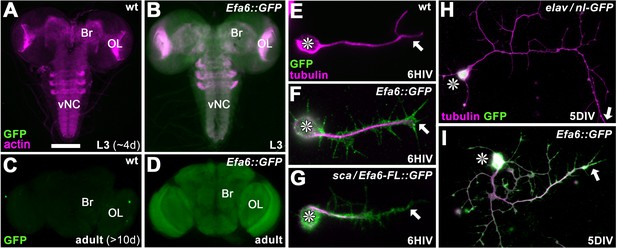
Efa6 is expressed throughout neurons at all developmental stages.
(A–B) Late larval CNSs at about 4d after egg lay (L3; A,B) and adult CNSs from 10d old flies (C,D) derived from control wild-type animals (wt) or the Efa6::GFP line (Efa6::GFP), stained for GFP and actin (Phalloidin, only larval preparations); OL, optic lobe; Br, central brain; vNC, ventral nerve cord. (E–I) Images of primary Drosophila neurons at 6HIV or 5DIV (as indicated bottom right), stained for tubulin (magenta) and GFP (green); control neurons are wild-type (wt) or express elav-Gal4-driven nuclear GFP (elav/nl GFP), further neurons are either derived from the endogenously tagged Efa6::GFP line or express Efa6-FL::GFP under the control of sca-Gal4 (sca/Efa6-FL::GFP); asterisks indicate cell bodies and arrows the axon tips. Scale bar in A represents 75 µm in A and B, 130 µm in C and D, 15 µm in E-H, 25 µm in I and E.
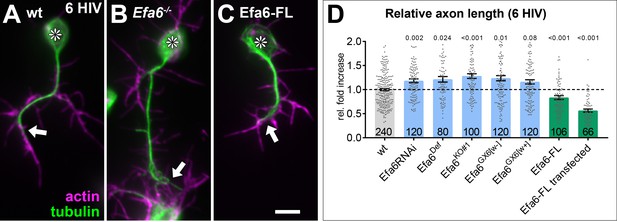
Efa6 regulates axonal length in primary Drosophila neurons.
Examples of primary Drosophila neurons at 6HIV (A–C), all stained for actin (magenta) and tubulin (green); neurons are either wild-type controls (A), Efa6-deficient (B), or expressing Efa6-FL::GFP (C); asterisks indicate cell bodies, arrows point at axon tips; the scale bar in C represents 10 µm. Quantification of axon lengths at 6HIV (D); different genotypes are colour-coded: grey, wild-type controls; blue, different Efa6 loss-of-function conditions; green, neurons over-expressing Efa6; data represent fold-change relative to wild-type controls (indicated as horizontal dashed ‘ctrl’ line); they are shown as single data points and a bar indicating mean ± SEM data; P values from Mann-Whitney tests are given above each column, sample numbers at the bottom of each bar represent individual neurons pooled from at least two replicates, that is experiments conducted on different days. For raw data see Figure 2—source data 1.
-
Figure 2—source data 1
Summary of the statistics from Figure 2.
- https://cdn.elifesciences.org/articles/50319/elife-50319-fig2-data1-v2.xlsx
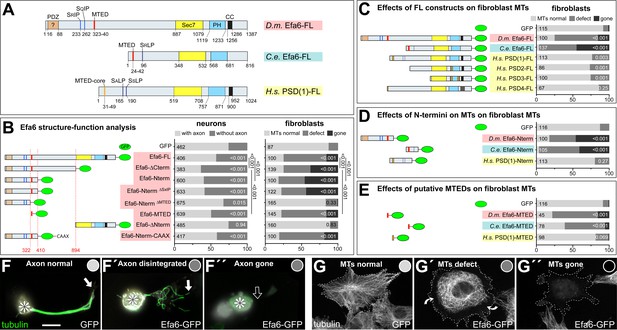
Efa6 domain and motif requirements for MT inhibition in neurons and fibroblasts.
(A) Schematics of Drosophila melanogaster (Dm) Efa6 (isoform C; CG31158), Caenorhabditis elegans (Ce) Efa6 (isoform Y55D9A.1a) and Homo sapiens (Hs) PSD1 (isoform 201/202; NP_002770.3), illustrating the positions (numbers indicate first and last residues) of the putative PDZ (PSD95-Dlg1-ZO1) domain [expected to anchor to transmembrane proteins (Ponting et al., 1997), but not mediating obvious membrane association in fibroblasts: Figure 3—figure supplement 6C,D], SxIP/SxLP motifs (SRIP, SQIP, SALP, SSLP), the MT elimination domain (MTED), SEC7 domain, plekstrin homology (PH) domain and coiled-coil domain (CC). (B) Schematics on the left follow the same colour code and show the DmEfa6 constructs used in this study (dashed red lines indicate the last/first residue before/behind truncations). Bar graphs on the right show the impact that transfection of these constructs had on axon loss in primary Drosophila neurons (dark grey in left graph) and on MT loss in fibroblasts (dark grey or black as indicated; for respective images see F and G below). Analogous fibroblast experiments as performed with Drosophila constructs were performed with full length constructs of C. elegans Efa6 and human PSDs (C), with N-terminal constructs (D) or synthetic MTEDs (E) of Dm and CeEfa6 and of human PSD1. Throughout this figure, construct names are highlighted in red for Drosophila, light blue for C. elegans and yellow for Homo sapiens; all graph bars indicate percentages of neurons with/without axons (light/dark grey) and of fibroblasts with normal, reduced or absent MTs (light, medium, dark grey, respectively); numbers in the left end of each bar indicate sample numbers indicating individual cells pooled from at least two replicates, on the right end the P values from Chi2 tests relative to GFP controls; numbers on the right of bars in B compare some constructs to Efa6-FL::GFP, as indicated by black lines. (F–F’’) Primary neurons expressing GFP or Efa6-FL::GFP transgenically and stained for tubulin (asterisks, cell bodies; white arrows, axon tips; open arrow, disintegrated or absent axon). (G–G'') Fibroblasts expressing Efa6-FL::GFP and stained for tubulin; curved arrows indicate areas where MTs are retracted from the cell periphery; grey dots in F-G'' indicate the phenotypic categories for each neuron and fibroblasts, as used for quantitative analyses in the graphs above. Scale bar at bottom of F refers to 10 µm in F and 25 µm in G. For raw data see Figure 3—source data 1.
-
Figure 3—source data 1
Summary of the statistics from Figure 3.
- https://cdn.elifesciences.org/articles/50319/elife-50319-fig3-data1-v2.xlsx
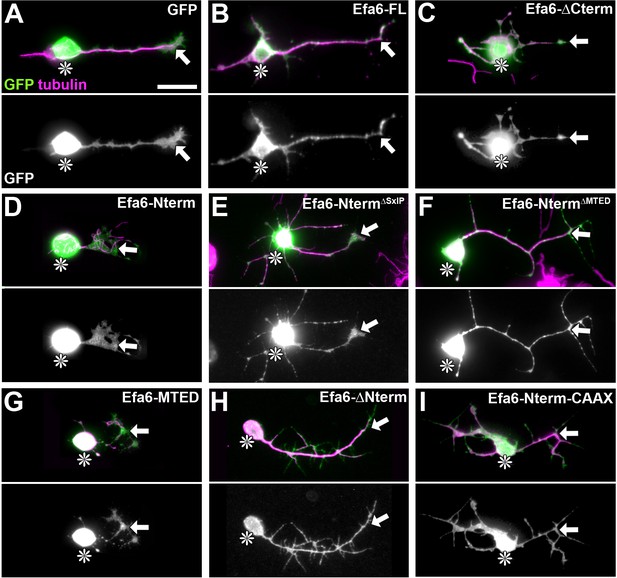
Localisation of Efa6 constructs in primary neurons.
Images show transfected primary Drosophila neurons at 18HIV stained for tubulin (magenta) and GFP (green and in greyscale below the colour image; nomenclature as explained in Figure 3B, but leaving out the ‘::GFP’-postfix). Cell bodies are indicated by asterisks, axon tips by arrows. The transfected constructs are indicated top right following the nomenclature explained in Figure 3B. Scale bar in A represents 10 µm for all figures shown.
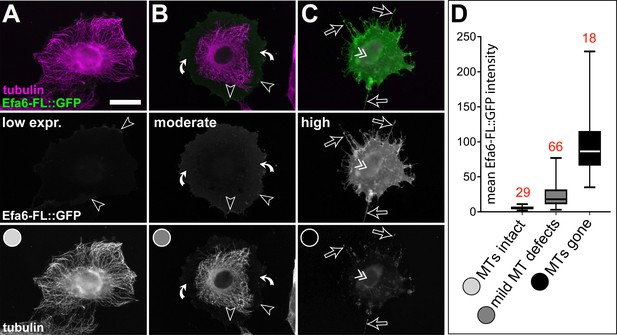
MT inhibition by Efa6-FL is concentration-dependent in fibroblasts.
(A–C) Representative images of fibroblasts expressing Efa6-FL::GFP and stained for tubulin (green and magenta, respectively; both shown as single channels in greyscale below the colour image). Images were taken 24 hr after transfection with Efa6-FL::GFP, assessed for GFP intensity (plotted on the ordinate in D). Examples for low, moderate and high expression are given in A, B and C, respectively, and then grouped with respect to their MT phenotypes into 'MTs intact' (light grey), 'moderate MT defects' (medium grey) or 'prominent MT networks gone' (black), as indicated by greyscale circles in the lowest row of A-C and the abscissa of D); red numbers above box plots indicate number of cells contained in each data set. Arrow heads point at GFP accumulation at membrane edges, white curved arrows indicate cell compartments from which MTs have retracted, open arrows point at retraction fibres and the double chevron indicates the nucleus position and signs of nuclear GFP localisation. Scale bar in A represents 10 µm in all images. For raw data see Figure 3—figure supplement 2—source data 1.
-
Figure 3—figure supplement 2—source data 1
Summary of the statistics from Figure 3—figure supplement 2.
- https://cdn.elifesciences.org/articles/50319/elife-50319-fig3-figsupp2-data1-v2.xlsx
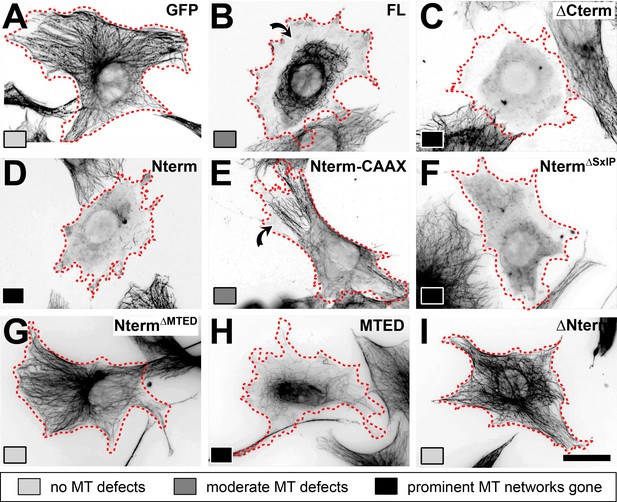
Representative MT phenotypes induced by the different constructs in transfected fibroblasts.
Fibroblasts 24 hr after transfection with different control (GFP) or Efa6-derived constructs as indicated top right in each image (nomenclature as explained in Figure 3B, but leaving out the ‘Efa6“-prefix and ‘::GFP’-postfix). Cells were stained for tubulin (black; images shown as inverted greyscale) and classified as 'no MT defects' (light grey), 'moderate MT defects' (medium grey) or 'prominent MT networks gone' (black), as indicated by greyscale boxes bottom left of each image; curved arrows indicate peripheral MT depletion. Each image represents the most prominent phenotype for each respective construct. Scale bar at the bottom right in H represents 25 µm in all images.
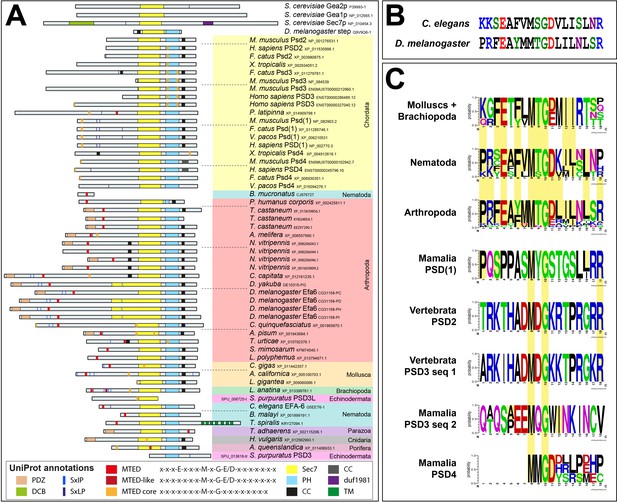
Efa6 N-terminal domains vary amongst different phyla.
(A) Domain annotations in 56 Efa6 orthologues via EMBL SMART and Uniprot. Phyla are colour-coded as in Figure 3 and Figure 3—figure supplement 5. Note that there is a strong variation of lengths and domain composition in particular of the N-terminus. The putative PDZ domain seems to be a feature mainly of insect versions of Efa6 and is absent from any analysed chordate orthologues; MTED and MTED-like sequences cannot be consistently identified in all Efa6 orthologues and are very divergent in chordate Efa6/PSD versions. SxIP/SxLP sites (flanked by positive charges as would be expected of functional motifs; Honnappa et al., 2009) are found in the N-terminal half of only a subset of Efa6 versions in nematodes (e.g. C. elegans), insects (in particular flies, for example D. melanogaster) and molluscs, and even fewer in chordates; in mouse, alpaca and cat SxIP/SxLP sites are flanked by negative charges. (B) To determine a potential MTED consensus sequence, 37 sequences of molluscs, nematodes, arthropods and putative MTED sequences of mammalian PSD1-4 were grouped according to phylum; consensus sequences were depicted using Berkley’s Weblogo online server (default colour scheme). Amino acid positions identical to D. melanogaster and C. elegans MTED are highlighted (faint yellow).
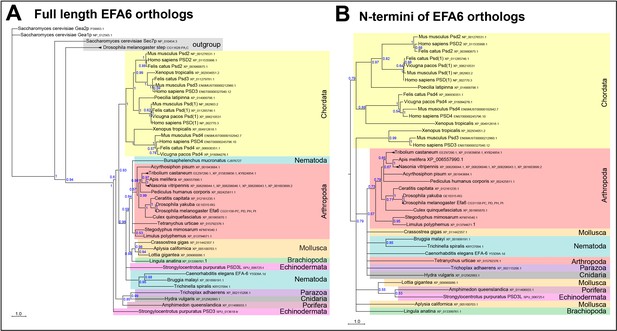
Phylogenetic tree analysis of Efa6.
Bayesian phylogenetic analysis of Efa6 orthologues, either full length (A) or their N-terminus (B). Sequences were aligned using Muscle or ClustalO and posterior probabilities and branch lengths calculated using MrBayes. Branch length scale is indicated; blue numbers show posterior probabilities of each branch split. As outgroup for the full length tree, we used Drosophila steppke (step; Hahn et al., 2013). In both full length and N-terminus analyses, chordates (cream colour) split off very early from Efa6 versions of other species, in line with an early speciation event separating both groups before the vertebrate multiplication events took place. Phyla are highlighted in different colours, gene symbols and/or accession numbers are given after the species names.
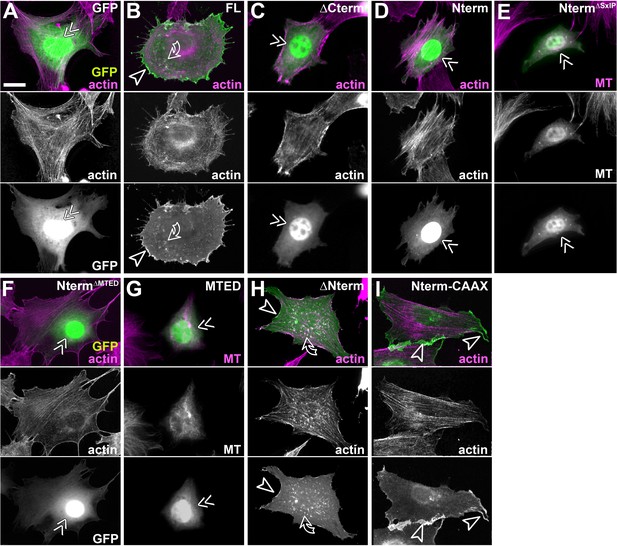
Efa6 constructs localisations in fibroblasts.
Images show fibroblasts 24 hr after transfection with control (GFP) or Efa6-derived constructs (nomenclature as explained in Figure 3B, but leaving out the ‘Efa6“-prefix and ‘::GFP’-postfix, as indicated top right); all cells express GFP-tagged Efa6 variants (green) and are stained for either for actin or MTs (magenta) as indicated bottom right; GFP and actin/MTs are shown as single channels in greyscale below the colour images, Double chevrons indicate nuclear localisation, arrow heads membrane localisation apparent at cell edges and curved arrows membrane ruffles. Scale bar in A represents 10 µm in all images.
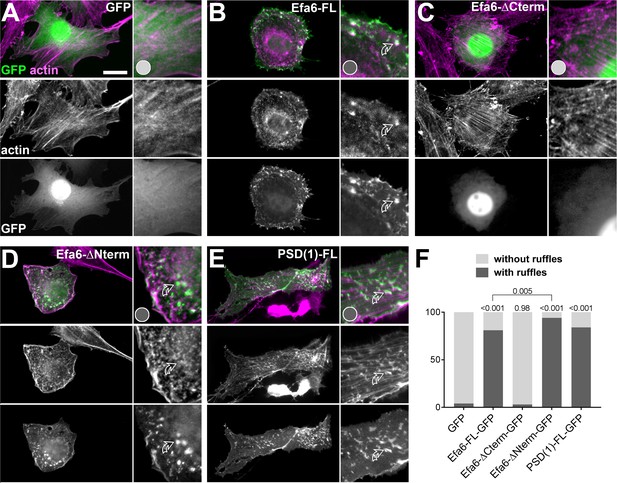
Conserved functions of the Efa6 C-terminus in membrane ruffle formation.
(A–E) Representative images of fibroblasts 24 hr after transfection with different constructs (indicated top right): control vector (GFP; A), Efa6-derived constructs (B-D; nomenclature as explained in Figure 3B, but leaving out the ‘::GFP’-postfix) or PSD1-FL::GFP (E). All cells were stained for GFP and actin (green and magenta, respectively; both shown as single channels in greyscale below the colour images). To the right of each image, a selected area is displayed with 2.5-fold magnification, showing dotted actin- and GFP-stained ruffles (curved open arrows) in B, D and E, but not A and C. (F) Ruffle formation was quantified and is shown as a bar graph indicating percentages of fibroblasts with/without membrane ruffles (dark/light grey); P values on top of bars are from Chi2 tests relative to GFP controls. As shown, ruffle phenotype was never observed with any N-terminal Efa6 variants but are reproduced with the Efa6-∆Nterm::GFP variant (comprising the C-terminal Sec7, PH and CC domains; Figure 3B). Scale bar in A represents 10 µM for all fibroblasts shown. For sample numbers and raw data see Figure 3—figure supplement 7—source data 1.
-
Figure 3—figure supplement 7—source data 1
Summary of the statistics from Figure 3—figure supplement 7.
- https://cdn.elifesciences.org/articles/50319/elife-50319-fig3-figsupp7-data1-v2.xlsx
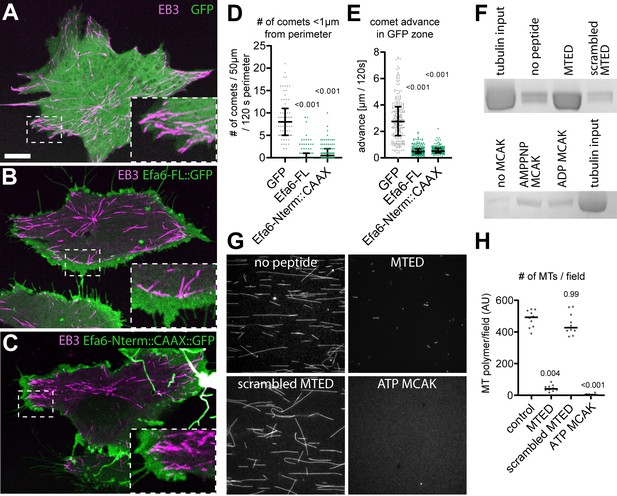
EFA6 peptide interacts directly with α/β-tubulin and inhibits microtubule growth.
(A–C) Fibroblasts co-expressing Eb3::RFP (magenta) together with either GFP (A), Efa6-FL::GFP (B) or Efa6-Nterm::GFP::CAAX (C; all shown in green); images are maximum intensity projections of all frames taken at 1 s intervals during a 120 s live imaging period; stippled areas are shown as 2.5fold magnified insets. Example movies are provided as Animations 1, 2 and 4. (D,E) Quantification of Eb3::RFP comet behaviours deduced from images comparable to those shown in A to C: in D, each data point represents the number of comets that were within a 1 µm range from the perimeter assessed over a 50 μm perimeter stretch, respectively (n = 73 for GFP, 90 for Efa6-FL::GFP and 90 for Efa6-Nterm::GFP::CAAX); in E, each data point represents the distance individual Eb3 comets reached into GFP-positive areas (in B and C assessed in areas of high GFP expression, in A in areas close to the perimeter; n = 224 for GFP, 238 for Efa6-FL::GFP and 197 for Efa6-Nterm::GFP::CAAX). (F) Pull-down of porcine brain tubulin using sepharose beads which were either uncoated, or coated with MTED, with a scrambled version of MTED or with MCAK (in the presence of ADP or the ATP analogue AMPPNP); proteins/peptides were randomly attached via cyanogen bromide coupling of lysines and/or N-terminal amines; quantification data are provided as Figure 4—source data 1. (G) Fluorescence images of rhodamine-labelled MTs grown in the presence of no peptide, MTED, scrambled MTED or MCAK together with ATP. (H) Quantification of the amount of MT polymer per field of view (n = 10 fields in each case; AU, arbitrary unit). Numbers above plots in D, E and F represent P-values determined via Kruskal–Wallis one-way ANOVA with post hoc Dunn’s test. Scale bar in A represents 10 µm in A-C and three in G. For raw data see Figure 4—source data 1.
-
Figure 4—source data 1
Summary of the statistics from Figure 4.
- https://cdn.elifesciences.org/articles/50319/elife-50319-fig4-data1-v2.xlsx
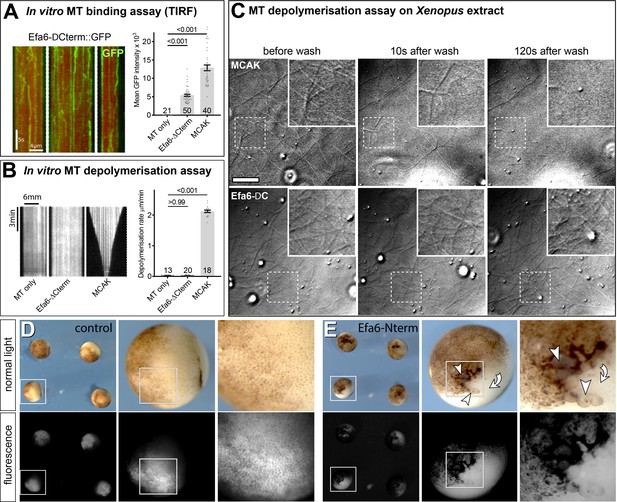
In vitro attempts to resolve the MT inhibition mechanism of Efa6.
(A) To determine whether Efa6 directly affects MT stability, we expressed Efa6-∆Cterm::GFP in Sf9 cells, purified the protein and observed its interaction with MTs using total internal reflection fluorescence (TIRF) microscopy in a low ionic strength buffer (BRB20, 75 mM KCl); the three images on the left show different examples of kymographs of MT lattices decorated with Efa6-∆Cterm::GFP, which displays a mixture of stationary molecules and diffusive interactions typical of non-translocating MT-associated proteins (Helenius et al., 2006; Hinrichs et al., 2012); bar charts (right) show quantification of the amount of interacting protein: background signal from MTs alone, Efa6-ΔCterm::GFP (20 nM), and the non-translocating kinesin MCAK::GFP (20 nM) as positive control; at the same protein concentration, over 2-fold more molecules of MKAC typically interact with MTs; individual data points represent mean intensity of individual MTs (sample numbers at bottom) and bars represent mean ± SEM; numbers above bars show P values obtained from Mann–Whitney Rank Sum statistical analyses. (B) Kymographs (left) show individual fluorescently-labelled GMPCPP-stabilised MTs in vitro (Patel et al., 2016), either alone (MTs only) or in the presence of 14 nM Efa6-ΔCterm::GFP (n = 20) or 40 nM MCAK::GFP; the bar chart (right) quantifies the induced depolymerisation rates (number of analysed MTs at bottom). Using two different purifications of Efa6-ΔCterm::GFP on three separate occasions, we saw no evidence of MT depolymerisation above the basal level of depolymerisation typically observed in these assays, whereas parallel control experiments with mitotic centromere-associated kinesin/MCAK showed MT depolymerisation rates typical of this kinesin. (C) To assess whether MT destabilisation might require additional cytoplasmic factors, we used Xenopus oocyte extracts: phase contrast images show MTs in Xenopus oocyte extracts (after they had been allowed to polymerise for 20 min) and then showing stills from before, 10 s after and 120 s after washing in 20 nM MCAK::GFP (as positive control) or 20 nM Efa6-ΔCterm::GFP; squares outlined by dashed white lines are shown as 2-fold magnified close-ups in the top right corner of each image; MTs clearly vanish upon treatment with MCAK, but counts of MTs did not reveal any obvious effects on MTs with Efa6-ΔCterm::GFP. (D,E) RFP controls and Efa6-Nterm::GFP expression constructs were injected into Xenopus embryos at the 4 cell stage and analysed 24 hr later; only the Efa6 construct caused a strong suppression of cell division, as indicated by the presence of very large cells (arrows) and pigmentation defects (curved arrows) at the site of injection, suggesting that Efa6-Nterm::GFP is functional when expressed in the Xenopus context. Scale bar (in top left image) represents 3 µm in C, and 1400 µm / 350 µm / 140 µm in left/middle/ right images of D and E, respectively. For raw data see Figure 4—figure supplement 1—source data 1.
-
Figure 4—figure supplement 1—source data 1
Summary of the statistics from Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/50319/elife-50319-fig4-figsupp1-data1-v2.xlsx
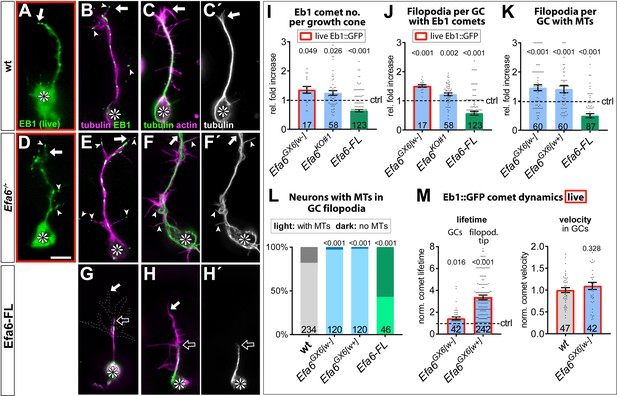
Efa6 regulates MT behaviours in GCs.
(A–H') Examples of primary neurons at 6HIV which are either wild-type controls (top), Efa6-deficient (middle) or expressing Efa6-FL::GFP (bottom); neurons were either imaged live for Eb1::GFP (green in A,D) or fixed and labelled for Eb1 and tubulin (B,E,G; as colour-coded) or actin and tubulin (C,F,H; as colour coded; tubulin shown as single channel image on the right); asterisks indicate cell bodies, white arrows the tips of GCs, open arrows the tips of MT bundles, arrow heads filopodial processes containing MTs or Eb1 comets; the GC in G is outlined with a white dashed line; scale bar in D represents 5 µm in all images. (I–M) Quantitative analyses of MT behaviours in GCs, as indicated above each graph. Different genotypes are colour-coded: grey, wild-type controls; blue, different Efa6 loss-of-function conditions; green, neurons over-expressing Efa-FL::GFP. The graph in L shows percentages of neurons without any MTs in shaft filopodia (dark shade) versus neurons with MTs in at least one filopodium (light shade; P values above bars assessed via Chi2 tests), whereas all other graphs show single data points and a bar indicating mean ± SEM, all representing fold-increase relative to wild-type controls (indicated as horizontal dashed ‘ctrl’ line; P values above columns are from Mann-Whitney tests). The control values in M (dashed line) equate to an Eb1 comet life-time of 2.10 s ± 0.24 SEM in filpododia and 5.04 s ± 0.60 SEM in growth cones, and a comet velocity of 0.136 μm/s ± 0.01 SEM. Throughout the figure, sample numbers are shown at the bottom of each bar where each data point represents one GC (I–K), one neuron (L) or one Eb1::GFP comet (M), respectively; in all cases data were pooled from at least two replicates; data obtained from live analyses with Eb1::GFP are framed in red. For raw data see Figure 5—source data 1.
-
Figure 5—source data 1
Summary of the statistics from Figure 5.
- https://cdn.elifesciences.org/articles/50319/elife-50319-fig5-data1-v2.xlsx
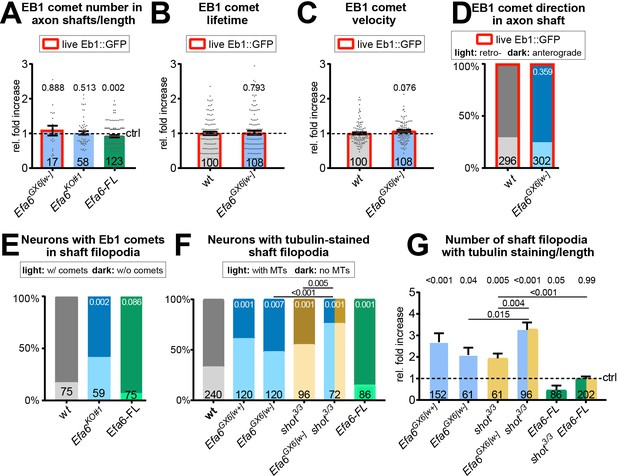
Loss of Efa6 promotes MT entry into axon shaft filopodia.
Quantitative analyses of MT behaviours in axon shafts, as indicated above each graph; bars are colour-coded: grey, controls; blue, different Efa6 mutant alleles; green, neurons over-expressing Efa-FL::GFP or Efa6::CAAX::GFP; orange, shot3 mutant allele; red outlines indicate live imaging data, all others were obtained from fixed specimens. (A–C,G) Fold-changes relative to wild-type controls (indicated as horizontal dashed ‘ctrl’ line) shown as single data points and a bar indicating mean ± SEM; P values were obtained via Mann-Whitney tests; control values (dashed line) in B and C equate to an Eb1 comet lifetime of 7.18 s ± 0.35 SEM and a velocity of 0.169 μm/s ± 0.01 SEM; in G the number of shaft filopodia per neuron was divided by the axon length of that same neuron. (D–F) Binary parameters (light versus dark shades as indicated) provided as percentages; P values are given relative to control or between different genotypes (as indicated by black lines); they were obtained via Chi2 tests. Numbers at the bottom of bars indicate sample numbers pooled from at least two replicates, respectively; data points reflect individual Eb1::GFP comets (A–D) or individual neurons (E–G). For raw data see Figure 6—source data 1.
-
Figure 6—source data 1
Summary of the statistics from Figure 6.
- https://cdn.elifesciences.org/articles/50319/elife-50319-fig6-data1-v2.xlsx
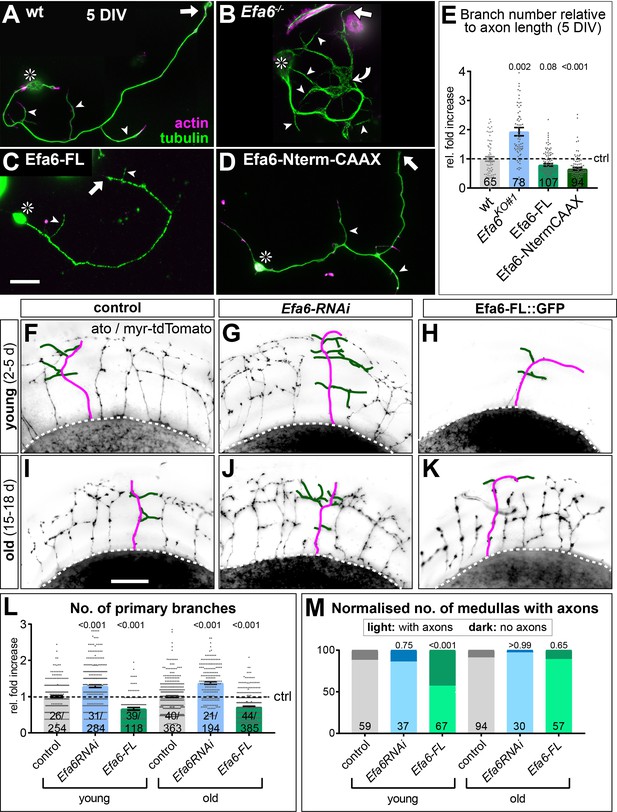
Efa6 regulates axon branching in primary Drosophila neurons and adult fly brains.
(A–D) Examples of primary Drosophila neurons at 5DIV, all stained for actin (magenta) and tubulin (green); neurons are either wild-type controls (A), Efa6-deficient (B), expressing Efa6-FL::GFP (C), or expressing Efa6-Nterm::CAAX::GFP (D); asterisks indicate cell bodies, arrows point at axon tips, arrow heads at axon branches, the curved arrow at an area of MT disorganisation; the scale bar in C represents 20 µm in A-D. (E) Quantification of axonal branch numbers; different genotypes are colour-coded: grey, wild-type controls; blue, Efa6 loss-of-function; green, neurons over-expressing Efa6 variants; data represent fold-change relative to wild-type controls (indicated as horizontal dashed ‘ctrl’ line); each neuron is shown as a single data point (sample number at bottom of bar) together with the mean ± SEM; P values from Mann-Whitney tests comparing to wild-type are given above each column. (F–K) Brains (medulla region of the optic lobe in oblique view) of young (2–5 d after eclosure; top) and old flies (15–18 d; bottom) driving UAS-myr-tdTomato via the ato-Gal4 driver in dorsal cluster neurons (example neurons are traced in magenta for axons and green for side branches); flies either carry ato-Gal4-and UAS-myr-tdTomato, alone (control, left), together with Efa6-RNAi (middle) or together with Efa6-FL::GFP (right). (L,M) Quantification of data for control (wt; grey), Efa6 knock-down (blue) and Efa6-FL::GFP over-expression (green): L) shows the number of primary branches per axon as fold-change normalised to controls (indicated as horizontal dashed ‘ctrl’ line); individual axons are shown as data points (sample number at bottom of bars indicate number of medullas before and number of axons after slash); bars indicate mean ± SEM accompanied by single data points. (M) displays the number of medullas (sample numbers at bottom of bars) which display axons (light colours) or lack axons (dark colours) shown as a percentages in young and old flies. P values above columns were obtained from Mann-Whitney (L) or Chi2 tests (M). Scale bar in I represents 60 µm in F-K. For raw data see Figure 7—source data 1.
-
Figure 7—source data 1
Summary of the statistics from Figure 7.
- https://cdn.elifesciences.org/articles/50319/elife-50319-fig7-data1-v2.xlsx
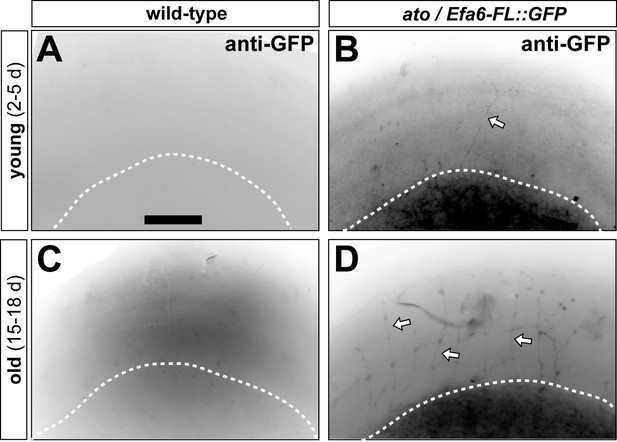
ato-Gal4-driven Efa-FL::GFP expression in adult brain tissue.
Images show the optic lobe in oblique view; white dashed line indicating the lower edge of the medulla) of young (2–5 d after eclosure; top) and old flies (15–18 d; bottom) which are either from wild-type controls or from flies co-expressing UAS-myr::tdTomato and UAS-Efa6-FL::GFP via the ato-Gal4 driver in dorsal cluster neurons. Specimens are stained for GFP and arrows point at stained axons; for morphological stainings of ato-Gal4 axons in these experiments see Figure 7F–K. Scale bar in A represents 60 µm in all images.
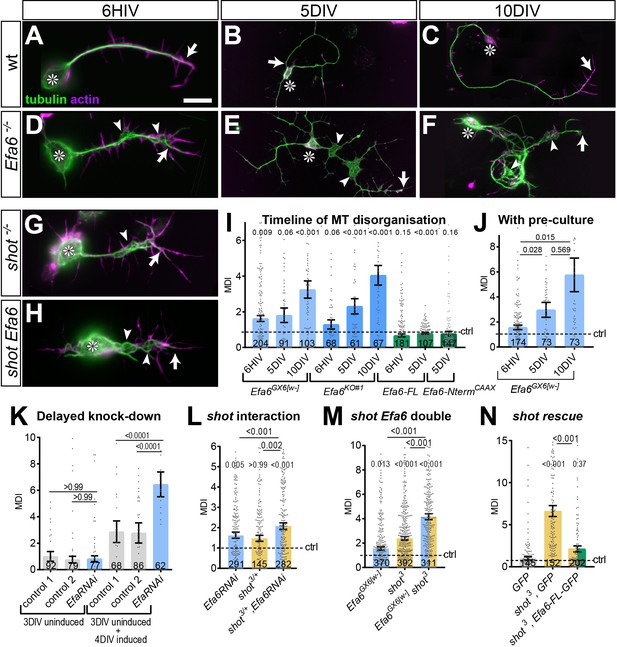
Efa6 helps to maintain axonal MT bundles in Drosophila neurons.
(A–H) Images of primary neurons at 6HIV (left), 5DIV (middle) and 10DIV (right), stained for tubulin (green) and actin (magenta), derived from embryos that were either wild-type (wt, A–C), Efa6 null mutant (D–F), homozygous for shot3 (G) or shot3 Efa6GX6[w-] double-mutant (shot Efa6, H; arrows point at axon tips, arrow heads at areas of MT disorganisation, and asterisks indicate the cell bodies; the scale bar in A represents 10 µm for 6HIV neurons and 25 µm for 5DIV and 10DIV neurons. (I–N) Quantitative analyses of MT disorganisation (measured as MT disorganisation index, MDI) in different experimental contexts (as indicated above graphs); different genotypes are colour-coded: grey, controls; blue, Efa6 loss-of-function; orange, shot3 in hetero-/homozygosis; green, neurons over-expressing Efa-FL::GFP or Efa6-Nterm::CAAX::GFP; in all cases, individual neurons are shown as single data points (sample numbers at bottom of bar), and the bars indicate mean ± SEM, all representing fold-change relative to wild-type controls (indicated as horizontal dashed ‘ctrl’ line); P values are given above each column and were obtained from Mann-Whitney tests (I, J) and Kruskal–Wallis one-way ANOVA with post hoc Dunn’s test (K–N) either relative to controls or between genotypes (indicated by black lines). In K, 'control 1' is tub-Gal80, elav-Gal4 alone and 'control 2' is UAS-Efa6RNAi alone. For raw data see Figure 8—source data 1.
-
Figure 8—source data 1
Summary of the statistics from Figure 8.
- https://cdn.elifesciences.org/articles/50319/elife-50319-fig8-data1-v2.xlsx
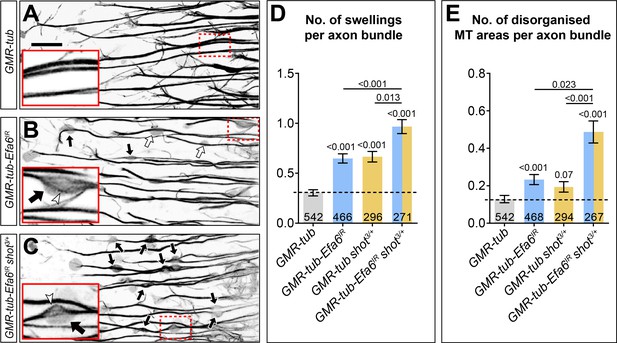
Efa6 is required for axonal MT bundle maintenance in adult fly brains.
(A–C) Medulla region of adult brains at 26–27 days after eclosure, all carrying the GMR31F10-Gal4 driver and UAS-GFP-α-tubulin84B (GMR-tub) which together stain MTs in a subset of lamina neuron axons that terminate in the medulla. The other specimens in addition co-express Efa6-RNAi either in wild-type background (GMR-tub-Efa6IR) or in shot3/+ heterozygous mutant background (GMR-tub-Efa6IR shot3/+). White/black arrows indicate axonal swellings without/with MT disorganisation; rectangles outlined by red dashed lines are shown as 2.5 fold magnified insets where white arrow heads point at disorganised MTs; the scale bar in A represents 15 µm in all images. (D, E) Quantitative analyses of all axonal swelling (D) or swellings with MT disorganisation (E); different genotypes are colour-coded (grey, control; blue, Efa6 loss-of-function; orange, shot3 heterozygous); bars show mean ± SEM, all representing fold-change relative to wild-type controls (indicated as horizontal dashed line). P values from Kruskal–Wallis one-way tests are given above each column, sample numbers (i.e. individual axon bundles) at the bottom of each bar. For raw data see table Figure 9—source data 1.
-
Figure 9—source data 1
Summary of the statistics from Figure 9.
- https://cdn.elifesciences.org/articles/50319/elife-50319-fig9-data1-v2.xlsx
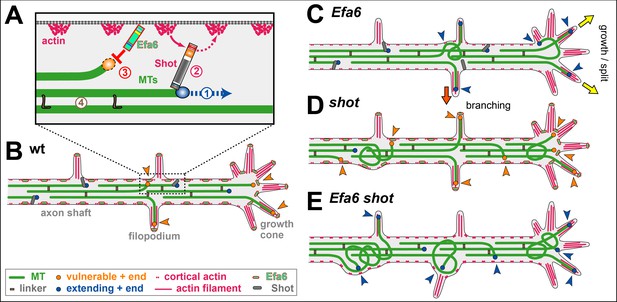
A model for axonal roles of Efa6.
(A) The model of local axon homeostasis (Hahn et al., 2019; Prokop, 2016) states that the maintenance of axonal MT bundles (green bars) is an active process. For example, the polymerisation (1) mediated by plus end machinery (blue circle) is guided by spectraplakins (here Shot) along cortical actin into parallel bundles (2), or MTs are kept together through cross-linkage (brown ‘L’; 4; Bettencourt da Cruz et al., 2005; Krieg et al., 2017); here we propose that MTs accidentally leaving the bundle become susceptible to inhibition through cortically anchored Efa6 (red ‘T’ and orange circle). (B) In normal neurons, MTs that polymerise within the axonal bundles (dark blue circles) are protected by Shot (grey lines) and MT-MT cross-linkers (brown rectangles), whereas MTs approaching the membrane (orange arrow heads) either by splaying out in GCs or leaving the bundle in the shaft (orange arrow heads) become susceptible (orange circles) to inhibition by Efa6 (light green/red dashes) both along the cortex and in filopodia. C) Upon Efa6 deficiency, MTs leaving bundles or entering GCs are no longer subjected to Efa6-mediated cortical inhibition (blue arrow heads) and can cause gradual build-up of MT disorganisation; when entering shaft filopodia they can promote interstitial branch formation (red arrow), when entering GC filopodia they can promote axon growth or even branching through GC splitting (yellow arrows). (D) Far more MTs leave the bundles in shot mutant neurons, but a good fraction of them can potentially be inhibited by Efa6 (increased number of orange arrow heads). (E) In the absence of both Shot and Efa6, more MTs leave the bundles, but there is no compensating cortical inhibition (increased number of blue arrow heads), so that the MT disorganisation phenotype worsens.
Tubulin pull down by MCAK attached to sepharose beads using different methods of attachment.
(A) MCAK covalently attached to beads via any lysine or N-terminal amine (Figure 4 of this publication). (B) MCAK attached to Protein A coated beads via anti-his antibody binding the C-terminal his-tag (Claire Friel, unpublished data).
Videos
MT behaviours in control fibroblasts expressing GFP.
Transfected fibroblasts co-expressing EB3::RFP (green) to visualise MT polymerisation, together with GFP (magenta) which localises throughout the cell. White boxed areas are shown as twofold magnified close-ups on the right. MTs clearly polymerise to the very perimeter of the transfected cell (quantifications are shown in Figure 4). Movies were taken at one frame per second. The scale bar corresponds to 25 μm in the main image.
MT behaviours in control fibroblasts expressing Efa6-FL::GFP.
Transfected fibroblasts co-expressing EB3::RFP (green) to visualise MT polymerisation, together with Efa6-FL::GFP (magenta) which localises primarily to the cell periphery. White boxed area is shown as twofold magnified close-up on the right, separated by channel. MTs clearly fail to polymerise into the Efa6-FL::GFP-enriched zone and hardly ever reach the cell perimeter (quantifications are shown in Figure 4). Movies were taken at one frame per second. The scale bar corresponds to 25 μm in the main image.
Efa6-Nterm::GFP tip-tracks in fibroblasts.
Upon Efa6-Nterm::GFP expression in mouse NIH3T3 fibroblast about 6 hr after transfection, comets were observed in a few instances. Pictures for the movie were taken at 2 s intervals. Scale bar indicates 10 µm.
MT behaviours in control fibroblasts expressing Efa6-Nterm::GFP::CAAX.
Transfected fibroblasts co-expressing EB3::RFP (green) to visualise MT polymerisation, together with Efa6-Nterm::GFP::CAAX (magenta) which localises primarily to the cell periphery. The close-up on the right is two-fold magnified and refers to the top left corner of the main image; it is separated by channel. MTs fail to polymerise into the Efa6-Nterm::GFP::CAAX-enriched zone and hardly ever reach the cell perimeter (quantifications are shown in Figure 4D,E). Movies were taken at one frame per second. The scale bar corresponds to 25 μm in the main image.
Eb1::GFP in a growth cone of a wild-type Drosophila primary neuron at 6 HIV.
Arrows indicate positions where individual Eb1::GFP comets terminate. Pictures of the movie were taken at 2 s intervals. Scale bar indicates 10 µm.
Eb1::GFP in a growth cone of a wild-type Drosophila primary neuron at 6 HIV.
Arrows indicate positions where individual Eb1::GFP comets terminate. Pictures of the movie were taken at 2 s intervals. Scale bar indicates 10 µm.
Eb1::GFP in a growth cone of an Efa6GX6[w-] mutant Drosophila primary neuron at 6 HIV.
The arrow heads follow individual Eb1::GFP comets illustrating either their trajectories adjacent to the membrane or prolonged dwell time at filopodial tips. Pictures for the movie were taken at 2 s intervals. Scale bar indicates 10 µm.
Eb1::GFP in a growth cone of an Efa6 GX6[w-] mutant Drosophila primary neuron at 6 HIV.
The arrow heads follow individual Eb1::GFP comets illustrating either their trajectories adjacent to the membrane or prolonged dwell time at filopodial tips. Pictures for the movie were taken at 2 s intervals. Scale bar indicates 10 µm.
Tables
Constructs and inserts used to generate expression and transgenic constructs.
The left column lists the final vector representing intermediate steps of cloning procedures or vectors employed for experiments on this study, the middle column the source of the vectors, and the right column the inserts of vectors on the left. Abbreviations: co = codon optimised; Dm = Drosphila melanogaster; Ce = Caenorhabditis elegans; Hs = Homo sapiens.
| final vector | Source | insert |
|---|---|---|
| pcDNA3-EGFP | Addgene | XhoI-EGFP-XbaI |
| pUAST-AscI-PacI-EGFP | this study | KpnI, AscI, PacI-EGFP-XbaI |
| pUAST-DmEfa6FL-EGFP (aa1-1387) | this study | KpnI, AscI-kozak-Efa6 (aa1-1387)-GSGSGS-EGFP-PacI, XbaI |
| P[acman]M-6-attB-UAS-1-3-4-DmEfa6FL-EGFP (aa1-1387) | this study | AscI-kozak-DmEfa6 (aa1-1387)-GSGSGS-EGFP-PacI |
| pcDNA3.1-DmEfa6FL-EGFP (aa1-1387) | this study | KpnI, AscI-kozak-DmEfa6 (aa1-1387)-GSGSGS-EGFP-PacI, XbaI |
| pUAST-DmEfa6∆Cterm-EGFP (aa1-894) | this study | KpnI, AscI-kozak-DmEfa6∆Cterm (aa1-894)-GSGSGS-EGFP-PacI, XbaI |
| P[acman]M-6-attB-UAS-1-3-4-DmEfa6∆Cterm-EGFP (aa1-894) | this study | AscI-kozak-DmEfa6∆Cterm (aa1-894)-GSGSGS-EGFP-PacI |
| pcDNA3.1-DmEfa6∆Cterm-EGFP (aa1-894) | this study | KpnI, AscI-kozak-DmEfa6∆Cterm (aa1-894)-GSGSGS-EGFP-PacI, XbaI |
| pUAST-DmEfa6-Nterm-EGFP (aa1-410) | this study | KpnI, AscI-kozak-DmEfa6-Nterm (aa1-410)-GSGSGS-EGFP-PacI, XbaI |
| P[acman]M-6-attB-UAS-1-3-4-DmEfa6-Nterm-EGFP (aa1-410) | this study | AscI-kozak-DmEfa6-Nterm (aa1-410)-GSGSGS-EGFP-PacI |
| pcDNA3.1-DmEfa6-Nterm-EGFP (aa1-410) | this study | KpnI, AscI-kozak-DmEfa6-Nterm (aa1-410)-GSGSGS-EGFP-PacI, XbaI |
| pUAST-DmEfa6-Nterm-CAAX-EGFP (aa1-410) | this study | KpnI, AscI-kozak-DmEfa6-Nterm (aa1-410)-GSGSGS-EGFP-CAAX[KRAS]-PacI, XbaI |
| P[acman]M-6-attB-UAS-1-3-4-DmEfa6-Nterm-CAAX-EGFP (aa1-410) | this study | AscI-kozak-DmEfa6-Nterm (aa1-410)-GSGSGS-EGFP-CAAX[KRAS]-PacI |
| pcDNA3.1-DmEfa6-Nterm-CAAX-EGFP (aa1-410) | this study | KpnI, AscI-kozak-DmEfa6-Nterm (aa1-410)-GSGSGS-EGFP-CAAX[KRAS]-PacI, XbaI |
| pUAST-DmEfa6-Nterm∆SxiP-EGFP (aa1-410) | this study | KpnI, AscI-kozak-DmEfa6-Nterm∆SxiP (aa1-410; SQIP > AAAA; SRIP > AAAA)-GSGSGS-EGFP-PacI, XbaI |
| pcDNA3.1-DmEfa6-Nterm∆SxiP-EGFP (aa1-410) | this study | KpnI, AscI-kozak-DmEfa6-Nterm∆SxiP (aa1-410; SQIP > AAAA; SRIP > AAAA)-GSGSGS-EGFP-PacI, XbaI |
| pUAST-DmEfa6-Nterm∆MTED-EGFP (aa1-300) | this study | KpnI, AscI-kozak-DmEfa6-Nterm∆MTED (aa1-300)-GSGSGS-EGFP-PacI, XbaI |
| pcDNA3.1-DmEfa6-Nterm∆MTED-EGFP (aa1-300) | this study | KpnI, AscI-kozak-DmEfa6-Nterm∆MTED (aa1-300)-GSGSGS-EGFP-PacI, XbaI |
| pUAST-DmEfa6∆Nerm-EGFP (aa851-1387) | this study | KpnI, AscI-kozak-DmEfa6∆Nerm (aa851-1387)-GSGSGS-EGFP-PacI, XbaI |
| pcDNA3.1-DmEfa6∆Nerm-EGFP (aa851-1387) | this study | KpnI, AscI-kozak-DmEfa6∆Nerm (aa851-1387)-GSGSGS-EGFP-PacI, XbaI |
| pUAST-DmEfa6-MTED-EGFP (aa322-341) | this study | KpnI, AscI-kozak-DmEfa6-MTED (aa322-341)-GSGSGS-EGFP-PacI, XbaI |
| pcDNA3.1-DmEfa6-MTED-EGFP (aa322-341) | this study | KpnI, AscI-kozak-DmEfa6-MTED (aa322-341)-GSGSGS-EGFP-PacI, XbaI |
| pcDNA3.1-CeEfa6-FL-EGFP (aa1-816) | this study | KpnI, AscI-kozak-CeEfa6 (aa1-816)-GSGSGS-EGFP-PacI, XbaI |
| pcDNA3.1-CeEfa6-Nterm-EGFP (aa1-152) | this study | KpnI, AscI-kozak-CeEfa6-Nterm (aa1-152)-GSGSGS-EGFP-PacI, XbaI |
| pcDNA3.1-CeEfa6-MTED-EGFP (aa24-42) | this study | KpnI, AscI-kozak-CeEfa6-MTED (aa24-42)-GSGSGS-EGFP-PacI, XbaI |
| pcDNA3.1-HsPSD1-FL-EGFP (aa1-1024) | this study | KpnI, AscI-kozak-HsPSD1 (aa1-1024)-GSGSGS-NotI-EGFP-PacI, XbaI |
| pcDNA3.1-HsPSD1-Nterm-EGFP (aa1-280) | this study | KpnI, AscI-kozak-HsPSD1-Nterm (aa1-280)-GSGSGS-NotI-EGFP-PacI, XbaI |
| pcDNA3.1-HsPSD1-MTED-EGFP (aa31-49) | this study | KpnI, AscI-kozak-HsPSD1-MTED (aa31-49)-GSGSGS-NotI-EGFP-PacI, XbaI |
| pcDNA3.1-HsPSD2-FL-EGFP (aa1-771) | this study | KpnI, AscI-kozak-HsPSD2 (aa1-771)-GSGSGS-NotI-EGFP-PacI, XbaI |
| pcDNA3.1-HsPSD3-EGFP (aa515-1047) | this study | KpnI, AscI-kozak-HsPSD3 (aa515-1047)-GSGSGS-NotI-EGFP-PacI, XbaI |
| pcDNA3.1-HsPSD4-FL-EGFP (aa1-1027) | this study | KpnI, AscI-kozak-HsPSD4 (aa1-1027)-GSGSGS-NotI-EGFP-PacI, XbaI |
| pcDNA3.1-co-HsPSD1-MTED-EGFP (aa31-49) | this study | KpnI, AscI-kozak-HsPSD1-MTED (aa31-49)-GSGSGS-EGFP-PacI, XbaI |
| pcDNA3.1-co-CeEfa6-MTED-EGFP (aa24-42) | this study | KpnI, AscI-kozak-CeEfa6-MTED (aa24-42)-GSGSGS-EGFP-PacI, XbaI |
| pcDNA3.1-co-DmEfa6-MTED-EGFP (aa322-341) | this study | KpnI, AscI-kozak-DmEfa6-MTED (aa322-341)-GSGSGS-EGFP-PacI, XbaI |
| pCS107-DmEfa6-Nterm-EGFP | this study | NotI-kozak-DmEfa6-Nterm (aa1-410)-GSGSGS-EGFP-StuI |
| pFastBac-His6-MCAK-EGFP-StrepII | not known | His6-MCAK::EGFP-StrepII |
| pFastBac-His6-DmEfa6∆Cterm-EGFP-StrepII (aa1-894) | this study | His6-DmEfa6∆Cterm::EGFP-StrepII (aa1-894) |





