Human MAIT cells respond to and suppress HIV-1
Figures
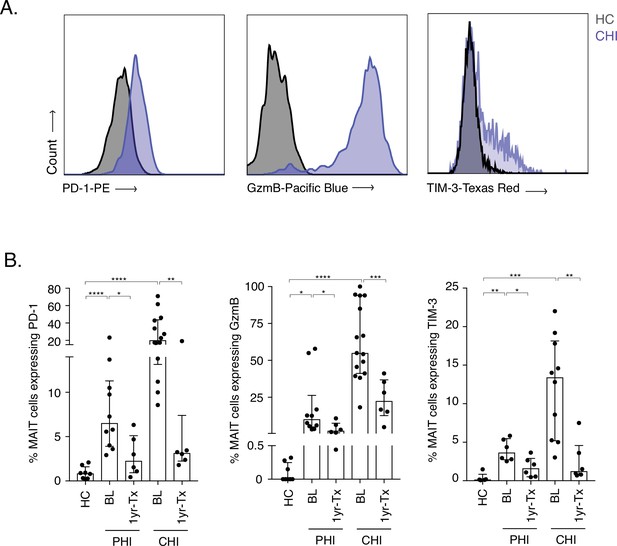
Increased activation and inhibitory marker expression on MAIT cells during HIV-1 infection.
(A) Representative histograms showing upregulation of the activation/inhibitory markers PD-1, Granzyme B (GzmB) and TIM-3 in MAIT cells in chronic HIV-1 infection (CHI) compared to a healthy control (HC). (B) Increased expression of PD-1, GzmB, and TIM-3 on CD8+ CD161++ and Vα7.2+ MAIT cells during PHI and CHI chronic at baseline (BL) and 1 year post-ART (1 yr-Tx). Data points are biological replicates, shown as mean and standard deviation. *p < 0.05, **p < 0.01, *** p < 0.001, **** p < 0.0001; two-tailed t-tests.
-
Figure 1—source data 1
Marker expression on MAIT cells during HIV-1 infection.
(B) GzmB, PD-1, and TIM-3 expression levels on MAIT cells during PHI and CHI HIV-1 infection. Fig. Suppl.(1) TIM-3 expression levels on MAIT cells in EC and VC.
- https://cdn.elifesciences.org/articles/50324/elife-50324-fig1-data1-v2.xlsx
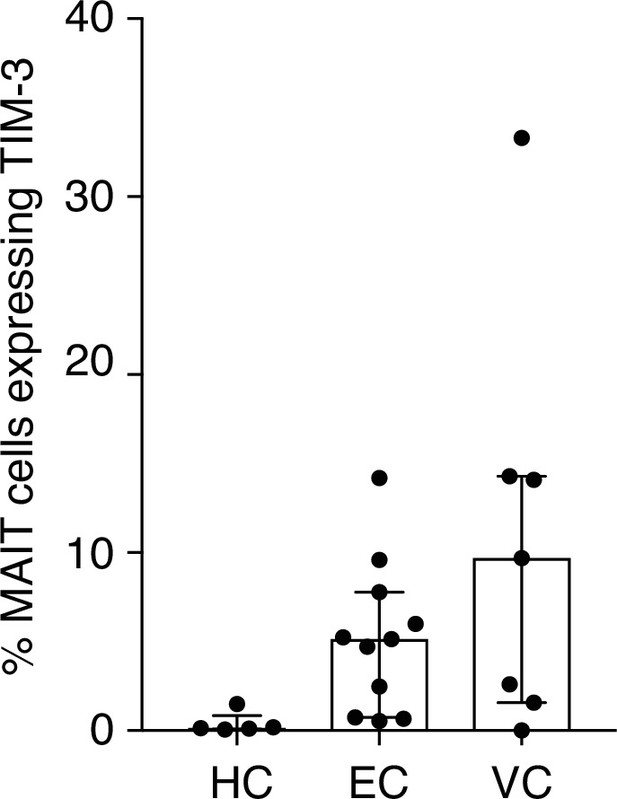
TIM-3 expression on MAIT cells in LTNP.
Elevated but not statistically significant expression of TIM-3 on MAIT cells from Elite vs.
Viraemic Controllers. Data points are biological replicates.
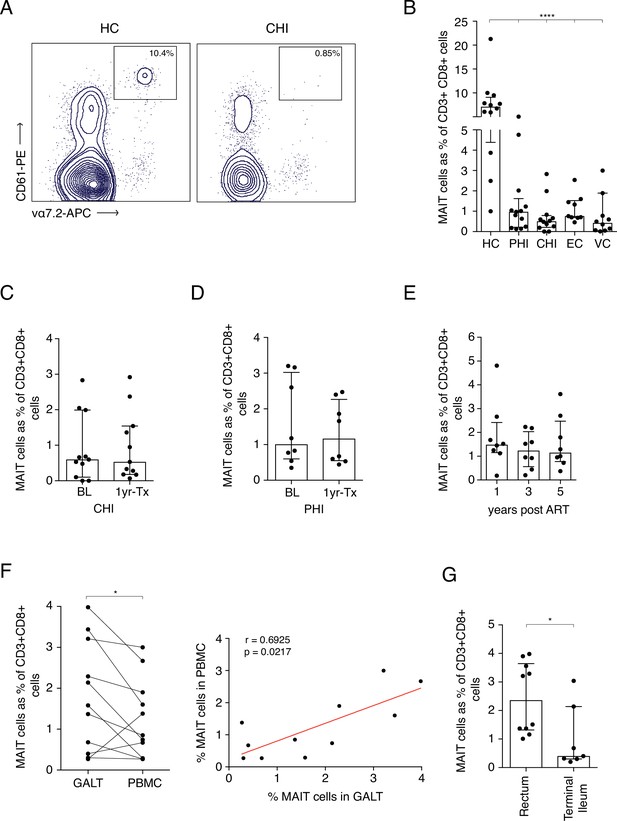
Frequency of MAITs cells in blood and intestine during HIV-1 infection.
(A) Representative dot-plot showing loss of CD8+ MAIT cells, gated on CD161++ and Vα7.2+, in CHI compared to HC. (B) Loss of MAIT cells in peripheral blood in HIV-1+ donors at different HIV-1 stages PHI, CHI, EC (Elite Controllers), and VC (Viraemic controllers). (C) No recovery of MAIT cells post-ART in CHI. (D) No recovery of MAIT cells post-ART in PHI. (E) No recovery of MAIT cells following long-term ART. (F) Higher percentage of MAIT cells in rectal and illeal tissue compared to blood in matched PHI-treated donors. (G) MAIT cell percentages in the rectum compared to terminal ileum of PHI-treated donors. Data points are biological replicates, shown as mean and standard deviation. Spearman’s correlation was used to calculate rho and p value. *p < 0.05, **p < 0.01, *** p < 0.001, **** p < 0.0001; two-tailed t-tests.
-
Figure 2—source data 1
Frequency of MAITs cells in blood and tissue during HIV-1 infection.
(B) MAIT cell frequencies in HC compared to PHI, CHI, EC, and VCs at baseline. (C) MAIT cell frequencies during CHI at baseline and 1 year post-ART. (D) MAIT cell frequencies during PHI at baseline and 1 year post-ART. (E) longitudinal MAIT cell frequencies during CHI at 1, 3, and 5 years post-ART. (F) Comparison of MAIT cell frequencies between GALT and PBMC. (G) Presence of MAIT cells in rectum and terminal ileum of PHI ART-treated subjects.
- https://cdn.elifesciences.org/articles/50324/elife-50324-fig2-data1-v2.xlsx
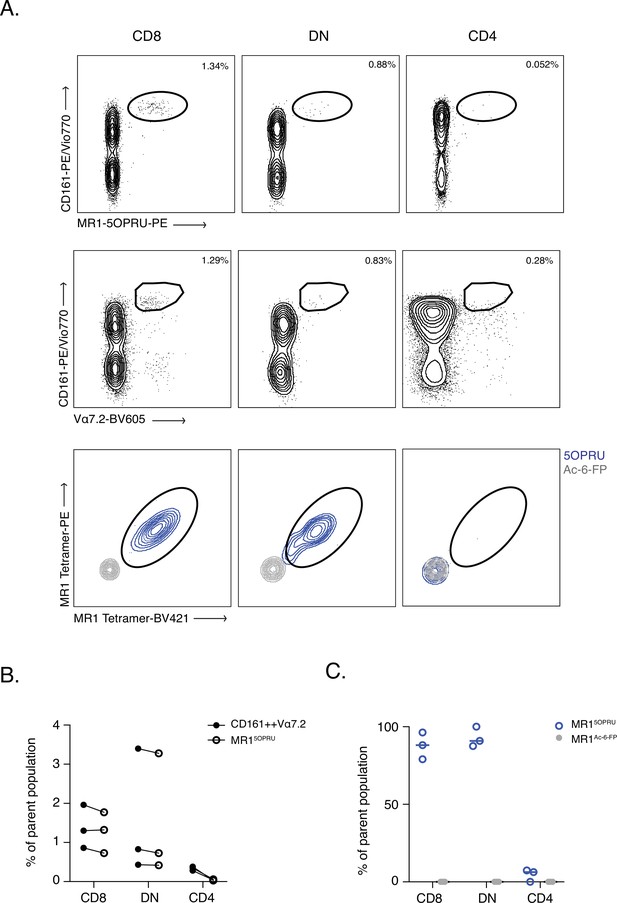
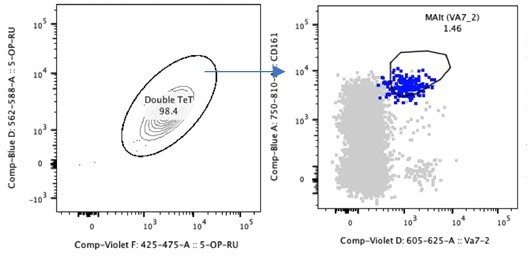
CD161++ Vα7.2+ identifies MAIT cells in tissue- comparable to MR1-5OPRU tetramers.
(A) Representative dot plot of CD161 and Vα7.2 versus MR1-5OPRU tetramer staining in T cell compartments. (B) Comparison of MAIT cell percentages identified using the two staining methods. (C) Proportion of MR1-5OPRU and MR1-Ac-6-FP tetramer-bound cells within CD8+, DN, and CD4+ populations. Data points are biological replicates.
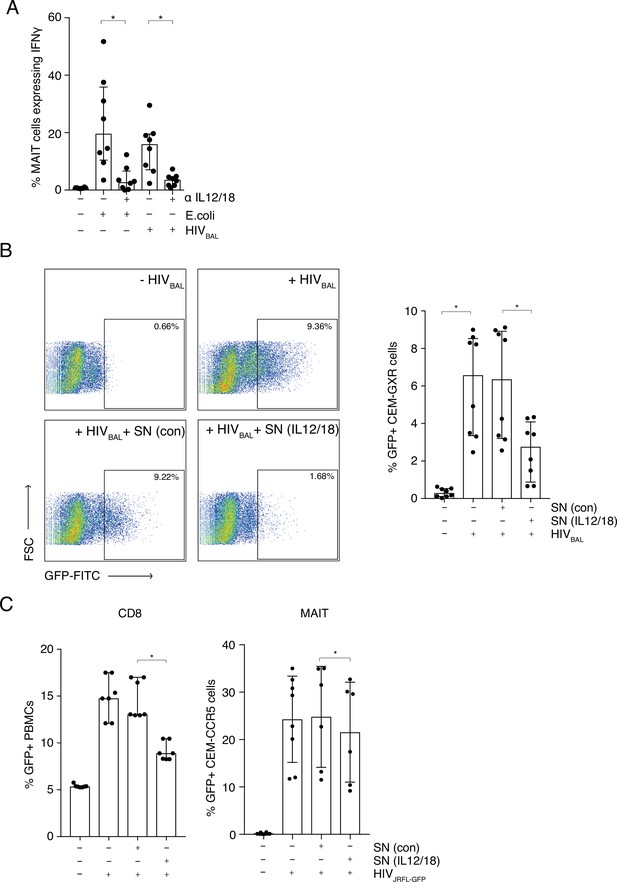
MAIT cells are activated by HIV-1 in an IL-12 and IL-18-dependent manner and display anti- HIV-1 activity.
(A) Bar plots showing the percentage of MAIT cells expressing IFN-γ upon in vitro stimulation with fixed E. coli or HIVBAL in the presence or absence of blocking antibodies directed against IL-12 and IL-18. (B) Reduced frequency of GFP positive CEM-GXR cells following infection with HIVBAL (MOI = 0.2) and pre-treatment with stimulated supernatant from MAIT cells. Shown are representative dot plots (left) and cumulative column bars (right). (C) Inhibition of HIVJRFL-GFP infection in primary human PBMCs or CEM-CCR5 cells by addition of control or IL-12/18-treated supernatants obtained from MACS-enriched CD8s (left) or FACS-sorted MAIT cells (right). *p < 0.05, paired t-tests. Data were pooled from three independent experiments; error bars indicate the standard deviation.
-
Figure 3—source data 1
MAIT cells are activated by HIV-1 in an IL-12 and IL-18-dependent manner.
(A) percentage of MAIT cells expressing IFN-γ upon in vitro stimulation with fixed E. coli or HIVBAL in the presence or absence of blocking antibodies directed against IL-12 and IL-18. (B) frequency of GFP positive CEM-GXR cells following infection with HIVBAL (MOI = 0.2) and pre-treatment with stimulated supernatant from MAIT cells. (C) HIVJRFL-GFP infection in primary human PBMCs or CEM-CCR5 cells by addition of control or IL-12/18-treated supernatants obtained from MACS-enriched CD8s (top) or FACS-sorted MAIT cells (bottom).
- https://cdn.elifesciences.org/articles/50324/elife-50324-fig3-data1-v2.xlsx
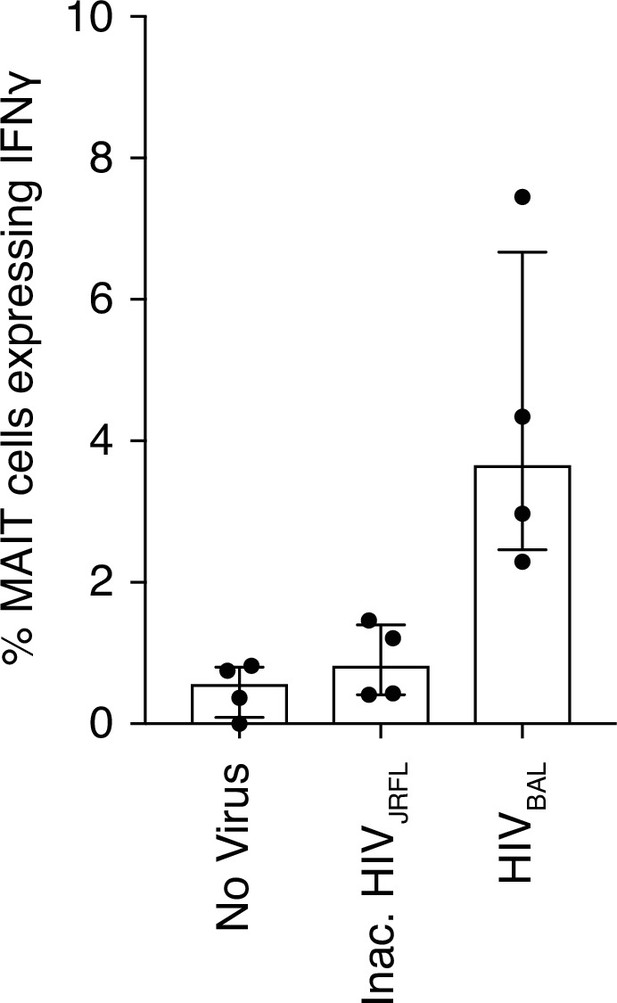
Inactivated HIV-1 does not stimulate MAIT cells.
(A) Increased IFN-γ expression from MAIT cells after infection with HIVBAL but not with an inactivated HIVJRFL virus. Data points are biological replicates.
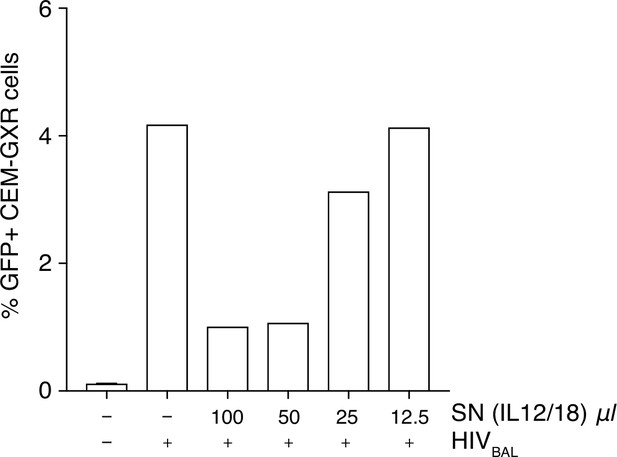
Inhibition of HIV-1 is increased with higher volumes of stimulated supernatant.
(A) Titration of IL-12/18 stimulated MAIT cell supernatants on CEM-GXR cells infected with HIVBAL. Plotted are the means from two biological replicates.
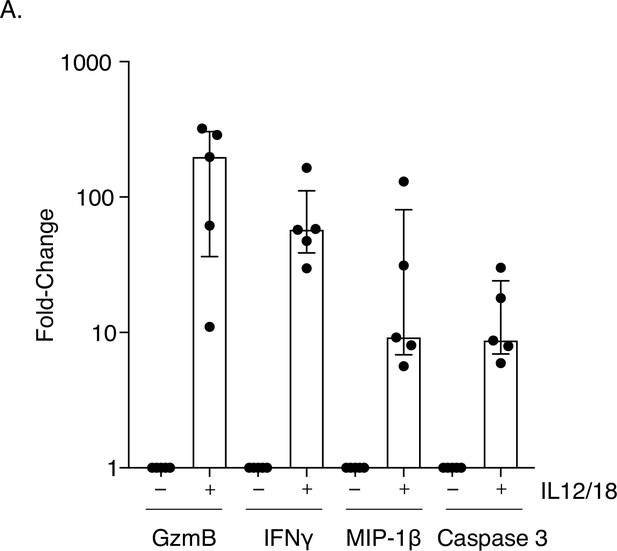
Increased expression of restriction factor and pro-apoptosis markers in MAIT cells stimulated with IL-12 and IL-18.
(A) Dot plots showing increased intracellular activation and apoptosis marker expression (caspase 3) in MAIT cells after IL-12/18 stimulation. Data points are biological replicates.
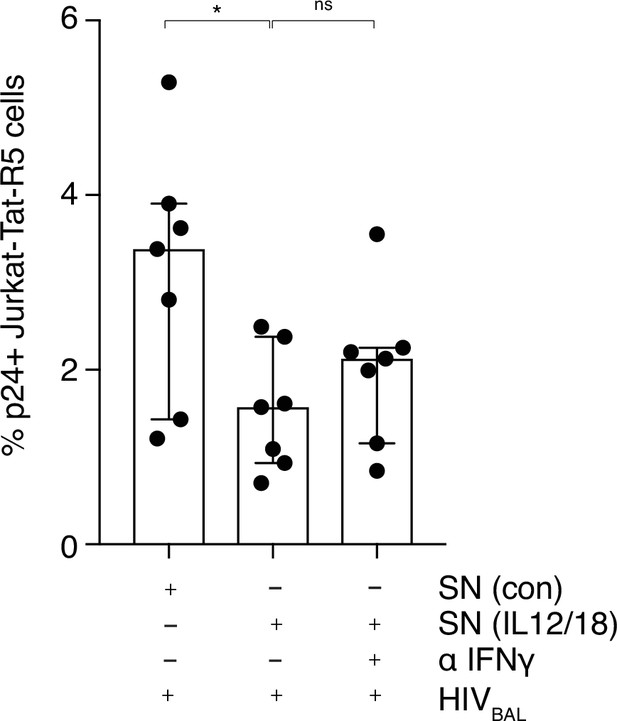
MAIT cell anti-viral activity is not dependent on IFNγ.
(A) Reduced p24 expression in Jurkat-Tat-R5 cells following incubation with stimulated supernatants from MAIT cells compared to unstimulated control supernatants. Addition of an IFNγ blocking antibody did not fully rescue the block to infection. Data points are biological replicates. Bar plot shown as mean and standard deviation. *p < 0.05; two-tailed t-tests. ns = not significant.
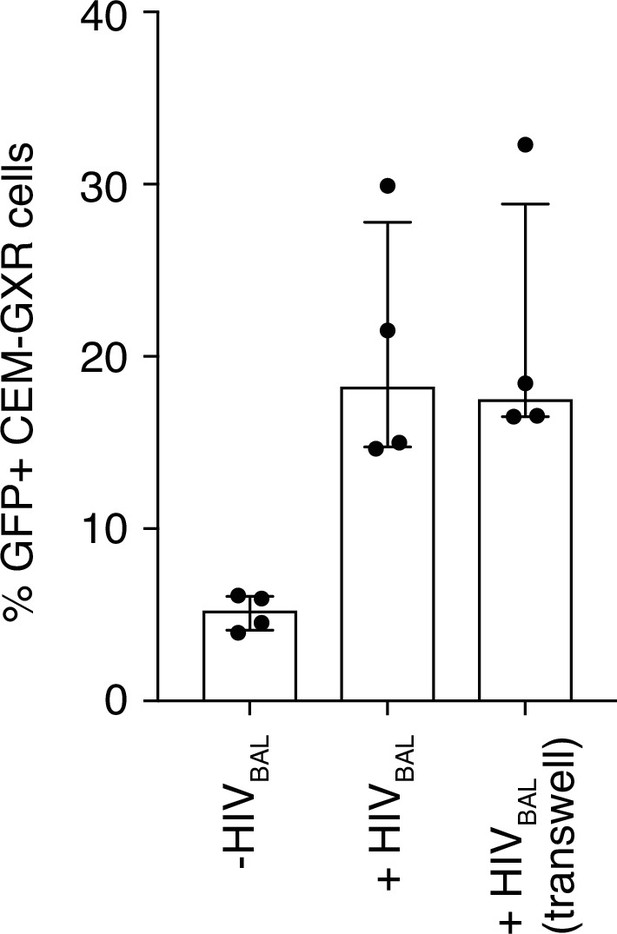
Inhibition of HIV-1 is not dependent on cell contact.
(A) MAIT cells and CEM-GXR cells in co-culture or physically separated in transwell plates were infected with HIVBAL and the number of GFP+ cells was measured. Data points are biological replicates. Bar plot shown as mean and standard deviation.
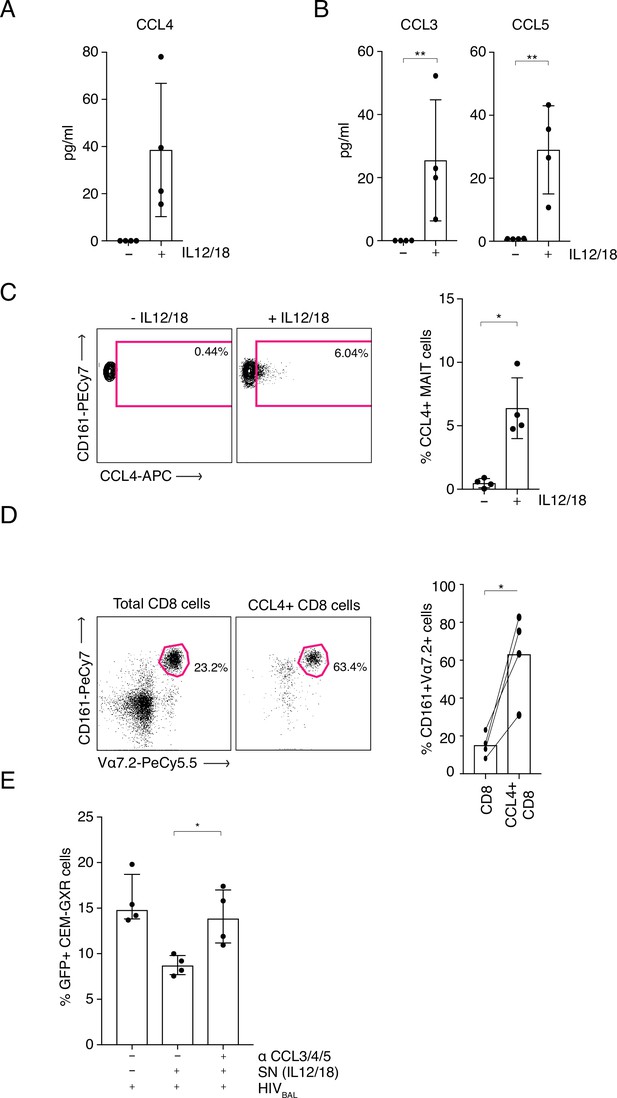
MAIT cell derived antiviral restriction factors are essential for suppressing HIV-1 in vitro.
(A) MAIT cells were FACS-sorted and the CCL4 (MIP-1β) concentration was measured in the supernatants by ELISA after 20 hr post stimulation with IL-12/18. (B) MAIT cells were FACS-sorted and the concentrations of CCL3 (MIP1α) and CCL5 (RANTES) were measured in the supernatants by cytometric bead array (CBA) after 20 hr post stimulation with IL-12/18. (C) Representative FACS-plots (left) and bar plots (right) depicting the expression of CCL4 by MAITs after incubation with IL-12/18 for 20 hr. MAIT cells were identified as CD161++ Vα7.2+ cells within MACS-enriched CD8s. (D) Representative FACS dot plots (left) and bar plots (right) showing the percentage of MAIT cells as identified by co-expression of Vα7.2 with high levels of CD161 within MACS-enriched CD8s and within all CCL4-expressing CD8 T cells from the same culture. CD8 T cells were stimulated with IL-12/18 for 20 hr. (E) Recovery of GFP-positive CEM-GXR cells following blocking of restriction factors (CCL3/4/5), after treatment with IL12/18 stimulated supernatant from CD8 cells and infection with HIVBAL. *p < 0.05, **p < 0.05, paired t-tests. Data were pooled from two independent experiments; error bars indicate the standard deviation.
-
Figure 4—source data 1
MAIT-cell-derived antiviral restriction factors are essential for suppressing HIV-1.
CCL4 (MIP-1β) concentration was measured in the supernatants by ELISA after 20 hr post stimulation with IL-12/18. (B) Concentrations of CCL3 (MIP1α) and CCL5 (RANTES) were measured in the supernatants by cytometric bead array (CBA) after 20 hr post stimulation with IL-12/18. (C) Expression of CCL4 by MAITs after incubation with IL-12/18 for 20 hr. MAIT cells were identified as CD161++ Vα7.2+ cells within MACS-enriched CD8s. (D) Percentage of MAIT cells as identified by co-expression of Vα7.2 with high levels of CD161 within MACS-enriched CD8s and within all CCL4-expressing CD8 T cells from the same culture. CD8 T cells were stimulated with IL-12/18 for 20 hr. (E) GFP-positive CEM-GXR cells following blocking of restriction factors (CCL3/4/5), after treatment with IL12/18 stimulated supernatant from CD8 cells and infection with HIVBAL.
- https://cdn.elifesciences.org/articles/50324/elife-50324-fig4-data1-v2.xlsx
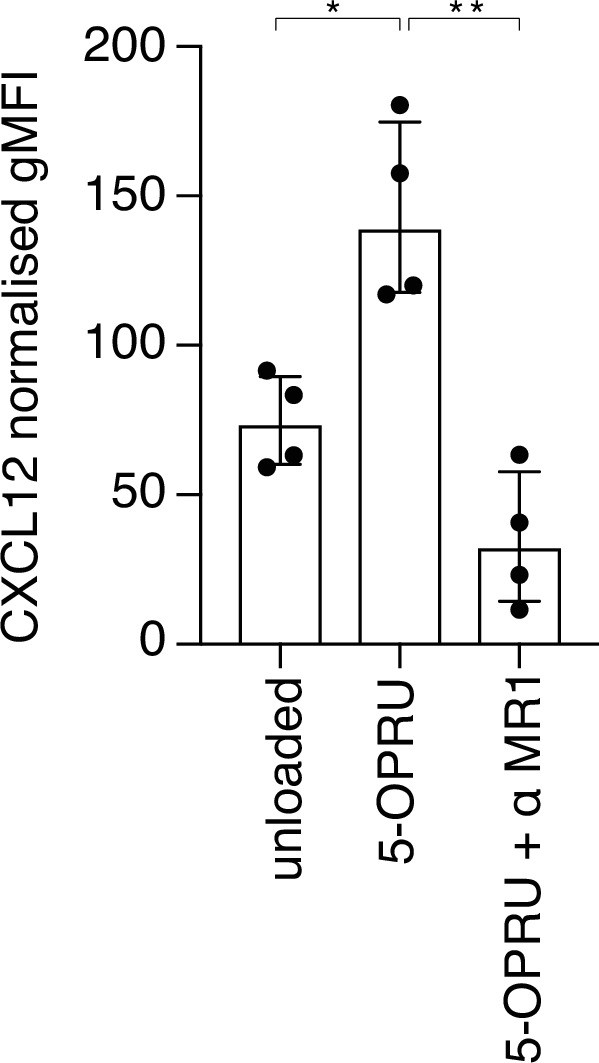
TCR-induced expression of CXCL12/SDF-1 by activated MAIT cells.
(A) Intracellular expression (gMFI) of CXCL12 is increased with addition of 5-OPRU and decreased with a blocking antibody against MR1 (α MR1). Data points are biological replicates. Bar plot shown as mean and standard deviation. *p < 0.05, **p < 0.01; two-tailed t-tests.
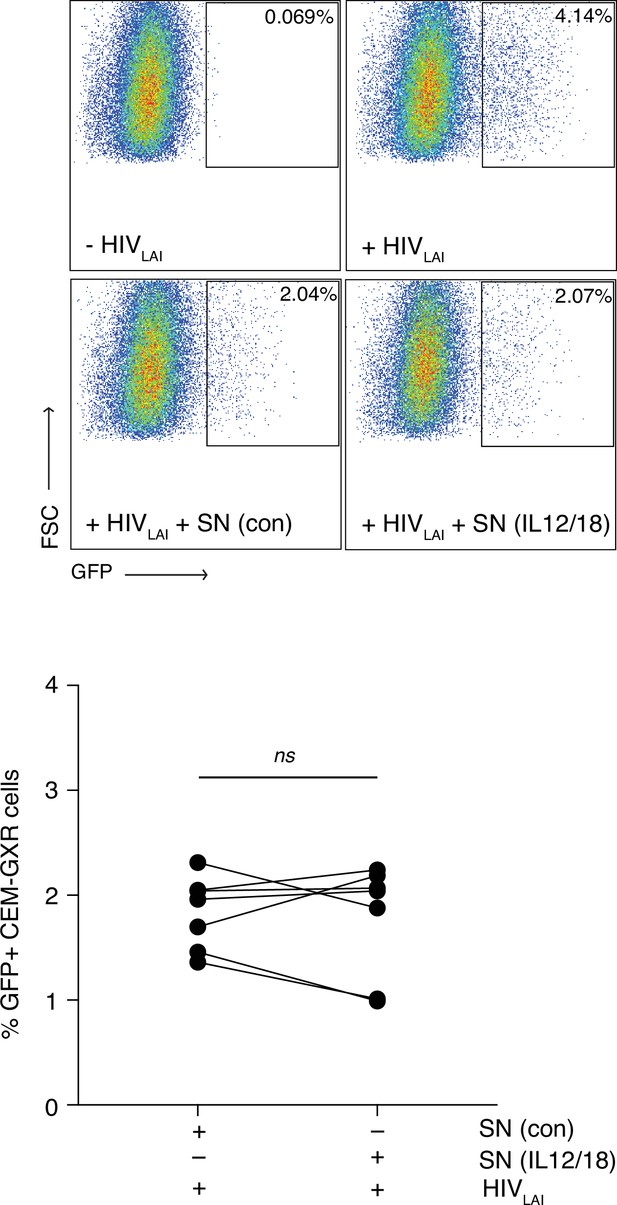
MAIT cells do not inhibit infection by a CXCR4 tropic virus.
(A) Representative FACS plots showing infection of CEM-GXR cells with the CXCR4 tropic virus HIVLAI following pre-treatment with unstimulated or IL-12/18-stimulated MAIT cell supernatants. B. Bar graph showing HIVLAI infection of CEM-GXR cells after incubation with unstimulated or IL-12/18-stimulated MAIT cell supernatants. Data points are biological replicates. ns = not significant, paired two-tailed t-test.
Tables
Participant cohort characteristics.
CD4 T cell count and HIV-1 (log) viral load.
| Patient cohort | Sample Number | CD4+ T cells (Count/μL) median (IQR) | Plasma viral load (log10 copies/mL) median (IQR) |
|---|---|---|---|
| Healthy Donors | 12 | - | - |
| PHI (SPARTAC) | 8 | 596 (437–755) | 5.04 (4.51–5.45) |
| PHI (HEATHER) | 12 | 524 (437–656) | 4.34 (3.19–4.88) |
| HEATHER GUT | 11 | - | < 1.3 |
| CHI | 12 | 360 (72–646) | 4.90 (3.55–5.59) |
| EC LTNP | 9 | 780 (615–1013) | 1.70 (1.60–1.70) |
| VC LTNP | 10 | 633 (442–800) | 5.18 (5.03–5.37) |