Dullard-mediated Smad1/5/8 inhibition controls mouse cardiac neural crest cells condensation and outflow tract septation
Figures
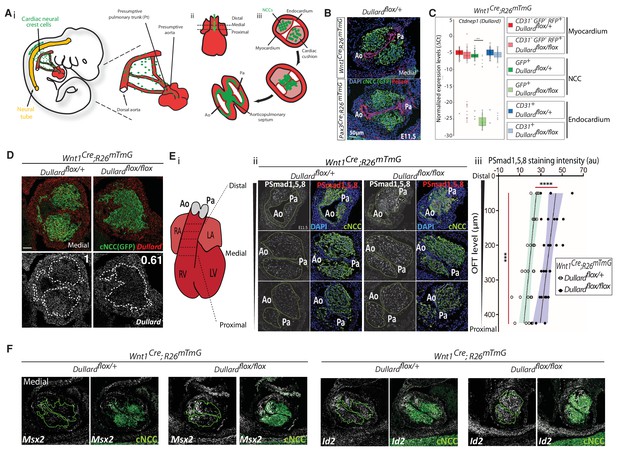
Dullard acts as a Smad1/5/8 activity inhibitor in cardiac NCC.
(A) Ai. Schematic representation of the migration routes the cardiac NCC (green) have taken to reach the heart region (red) in a E10.5 mouse embryo. Aii. Schematics of the embryonic heart at E11.5 showing the distal-proximal axis of the OFT. Aiii. Schematic representation of transverse sections through the OFT showing discrete stages of NCC condensation and endocardium septation along the OFT distal-proximal axis. (B) Pecam and GFP immunolabelling and DAPI staining on transverse sections throughout the medial OFT of E11.5 Wnt1Cre or Pax3Cre; Dullardflox/+; Rosa26mTmG embryos. (C) Normalized expression levels of Dullard assayed by q-RT-PCR on single cells isolated after immuno-marking endothelial CD31+ cells from E11.5 Wnt1Cre; Dullardflox/+ and Wnt1Cre; Dullardflox/flox; Rosa26mTmG hearts (dots: value for a single cell; boxplot: mean ± s.e.m.). The primers used to amplify Dullard specifically binds to exons 2 and 3, which are excised by the Cre recombinase. (D) Dullard mRNA distribution detected using RNAscope probes, in transverse sections of E11.5 control and mutant OFTs, assessed by RNAscope. Dullard mRNA levels were significantly reduced in mutant cardiac cushions compared to controls; however, mRNA signals were still detected given the binding of Z pair probes to non-recombined exons 5 to 8 and UTR region. (E) Ei. Schematics of E11.5 heart showing the position of the transverse sections used to quantify the levels of the phosphorylated forms of Smad1/5/8 in iii. Eii. Immunolabelling for P-Smad1/5/8 and GFP, and DAPI staining on transverse sections across the OFT at three distinct distal-proximal levels in E11.5 embryos with the indicated genotype. Eiii. Quantification of P-Smad1/5/8 levels in cardiac NCC along the OFT distal-proximal axis of E11.5 embryos with the indicated genotype (dots: values obtained on a given section; n > 4 embryos per genotype recovered from at least three liters; the black line is the linear regression, the coloured areas delineate the 95% confidence intervals, ***: p-value<0001 for a two-way Anova statistical test). (F) Msx2 and Id2 mRNA distribution detected using RNAscope probes (grey) and immunostaining of GFP (green) in transverse sections of E11.5 control and mutant OFTs (n = 2 embryos). On all A-F panels: green dotted lines delineate the area colonised by cardiac NCC. Ao: aortic artery, Pa: pulmonary artery.
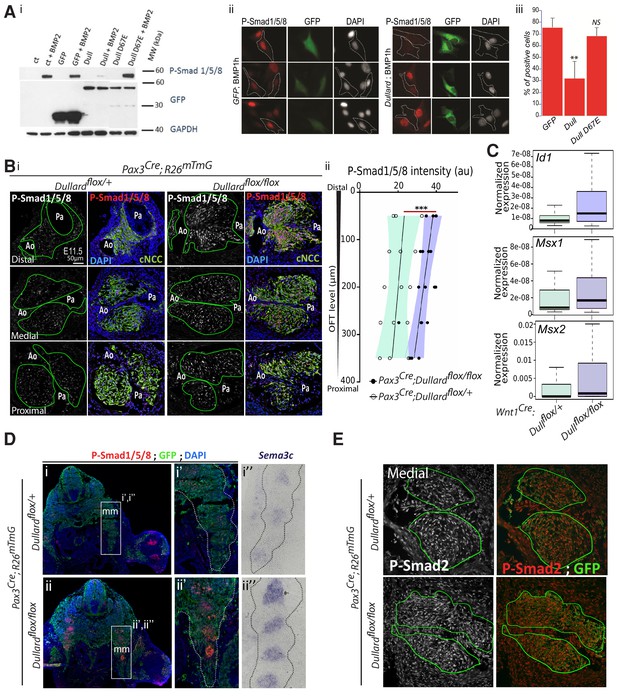
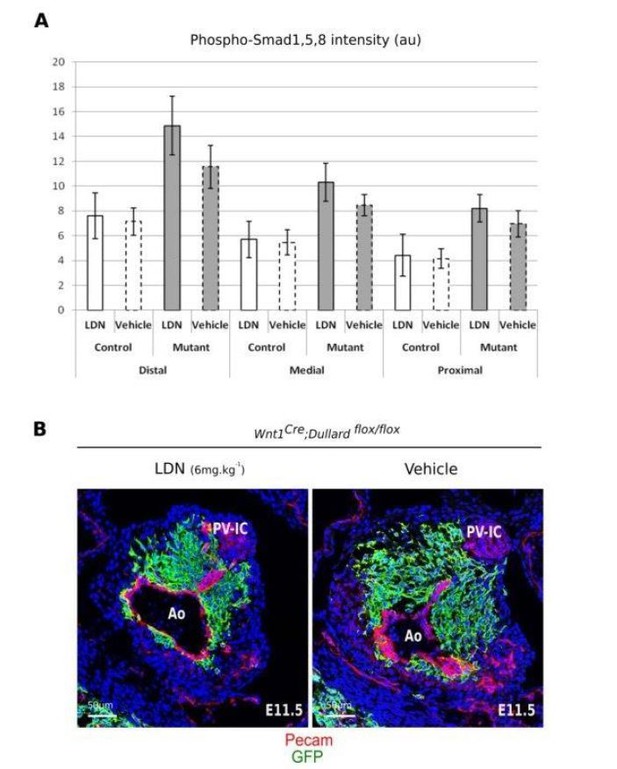
Dullard phosphatase is a negative regulator of BMP signalling in several mammalian cells.
(A) Ai. Western blot detecting the phosphorylated forms of Smad1/5/8, GFP or Gapdh in C2C12 muscle cells with or without BMP2 treatment for 1 hr. These cells were non-transfected (ct) or transfected with either a GFP expressing plasmid (GFP), a GFP tagged version of the wild-type Dullard (Dull) or of Dullard carrying D67E mutation in its phosphatase domain (Dull D67E). Dullard inhibits BMP2-mediated phosphorylation of Smad1/5/8; this inhibition is dependent on the functionality of its phosphatase domain. Aii. Immunofluorescence for P-Smad1/5/8 and GFP and DAPI staining in C2C12 transfected with GFP or GFP-Dullard and exposed for 1 hr to BMP2 showing that only cells transfected with Dullard do not show nuclear phosphorylated Smad1/5/8. Aiii. Quantification of the number of P-Smad1/5/8 positive C2C12 cells exposed to BMP2 for 1 hr and transfected with GFP (ct), Dullard or Dullard carrying the D67E mutation (n = 3 independent experiments; Student t-test **: p-value<0.01.; N.S.: non-significant). (B) Bi. Immunolabelling for P-Smad1/5/8 and GFP and DAPI staining on transverse sections across the OFT at three distinct distal-proximal levels in E11.5 of control Pax3Cre; Dullardflox/+; Rosa26mTmG and mutant Pax3Cre; Dullardflox/flox; Rosa26mTmG hearts. Pale green dotted lines delineate the area colonized by cardiac NCC. Bii. Quantification of P-Smad1/5/8 levels in cardiac NCC along the distal-proximal axis of the OFT of E11.5 embryos with the indicated genotype (dots: values obtained on a given section; n > 4 embryos per genotype recovered from at least three liters; the black line is the linear regression, the coloured areas delineate the 95% confidence intervals, ***: p-value<0001 for a two-way Anova statistical test). Ao: aortic artery, Pa: pulmonary artery. (C) Normalized expression levels of Id1, Msx1 and Msx2 assayed by q-RT-PCR on single GFP+ cardiac NCC isolated from E11.5 Wnt1Cre; Dullardflox/+ and Wnt1Cre; Dullardflox/flox; Rosa26mTmG hearts (boxplot: mean ± s.e.m.). (D) Immunolabelling of P-Smads1/5/8 and Sema3c transcripts detected by ISH on transverse sections of E11.5 control and Dullard mutant at brachial levels. i’’ to i’’’ and ii’’ to ii’’’ are blown up on muscle masses (mm). (E) Immunolabelling for the phosphorylated form of Smad2 and GFP and DAPI staining on transverse sections across the OFT at medial levels in E11.5 of control Pax3Cre; Dullardflox/+; Rosa26mTmG and mutant Pax3Cre; Dullardflox/flox; Rosa26mTmG hearts. Pale green dotted lines delineate the area colonised by cardiac NCC. P-Smad2 levels are similar in control and dullard mutant cardiac NCC. Ao: aortic artery, Pa: pulmonary artery.
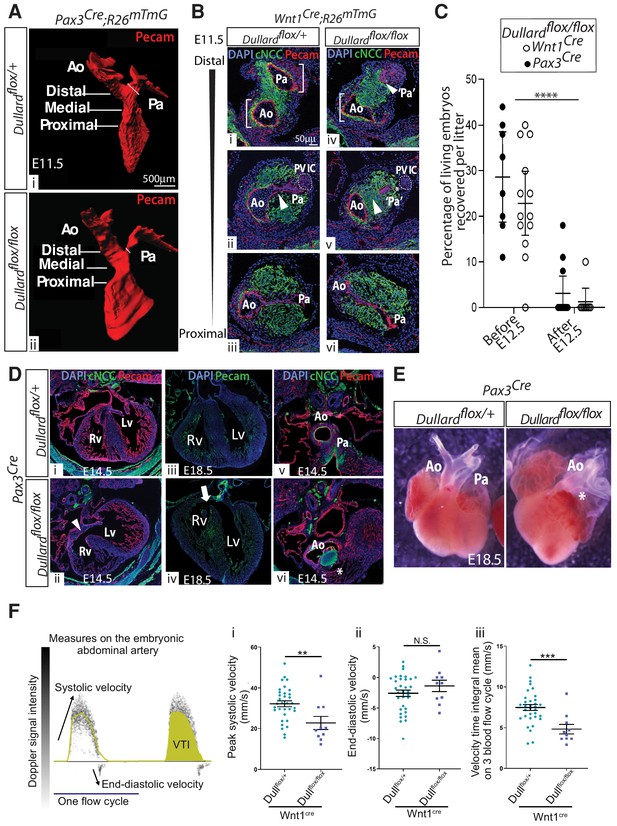
Dullard deletion in cardiac NCC causes asymmetric and premature OFT septation similar to Fallot’s tetralogy.
(A) Three-dimensional rendering of the Pecam+ endocardium of E12 Pax3Cre; Dullardflox/+ and Pax3Cre; Dullardflox/flox embryos after 3Disco clearing and lightsheet acquisition (n = 3 per genotype). The fine oblique white line marks the Pa width. The OFT levels along its distal-proximal axis analyzed in B are also indicated. (B) Immunolabelling for Pecam (red), GFP (green) and DAPI (blue) on transverse sections along the distal-proximal axis of the OFT in E11.5 embryos with the indicated genotypes (n > 10 embryos collected from more than three liters). Brackets in i and iv highlight the symmetric and asymmetric Ao and Pa poles in control and mutant embryos, respectively. Arrowheads in ii and v point at the unruptured and ruptured endocardium in control and mutant embryos, respectively. (C) Percentage of living Dullard mutant embryos before E12.5 and after E12.5, carrying the indicated Cre driver. (D) Immunolabelling for Pecam, GFP and DAPI staining on sections through the hearts of E14.5 (i,ii,v,vi) and E18.5 (iii,iv) embryos with the indicated genotypes (n = 2 embryos per genotype). Arrowheads in ii and arrow in iv point at a septation defect, the star in vi indicates the lack of Pa. (E) Whole dissected E18.5 hearts coming from embryos with the indicated genotype (n = 2 per genotype). (F) Two cycles of blood flow measured at the level of the abdominal artery of E11.75 control embryos and indication of the parameters analysed. VTI: velocity time integral. Parameters (i-iii) of the blood flow velocity measured in the abdominal artery of E11.75 control (turquoise dots) and Wnt1Cre; Dullardflox/flox embryos (purple squares)(dots and squares: mean of two to five measures obtained on a single embryo, bars: mean ± s.e.m; differences evaluated using a Mann-Whitney test: N.S. non-significant, *: p<0.05, **: p<0.01, ***: p<0.001). i. peak systolic velocity, ii. end-diastolic velocity, iii. mean of three velocity time integrals (n = 10 mutants and n = 32 controls). Ao: aortic artery, Pa: pulmonary artery, PV IC: pulmonary valve intercalated-cushion.
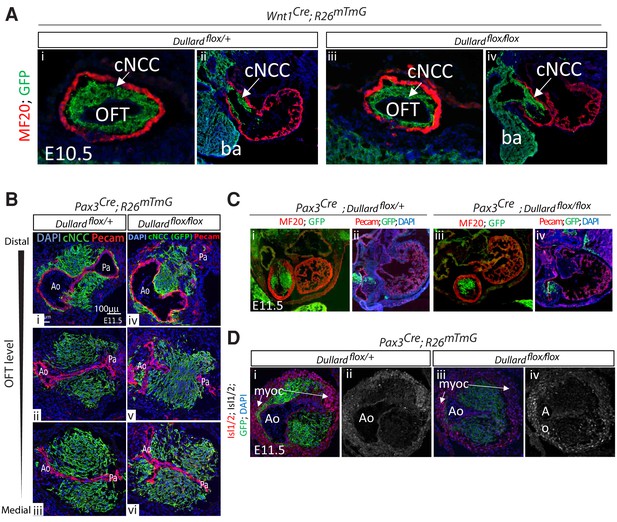
Morphological defects in Dullard mutants become striking from E11.5 onwards.
(A) MF20 (myosin heavy chain) and GFP immunostaining and DAPI staining on transverse sections (i,iii) and coronal sections (ii,iv) in E10.5 embryos with the indicated genotype. (B) Immunolabelling for Pecam (red) and GFP (green) and DAPI (blue) staining on transverse sections along the distal-proximal axis of the OFT in E11.5 embryos with the indicated genotypes (n > 10 embryos collected from more than three liters). (C) Immunolabelling of MF20, Isl1/2, Pecam and GFP, as well as DAPI staining, on transverse sections of E11.5 hearts of embryos with the indicated genotypes. Ao: Aorta; Pa: pulmonary artery; ba: branchial arch; myoc: myocardium; Lv: left ventricle; Rv: right ventricle.
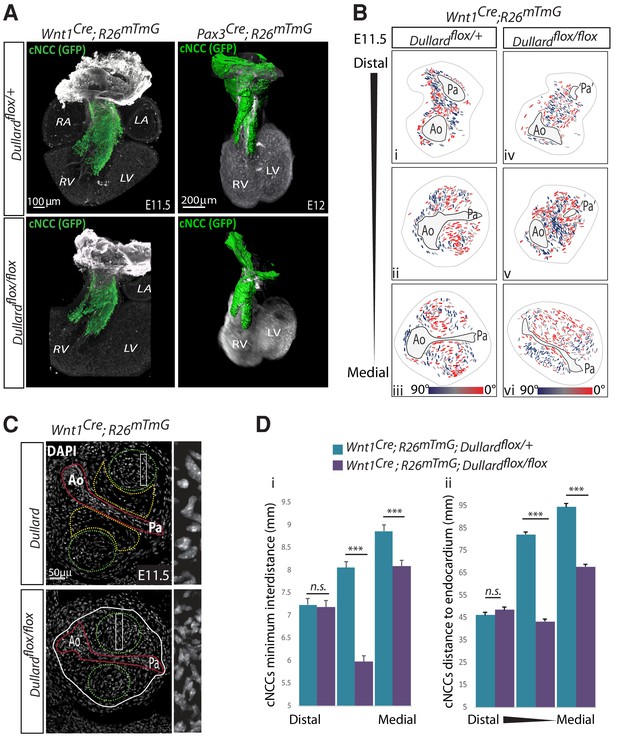
Dullard does not affect NCC migration, but prevents NCC premature condensation.
(A) Three-dimensional rendering of cardiac NCC (green) over Pecam (white) after BABB clearing and confocal acquisition (Wnt1cre samples) or 3disco clearing and lightsheet microscopy (Pax3cre samples) of whole E11.5 hearts isolated from embryos with the indicated genotype (n = 2 per genotype). (B) Coloured coded orientation of the major axis of NCC cells relative to Ao-Pa axis colour-coded as indicated in the section shown in Figure 2Bi–vi. (C) DAPI staining on transverse sections through the medial part of the OFT of E11.5 embryos with the indicated genotype. Magnified regions on the right are indicated by white rectangles in NCC cushions. The entire OFT is circled with a while line. The endocardium is delineated in red, the condensed and round NCC in green, the loose and elongated NCC in yellow. (D) Minimum distances between NCCs (Di) and distances between NCCs and the endocardium (Dii) quantified along the distal-proximal axis of the OFT in E11.5 embryos with the indicated genotypes (n = 3 embryos from distinct liters were analyzed for each genotype and OFT level, bars: mean ±s.d.; ***: p-value<0.0001 for Student statistical t-test). Ao: Aorta; Pa: pulmonary artery; Lv: left ventricle; PV-IC: Pulmonary valve intercalated-cushion; Rv: right ventricle.
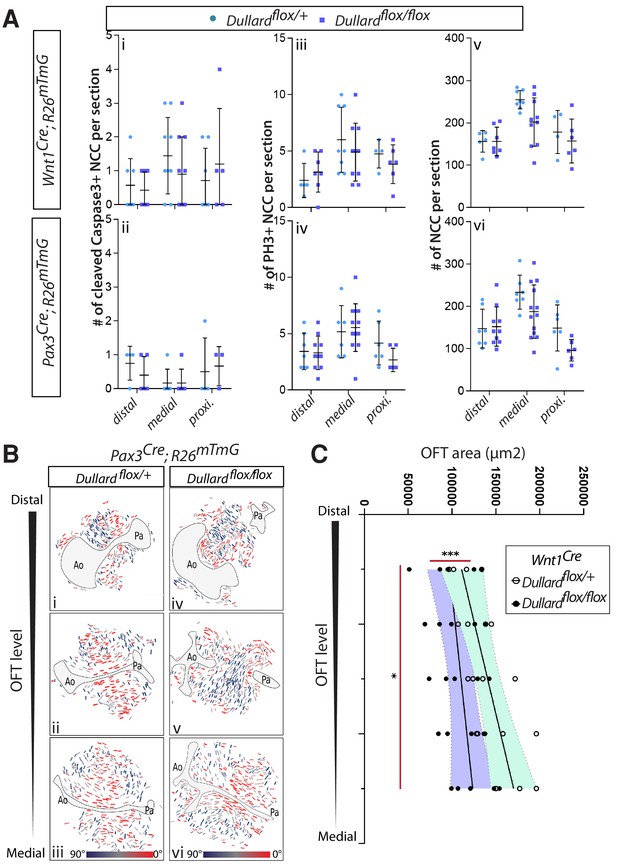
Morphogenetic defects in Dullard NCC mutants.
(A) Quantification of the total number of activated caspase 3+ apoptotic NCC (i,ii), PH3+ mitotic cardiac NCC (iii,iv) and GFP+ cardiac NCC (v,vi) per OFT section at the indicated distal-proximal OFT axis levels (dots: value per section; bars: mean ±s.d.; n.s.: non-significant differences evaluated using a Mann-Whitney test). (B) Orientation of the major axis of NCC cells relative to Ao-Pa axis colour coded as indicated in the section shown in Figure supplement 2B. (C) Quantification of the OFT surface area measured at distinct distal-proximal axis levels in control and Dullard mutants at E11.5 (dots: values obtained on a given section; n > 4 embryos per genotype recovered from at least three liters; the black line is the linear regression, the coloured areas delineate the 95% confidence intervals, ***: p-value<0001 for a two way-Anova statistical test).
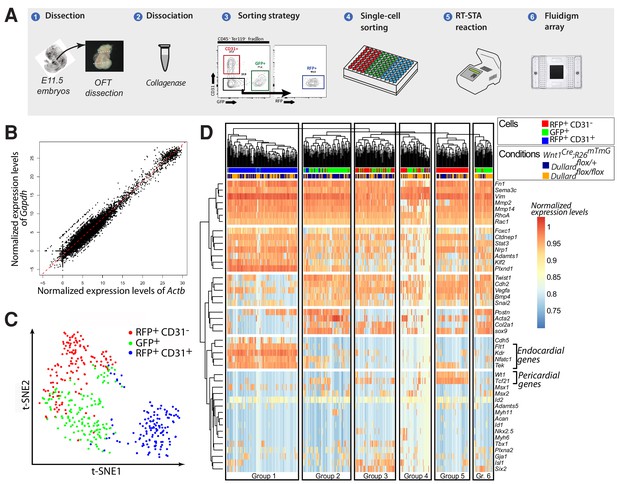
Single-cell transcriptional analyses of all OFT cells at E11.5.
(A) Experimental steps performed to profile gene expression in OFT single-cells sorted from five Wnt1Cre; Dullardflox/+; Rosa26mTmG and five Wnt1Cre; Dullardflox/flox; Rosa26mTmG E11.5 embryos. At least 70 cells were isolated per gate and genotype (GFP+, CD31+, RFP+). (B) Graph showing the distribution of all cells analysed (dots) as a function of normalised expression values of the house keeping genes Actb and Gapdh and a linear regression (red line). (C) t-SNE plot showing the distribution of 44-genes-based transcriptomes of 433 OFT cells expressing the indicated markers isolated from both Wnt1Cre; Dullard+/flox; Rosa26mTmG and Wnt1Cre; Dullardflox/flox; Rosa26mTmG E11.5 embryos. (D) Unsupervised clustering heatmap of the 433 OFT isolated cells from Wnt1Cre; Dullard+/flox; Rosa26mTmG and Wnt1Cre; Dullardflox/flox; Rosa26mTmG based on the gene expression level of the 44 genes included in the panel (Supplementary file 1). Six different groups of cells can be discriminated, among which the endocardial (Group 1) cells expressing high levels of Flt1, Kdr, Nfatc1 and Tek, and the epicardial (Group 5) cells expressing high levels of Wt1, Tcf21..
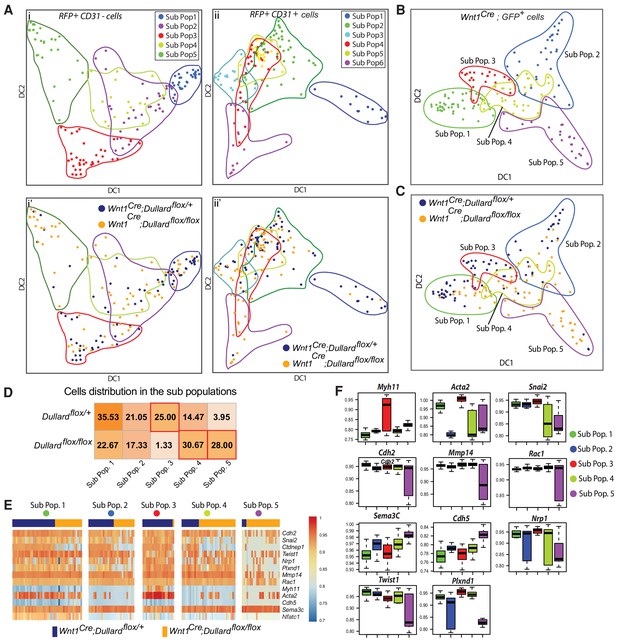
Impact of Dullard deletion in NCC on the transcriptomic variations within the distinct cellular subtypes of E11.5 hearts.
(A) Projected position of 44 genes-based transcriptomes assessed in mutant and control CD31+; RFP+ and CD31-; RFP+ OFT cells on diffusion maps made using the first two Diffusion Component 1 (DC1) and 2 (DC2) (Figure 5—figure supplement 1B). Sub-populations defined with hierarchical clustering are presented in i and ii, while the genotype of cells is illustrated in i’ and ii’. (B, C) Projected position of 44 genes-based transcriptomes assessed in mutant and control cardiac GFP+ NCC on diffusion maps made using the first two Diffusion Component 1 (DC1) and 2 (DC2) (see Figure 5—figure supplement 2B). The five subpopulations defined in Figure 3A are highlighted. (D) Percentage of cardiac NCC in each subpopulation. (E) Heatmaps showing the levels of expression of selected genes in all GFP+ NCC in the five subpopulations identified using unsupervised hierarchical clustering (Figure 5—figure supplement 2A) coming from control (blue) or mutant (orange) E11.5 embryos. (F) Boxplot representation of the expression levels of genes differentially expressed between the five NCC subpopulations (Sub-Pop 1: 44 cells, Sub-Pop 2: 29 cells, Sub-Pop3: 20 cells, Sub-Pop 4: 34 cells, Sub-Pop 5: 24 cells) (mean ±s.d.).
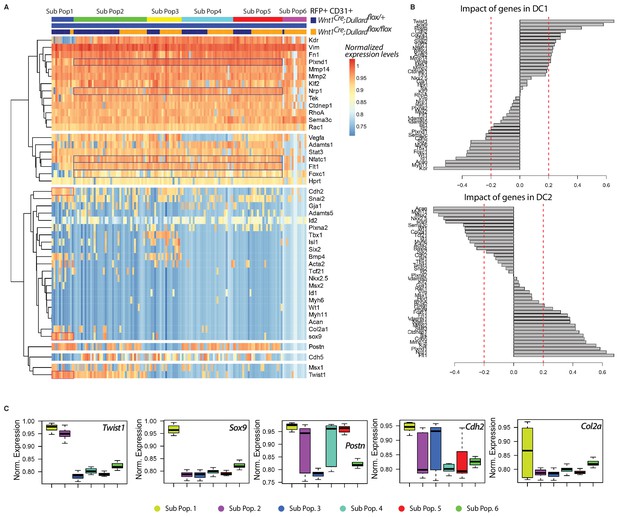
Transcriptional profiling of CD31+; RFP+ OFT cells.
(A) Unsupervised clustering heatmap of 139 CD31+; RFP+ endocardium cells isolated from Wnt1Cre; Dullard+/flox; Rosa26mTmG and Wnt1Cre; Dullardflox/flox; Rosa26mTmG based on the gene expression level of the 44 genes included in the panel (Supplementary file 1). Six different subpopulations can be identified, two of which (Sub-Pop 1 and 6) are enriched for either mutant (Sub-Pop 6) or control cells (Sub-Pop 1). (B) Genetic composition of the diffusion components (DC) 1 and 2 for CD31+; RFP+ endocardium cells on which are based the diffusion maps presented in Figure 5A. (C) Boxplot representation of the expression levels of genes highlighted in DC1 and 2 shown in B in the 6 subpopulations of CD31+; RFP+ endocardium cells defined in A (mean ±s.d.).
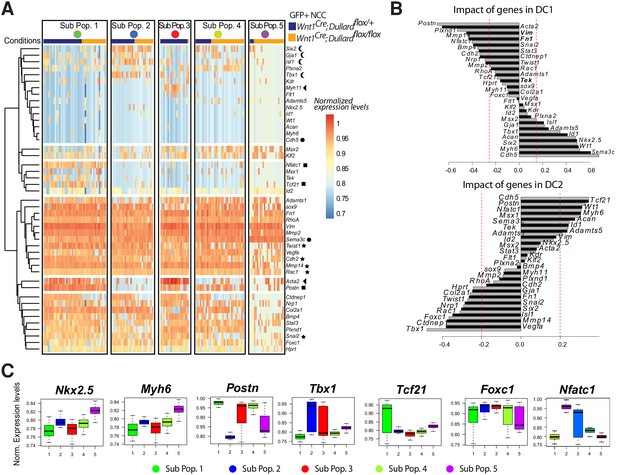
Single-cell transcriptional profiling of GFP+ NCC at E11.5.
(A) Unsupervised clustering heatmap of 151 GFP+ NCC isolated from Wnt1Cre; Dullard+/flox; Rosa26mTmG and Wnt1Cre; Dullardflox/flox; Rosa26mTmG based on the gene expression level of the 44 genes included in the panel (Supplementary file 1). Five different subpopulations can be identified, two of which (Sub-Pop 1 and 2) are not Dullard dependent, while the three others (Sub-Pop3 to 5) are enriched for either mutant (Sub-Pop 4,5) or control cells (Sub-Pop 3). Stars indicate genes involved in the mesenchymal state. The circles marks instead the genes associated with an epithelial fate. The arrows points at genes encoding for smooth muscle specific genes marking the subpopulation 3. Crescents point to typical cardiac genes, whereas squares point to genes involved in valve formation. (B) Genetic composition of the diffusion components (DC) 1 and 2, on which are based the diffusion maps presented in Figure 5B and C. (C) Boxplot representation of the expression levels of genes highlighted in DC1 and 2 in B and not presented in Figure 5F, in the GFP+ NCC cells from both genotypes (mean ±s.d.).
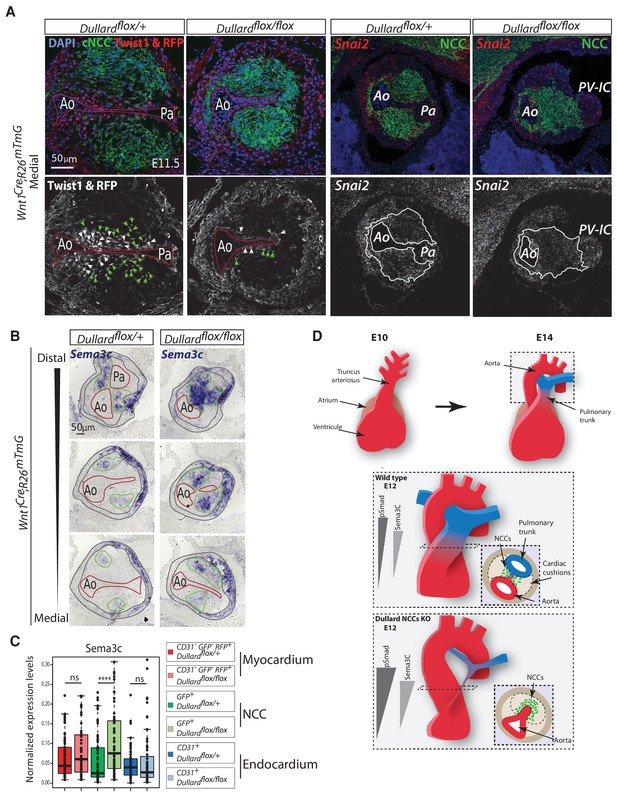
Dullard prevents NCC from acquiring epithelial-like traits and prolongs the expression of mesenchymal drivers.
(E) Twist1 immunolabelling and RFP signal (red; grey), Snai2 mRNA distribution assessed by RNAscope (red; grey), GFP (green) immunolabelling and DAPI staining on transverse sections through the medial OFT of E11.5 Wnt1Cre; Dullardflox/+; Rosa26mTmG and Wnt1Cre; Dullardflox/flox; Rosa26mTmG embryos. The red lines mark the endocardium, green arrowheads point at NCC, while the white ones indicate the endocardium. The white lines delineate the cardiac NCC cushions. RFP and GFP mark cell membrane whereas Twist1 is cytoplasmic or nuclear. (B) Sema3c expression assessed by ISH on transverse sections from distal to medial OFT levels of E11.5 Wnt1Cre; Dullardflox/+ and Wnt1Cre; Dullardflox/flox embryos. The endocardium is delineated with a red line, the cardiac NCC areas with a green line and the myocardium with grey lines. (C) Normalized expression levels of Sema3c assayed by q-RT-PCR on single cells isolated after immuno-marking endothelial CD31+ cells from E11.5 Wnt1Cre; Dullardflox/+ and Wnt1Cre; Dullardflox/flox; Rosa26mTmG hearts (dots: value for a single cell; boxplot: mean ± s.e.m.). (D) Model for the molecular and cellular cues controlling OFT septation. Upper panel: Morphogenesis of the single truncus arteriosus at E10.5 into fully formed great arteries at E14.0. Middle panel: Shape of P-Smad1/5/8 and Sema3c gradients along the OFT distal-proximal axis in a control situation. NCC in the cardiac cushions condense toward the endocardium between the aorta and the pulmonary trunk. Lower panel: Premature condensation of NCC at the pulmonary trunk in absence of Dullard in NCC, is associated with increased levels of Sema3c and BMP signalling.
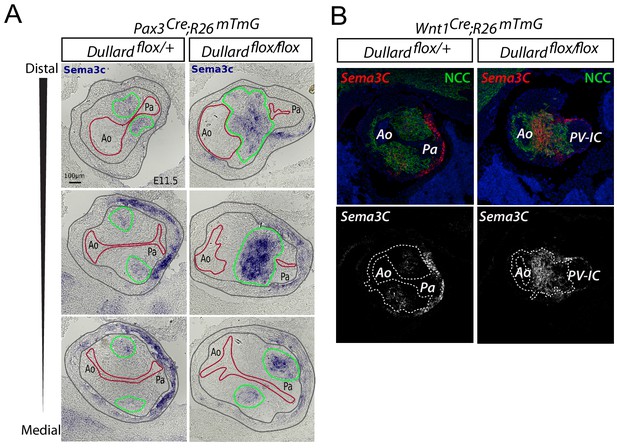
Dullard loss in cardiac NCC alters the expression of Sema3c specifically in NCC.
(A) Sema3c expression assessed by ISH on transverse sections from distal to medial OFT levels of E11.5 Pax3Cre; Dullardflox/+ and Pax3Cre; Dullardflox/flox embryos. The endocardium is delineated with a red line, the cardiac NCC areas with a green line and the myocardium with grey lines. (B) Sema3c mRNA distribution assessed by RNAscope, GFP immunolabelling and DAPI staining in transverse sections of E11.5 control Wnt1Cre; Dullardflox/+and mutant Wnt1Cre; Dullardflox/flox OFTs at distal levels.
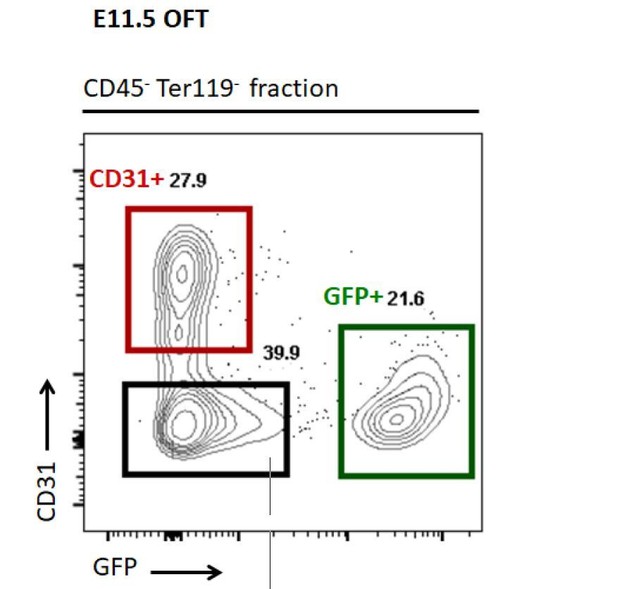
FACS plot showing that CD31+ cells are segregating from GFP+ NCC cells and the gating (coloured squares) used to collect cells for the q-RT-PCR screening.
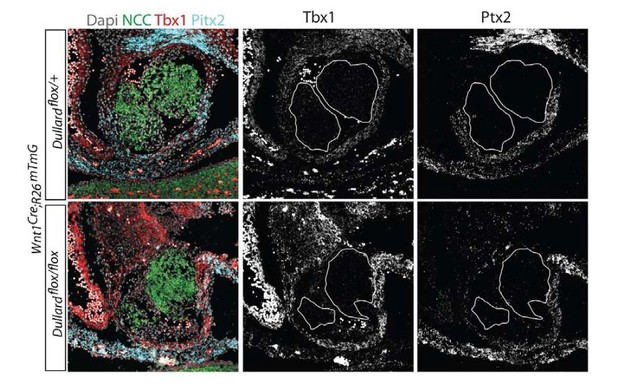
RNAscope for Tbx1 and Pitx2 showing there are no differences between Control and mutants PFT for their expression levels.
Videos
Three-dimensional rendering of cardiac NCC (green) over Pecam (white) after BABB clearing and Lightsheet acquisition of Pax3Cre; Dullardflox/+; Rosa26mTmG and Pax3Cre; Dullardflox/flox; Rosa26mTmG E12 embryos.
No defect in the OFT colonisation of mutant cardiac NCC is observed.
Three-dimensional rendering of cardiac NCC (green) over Pecam (white) after BABB clearing and Lightsheet acquisition of Pax3Cre; Dullardflox/+; Rosa26mTmG and Pax3Cre; Dullardflox/flox; Rosa26mTmG E12 embryos.
No defect in the OFT colonisation of mutant cardiac NCC is observed.
Three-dimensional rendering of cardiac NCC (green isosurface) over Pecam (red isosurface) and Dapi (white) of Wnt1Cre; Dullardflox/+; Rosa26mTmG and Wnt1Cre; Dullardflox/flox; Rosa26mTmG E11.5 embryos.
No defect in the OFT colonisation of mutant cardiac NCC is observed, and reduction of the pulmonary artery is visible in the mutant.
Three-dimensional rendering of cardiac NCC (green isosurface) over Pecam (red isosurface) and Dapi (white) of Wnt1Cre; Dullardflox/+; Rosa26mTmG and Wnt1Cre; Dullardflox/flox; Rosa26mTmG E11.5 embryos.
No defect in the OFT colonisation of mutant cardiac NCC is observed, and reduction of the pulmonary artery is visible in the mutant.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | C57BL/6JRj | Janvier Labs | ||
| Genetic reagent (Mus musculus) | Wnt1Cre | PMID: 9843687 | MGI:2386570 | Dr A Pierani (Imagine Institute) |
| Genetic reagent (Mus musculus) | Pax3Cre | PMID: 15882581 | MGI: 3573783 | Dr F Relaix (Créteil University) |
| Genetic reagent (Mus musculus) | Rosa26mTmG | PMID: 17868096 | MGI: 3716464 | Dr F Relaix (Créteil University) |
| Genetic reagent (Mus musculus) | DullardFlox | PMID: 23360989 | other | Dr R Nishinakamura (Kumamoto University) |
| Antibody | Anti-GFP (Chicken) | Aves Labs | GFP-1020 RRID:AB_10000240 | IF(1:500) |
| Antibody | anti-PECAM (Rat monoclonal) | Santa-Cruz Biotechnology | Sc-18916 RRID:AB_627028 | IF(1:200) |
| Antibody | Anti-phosphoSmad1/5/8 (Rabbit monoclonal) | Cell Signalling Technology | 13820S RRID:AB_2493181 | IF (1:500) |
| Antibody | Anti-Myosin Heavy Chain (mouse monoclonal) | DSHB | MF20 RRID:AB_2147781 | IF (1:300) |
| Antibody | Anti-phospho-histone H3 (rabbit polyclonal) | Cell Signalling Technology | 9701 RRID:AB_331535 | IF (1:500) |
| Antibody | Anti-cleaved Caspase 3 (Rabbit monoclonal) | Cell Signalling Technology | 9664 RRID:AB_2070042 | IF (1:500) |
| Antibody | Anti-Isl1 (Rabbit polyclonal) | Abcam | Ab20670 RRID:AB_881306 | IF (1:500) |
| Antibody | AF 488 donkey IgG anti-chicken IGG (H+L) | Interchim | 703-545-155 RRID:AB_2340375 | IF (1:500) |
| Antibody | Alexa Fluor 555 Goat Anti-Rabbit IgG (H+L), highly cross-adsorbed | Life Technologies | A-21429 RRID:AB_2535850 | IF (1:500) |
| Antibody | AF 647 Donkey IgG Anti Rabbit IGG (H+L) | Interchim | 711-605-152 RRID:AB_2492288 | IF (1:500) |
| Antibody | Alexa Fluor 647 Goat Anti-Rat IgG (H+L) | Life Technologies | A21247 RRID:AB_141778 | IF (1:500) |
| Antibody | Alexa Fluor 647 Goat Anti-Mouse IgG (H+L) Antibody, highly cross-adsorbed | Life Technologies | A-21236 RRID:AB_2535805 | IF (1:500) |
| Antibody | Alexa Fluor 488 Goat Anti-Mouse IgG (H+L) Antibody, highly cross-adsorbed | Life Technologies | A-11029 RRID:AB_2534088 | IF (1:500) |
| Antibody | Alexa Fluor 488 Goat Anti-Rabbit IgG (H+L) Antibody | Life Technologies | A-11008 RRID:AB_143165 | IF (1:500) |
| Antibody | Anti-Phospho Smad 2/3 (Rabbit monoclonal) | Ozyme | 8828 s RRID:AB_2631089 | IF (1:200) |
| Sequence-based reagent | Sema3C | PMID: 16397144 | other | RNA probe from Dr S Zaffran (Aix Marseille University) |
| Sequence-based reagent | Dullard | Advanced Cell Diagnostics | #456911 | RNA probe |
| Sequence-based reagent | Sema3C | Advanced Cell Diagnostics | #441441 | RNA probe |
| Sequence-based reagent | Twist1 | Advanced Cell Diagnostics | #414701 | RNA probe |
| Sequence-based reagent | Snai2 | Advanced Cell Diagnostics | #451191 | RNA probe |
| Sequence-based reagent | Msx2 | Advanced Cell Diagnostics | #421851 | RNA probe |
| Sequence-based reagent | Id2 | Advanced Cell Diagnostics | #445871 | RNA probe |
| Antibody | Anti-Ter119 Pe-Cy7 (Mouse monoclonal) | Sony | 1181110 | Facs (1:300) |
| Antibody | Anti-CD45 APC-Cy7 (Mouse monoclonal) | BD Pharmingen | 557659 RRID:AB_396774 | Facs (1:300) |
| Antibody | Anti-CD31 APC (Mouse monoclonal) | BD Pharmingen | Clone MEC 13.3 Catalog No. 561814 RRID:AB_10893351 | Facs (1:300) |
| Commercial assay or kit | 7AAD PE-Cy7 | BD Pharmingen | 559925 | Facs (1:800) |
| Commercial assay, kit | CellsDirect One-Step qRTPCR Kit | Invitrogen | 11753100 | |
| Commercial assay, kit | 48.48 Sample/Loading Kit— 10 IFCs | Fluidigm Corporation | BMK-M10- 48.48 | |
| Cell Line (Mus musculus) | C2C12 | ATCC, PMID: 28966089 | CRL-1772, RRID:CVCL_0188 | |
| Peptide, recombinant protein | Human BMP-2 | Thermo Fisher Scientific | #PHC7145 | 50 ng/ml |
| Software, algorithm | R, pHeatmap (v1.0.10) | R foundation | R package (v3.2.2), RRID:SCR_016418 | |
| Software, algorithm | R, phenograph (v0.99.1) | R foundation | R package (v3.2.2), RRID:SCR_016919 | |
| Software, algorithm | R, ggplot2 (v3.1.0) | R foundation | R package (v3.2.2), RRID:SCR_014601 | |
| Software, algorithm | R, Destiny (v2.6.1) | R foundation | R package (v3.6) | https://github.com/theislab/destiny |
| Recombinant DNA reagent | pEGFP-Dullard (plasmid) | This paper | GFP-Dullard expression plasmid | |
| Recombinant DNA reagent | pEGFP-Dullard D67E (plasmid) | This paper | GFP-Dullard D67E expression plasmid | |
| Recombinant DNA reagent | pEGFP-N1 | Clontech | 6085–1 | |
| Commercial assay or kit | Gateway LR Clonase | ThermoFisher | 11791100 | |
| Commercial assay or kit | Taq Polymerase, Superscript III | Thermofisher | 11732020 | |
| Sequence-based reagent | Dull_FL_C1_FW | This paper | PCR primers | GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGATGCGGACGCAGTGT |
| Sequence-based reagent | Dull_FL_C1_Rev | This paper | PCR primers | GGGGACCACTTTGTACAAGAAAGTGGGTCTCACCAGAGCCTATGTTGGTG |
| Sequence-based reagent | Dull_D67E_FW | This paper | PCR primers | GATCCTGGTGCTGGAACTGGACGAAACCCTG |
| Sequence-based reagent | Dull_D67E_Rev | This paper | PCR primers | CAGGGTTTCGTCCAGTTCCAGCACCAGGATC |
Additional files
-
Supplementary file 1
List of TaqMan gene expression assays (20x, Life Technologies) used for single-cell qPCRs experiments.
- https://cdn.elifesciences.org/articles/50325/elife-50325-supp1-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/50325/elife-50325-transrepform-v2.pdf