Viral miRNA adaptor differentially recruits miRNAs to target mRNAs through alternative base-pairing
Figures
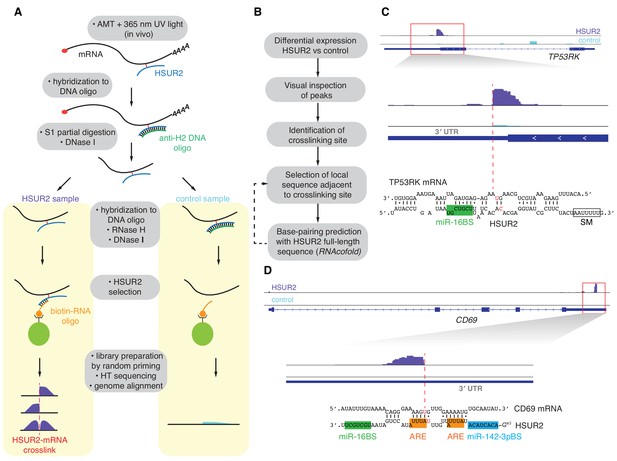
iRICC defines sequences that mediate HSUR2-mRNA interactions.
(A) Schematic overview of the iRICC protocol. Critical steps: in vivo ATM crosslinking, protection of HSUR2 and partial RNA digestion with S1 nuclease, depletion of HSUR2 in control sample by RNase H digestion, HSUR2 pulldown, and strand-specific library preparation for high-throughput sequencing. Since crosslinking is not reversed, reverse transcriptase stalls or starts around the site of crosslinking. A sharp drop of aligned reads results in peaks with abrupt edges indicating the site of crosslinking between HSUR2 and target RNAs. (B) Outline of analysis to determine sequences involved in interactions between HSUR2 and target RNAs. (C) Zoomed-in view to the TP53RK gene for HSUR2 (purple) and Control (Cyan) samples. Dashed red line denotes abrupt drop of aligned reads indicating the site of crosslinking. Predicted base-pairing between HSUR2 and TP53RK sequences adjacent to the site of crosslinking is shown. Putative psoralen-crosslinked nucleotides are shown in red. Binding sites for miR-16 (green box) and Sm proteins (empty box) are indicated. One representative experiment (out of three performed) is shown. (D) Same as in (C) for CD69 gene showing the ARE-like sequence (orange boxes) and binding site for miR-142–3p (blue box) in HSUR2.
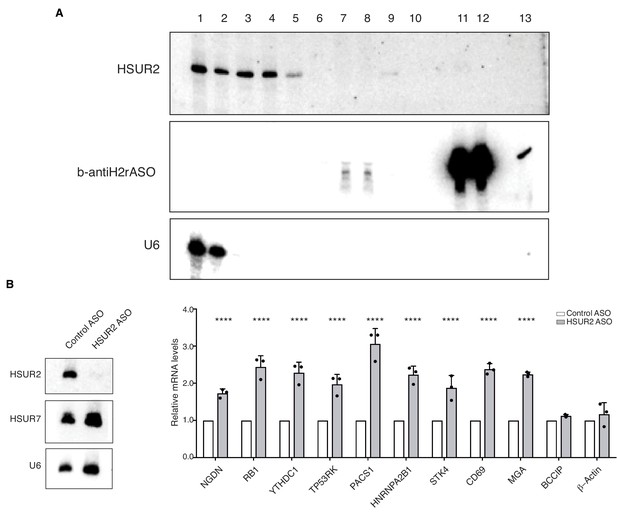
iRICC identifies HSUR2 target mRNAs.
(A) AMT-mediated crosslinking and purification of HSUR2 for iRICC. Northern blot analyses of purification of HSUR2 for iRICC. RNA was isolated from different fractions and beads (lanes 11 and 12), crosslinks were reversed, and analyzed by Northern blot for HSUR2, b-antiH2rASO, or U6 snRNA as a crosslinking and purification control. Lane 1: Total RNA (1%), lane 2: polyA flow-through (1%), lane3: polyA-bound (10%), lane 4: S1/DNAse1 treatment (10%), lane 5: RNAseH/DNAse treatment in HSUR2 sample (10%), lane 6: RNAseH/DNAse treatment in control sample (10%), lane 7: b-antiH2rASO flow-through in HSUR2 sample (25%), lane 8: b-antiH2rASO flow-through in control sample (25%), lane 9: b-antiH2rASO-bound in HSUR2 sample (100%), lane 10: b-antiH2rASO-bound in control sample (100%), lane 11: b-antiH2rASO beads from HSUR2 sample (100%), lane 12: b-antiH2rASO beads from control sample (100%), lane 13: b-antiH2rASO (five pmoles). (B) iRICC identifies functional targets of HSUR2. Northern blot analyses of HSUR2, HSUR7, and U6 snRNA on total RNA prepared from cj319-WT marmoset T cells transiently transfected with either Control ASO or HSUR2 ASO. Chart shows HSUR2 target mRNA levels in cj319-WT cells after transfection with Control or HSUR2 ASO. Dots represent mean values of independent experiments (n = 3) with error bars representing s.d. Two-sided Student's t-tests corrected for multiple comparisons using the Holm-Sidak method (alpha = 0.05). ****p<0.0001.
-
Figure 1—figure supplement 1—source data 1
Source data for Figure 1—figure supplement 1.
- https://doi.org/10.7554/eLife.50530.004
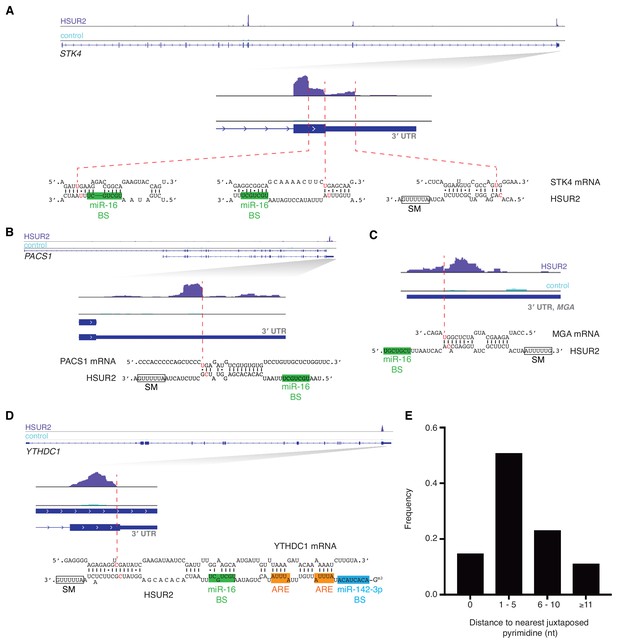
iRICC defines sequences involved in HSUR2-mRNA interactions.
(A–D) Same as in Figure 1C for STK4 (A), PACS1 (B), MGA (C), and YTHDC1 (D) genes. Binding sites for miR-142–3p (blue box), miR-16 (green box), Sm proteins (empty box) and ARE-like sequences (orange boxes) are indicated. One representative experiment (out of three performed) is shown. (E) Distance from peak drop or peak edge to nearest putative psoralen-crosslinked pyrimidines (n = 170).
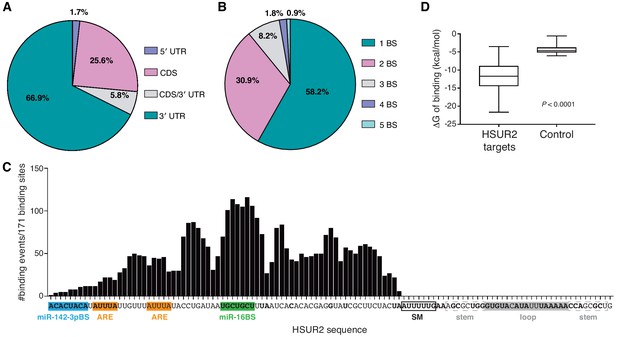
HSUR2 binding properties.
(A–B) Location (A) and number (B) of HSUR2 binding sites in mRNAs (n = 171). (C) HSUR2 does not display a seed sequence that is involved in all interactions with target mRNAs. The graph shows the number of times that each HSUR2 nucleotide is involved in base-pairing with a target mRNA (171 HSUR2-mRNA interactions analyzed, see Supplementary file 2). Bold nucleotides are conserved among the different strains of HVS and also in H. ateles (Cazalla et al., 2010). ARE-like sequences, miRNA binding sites, and SM binding sequence are shown. (D) Average free energy of binding (ΔG) of interactions between HSUR2 and HSUR2-binding sites (HSUR2 targets, n = 171) or with 250 length-matched, randomly selected 3′UTR sequences (Control).
-
Figure 2—source data 1
Source data for Figure 2.
All identified interactions involve sequences located between the 5’ end and the binding site for Sm proteins in HSUR2 (Figure 2C). Surprisingly, conserved sequences (Cazalla et al., 2010) in the 3’ terminal loop of HSUR2 are not found to participate in base-pairing with target mRNAs. Even though HSUR2 binds to target sequences with a significantly lower Gibbs free energy difference (ΔG) when compared to random sequences of similar length (Figure 2D), HSUR2 does not utilize a dedicated single sequence to do so (Figure 2C and Supplementary file 2). Thorough sequence analysis of 171 HSUR2 binding sites (see Materials and methods), with lengths ranging from 11 to 66 nts (Figure 2—figure supplement 1B and Supplementary file 2), failed to identify a sequence motif shared by most targets. HSUR2 sequences involved in interactions with target mRNAs differ in length (Figure 2—figure supplement 1B) and identity for each target (Figure 1C,D and Figure 1—figure supplement 2C; see also Supplementary file 2). Even in cases in which HSUR2 utilizes a similar subregion to base-pair with two different targets, the arrangement of base pairs always differs between the two interactions (Figure 1—figure supplement 2A–C). Altogether, these findings suggest that unlike other classes of small ncRNAs that participate in RNA-based regulatory systems (Gorski et al., 2017), HSUR2 does not feature a subregion or seed sequence that binds to target mRNAs, but instead uses different sequences to interact with different RNA partners, a characteristic shared with other Sm-class ncRNAs (Cotten et al., 1988; Nilsen, 1994; Roca and Krainer, 2009; Staley and Guthrie, 1998).
- https://doi.org/10.7554/eLife.50530.009
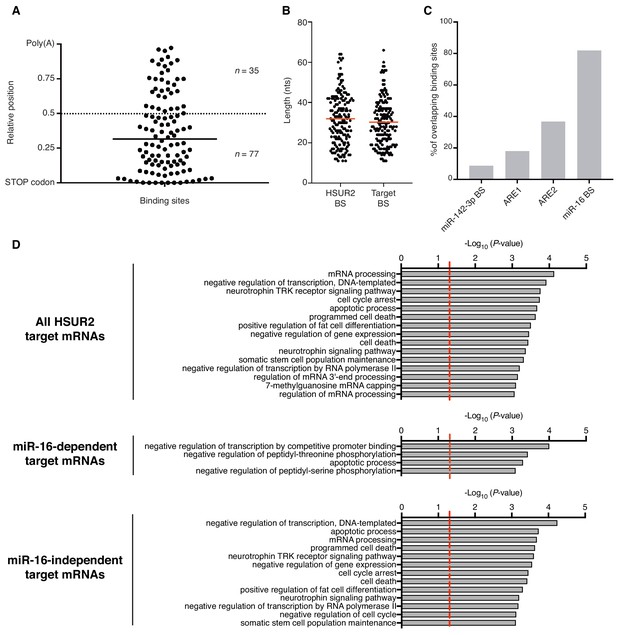
Analysis of HSUR2 binding sites.
(A) HSUR2 binding sites in the 3′UTR of target mRNAs (n = 112). Bar indicates median relative distance to the STOP codon. Wilcoxon Signed Rank test (alpha = 0.05) relative to theoretical median set at 0.5 (dashed line, p<0.0001). (B) Length distribution of HSUR2 and mRNA sequences involved in interactions (n = 171). Red bars represent median length of HSUR2 and target binding sites. (C) Overlap between regulatory elements and mRNA-binding sequences in HSUR2 (n = 171). (D) Gene ontology (GO)-term enrichment analyses for all, miR-16-dependent, and miR-16-independent HSUR2 targets. Dashed red lie demarcates significance threshold (p<0.05).
-
Figure 2—figure supplement 1—source data 1
Source data for Figure 2—figure supplement 1.
- https://doi.org/10.7554/eLife.50530.008
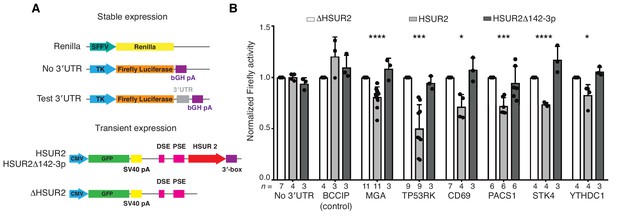
iRICC-seq identifies functional HSUR2 binding sites.
(A) Schematic representation of plasmid constructs used for stable expression of luciferase reporters and transient expression of HSUR2. U937 cells were sequentially transduced with lentiviruses carrying the Renilla luciferase gene under the spleen focus-forming virus (SFFV) promoter and Firefly luciferase gene driven by the thymidine kinase (TK) promoter with no 3′UTR, full-length 3′UTR of BCCIP (control) or partial 3′UTR of HSUR2-target mRNAs containing single binding sites identified by iRICC-seq. Plasmids carrying GFP and either HSUR2 promoter alone (ΔHSUR2), wild-type HSUR2 (HSUR2), or a mutant version of HSUR2 that cannot bind miR-142–3p and does not promote mRNA destabilization (HSUR2Δ142–3p) were used for transient transfection. bGH: bovine growth hormone; PSE: proximal sequence element; DSE: distal sequence element. (B) Luciferase assays confirm binding sites for HSUR2. Luciferase-expressing U937 cells were transiently transfected with the indicated plasmids. Dots represent mean values of independent experiments with error bars representing s.d. Two-sided one sample Student’s t-test vs. control (ΔHSUR2) set at 1.0. Sample size (n) is indicated below the chart in each case. *p<0.05, ***p<0.001, ****p<0.0001.
-
Figure 3—source data 1
Source data for Figure 3.
- https://doi.org/10.7554/eLife.50530.011
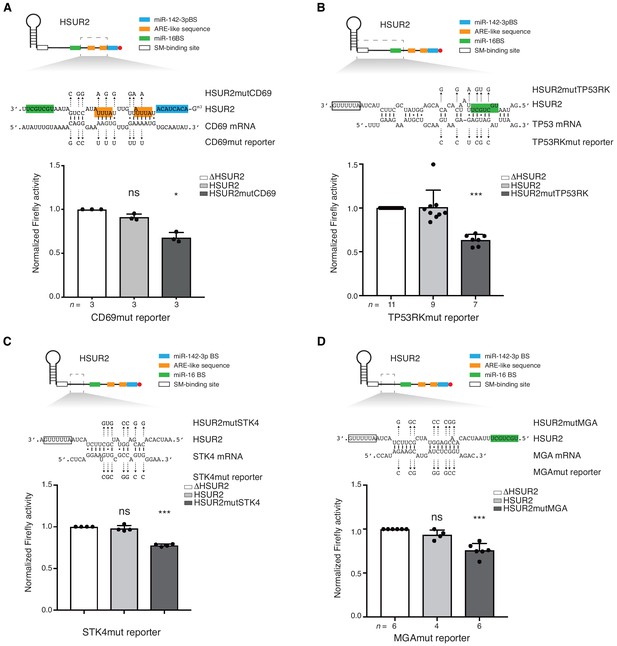
Base-pairing is required for HSUR2-mediated mRNA regulation.
(A) Same as in Figure 3B but with cells stably expressing Firefly luciferase followed by partial CD69 3′UTR carrying point mutations (shown) predicted to disrupt interaction with HSUR2. Cells were transiently transfected with control plasmid (ΔHSUR2), plasmid expressing wild-type HSUR2, or a mutant version of HSUR2 with point mutations that restore binding to the mutant CD69 3′UTR. (B–D) Same as in (A) with the TP53RK (B), STK4 (C), and MGA (D) luciferase reporters. Dots represent mean values of independent experiments with error bars representing s.d. Two-sided one-sample Student’s t-test vs. control (ΔHSUR2) set at 1.0. Sample size (n) is indicated below the chart in each case. *p<0.05, ***p<0.001.
-
Figure 4—source data 1
Source data for Figure 4.
- https://doi.org/10.7554/eLife.50530.015
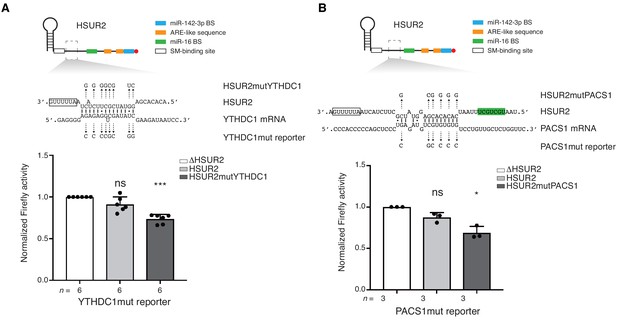
iRICC identifies functional HSUR2 binding sites.
(A–B) Same as in Figure 4 for HSUR2 binding sites in YTHDC1 (A) and PACS1 (B) partial 3′UTRs. Dots represent mean values of independent experiment with error bars representing s.d. Sample size (n) is indicated below the chart in each case. Two-sided one-sample Student's t-test vs. control set at 1.0. *p<0.05, ***p<0.001.
-
Figure 4—figure supplement 1—source data 1
Source data for Figure 4—figure supplement 1.
- https://doi.org/10.7554/eLife.50530.014
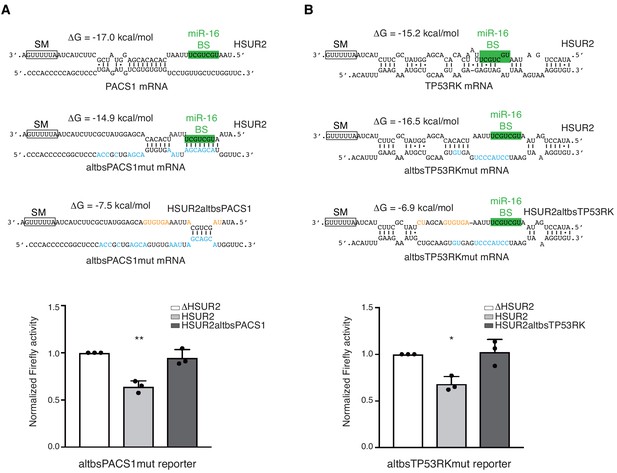
HSUR2 binds target mRNAs through a flexible arrangement of base-pairs.
(A) Same as in Figure 3B, but with a mutant version of partial PACS 3′UTR with mutations (blue residues) that result in alternative base-pairing with wild-type HSUR2. Predicted base-pairing between wild-type HSUR2 and wild-type PACS1 mRNA, wild-type HSUR2 and mutant PACS1 mRNA (altbsPACS1mut), and mutant PACS1 mRNA and mutant HSUR2 (HSUR2altbsPACS1) with mutations (orange residues) that disrupt interaction with altbsPACS1mut shown in orange. (B) Same as in (A), but with partial TP53RK 3′UTR. Dots represent mean values of independent experiments (n = 3) with error bars representing s.d. Two-sided one-sample Student's t-test vs. control (ΔHSUR2) set at 1.0. *p<0.05, **p<0.01.
-
Figure 5—source data 1
Source data for Figure 5.
- https://doi.org/10.7554/eLife.50530.017
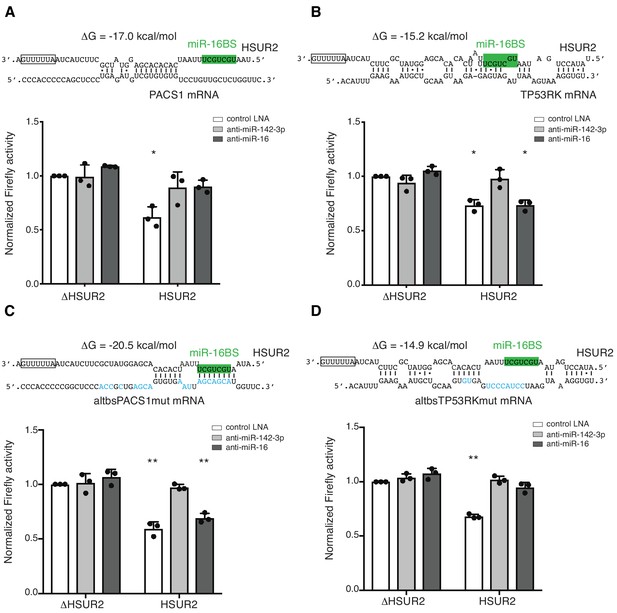
Base-pairing with target mRNA dictates the use of miR-16 in HSUR2-mediated mRNA repression.
(A) U937 cells stably expressing the Firefly luciferase gene with partial PACS1 3′UTR (Figure 2a) were transiently co-transfected with a plasmid carrying GFP and either HSUR2 promoter alone (ΔHSUR2) or wild-type HSUR2 (HSUR2) and a control LNA inhibitor, or an LNA inhibitor with complementarity to miR-142–3p or miR-16. (B–D) Same as in (A) but with cells stably expressing the Firefly luciferase gene with partial TP53RK 3′UTR (B), mutant PACS1 (altbsPACS1mut) 3′UTR (C), or mutant TP53RK (altbsTP53RKmut) 3′UTR (D). Modified residues are shown in blue. Dots represent mean values of independent experiments (n = 3) with error bars representing s.d. Two-sided one-sample Student’s t-test vs. control (ΔHSUR2) set at 1.0. *p<0.05, **p<0.01.
-
Figure 6—source data 1
Source data for Figure 6.
- https://doi.org/10.7554/eLife.50530.022
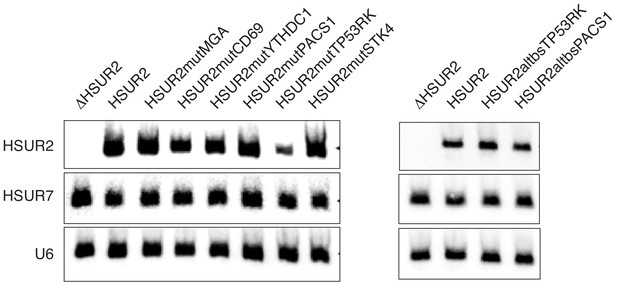
Expression of mutant versions of HSUR2 used in this study.
Northern blot analysis of total RNA isolated from HEK293T/17 cells transiently co-transfected with a plasmid expressing HSUR7 (transfection control) and a plasmid carrying GFP and either HSUR2 promoter alone (ΔHSUR2), wild-type HSUR2 (HSUR2), or the indicated mutant version of HSUR2. U6 snRNA provides a loading control.
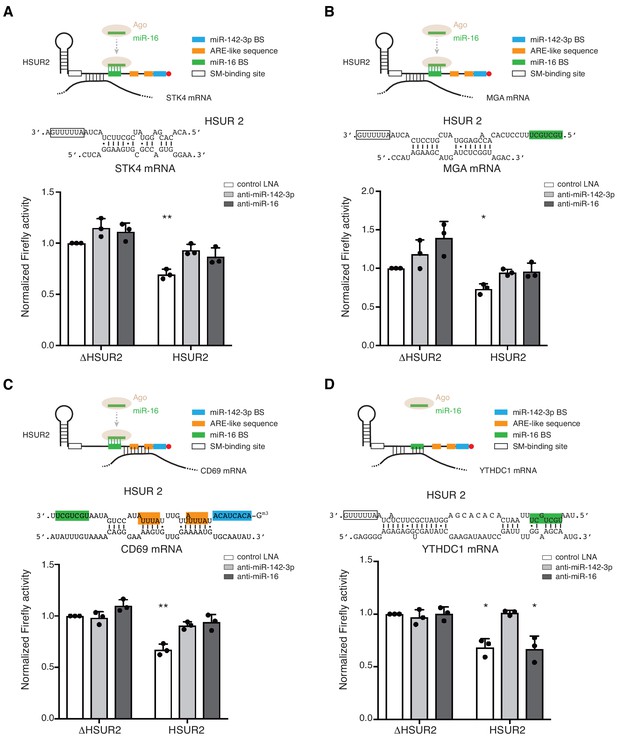
Base-pairing with target mRNA determines the use of miR-16 in HSUR2-mediated mRNA repression.
(A–D) Same as in Figure 6 for HSUR2 binding sites in STK4 (A), MGA (B), CD69 (C), and YTHDC1 (D) partial 3′UTRs. Dots represent mean values of independent experiments (n = 3) with error bars representing s.d. Two-sided one-sample Student's t-test vs. control (ΔHSUR2 + control LNA) set at 1.0. *p<0.05, **p<0.01.
-
Figure 6—figure supplement 2—source data 1
Source data for Figure 6—figure supplement 2.
- https://doi.org/10.7554/eLife.50530.020
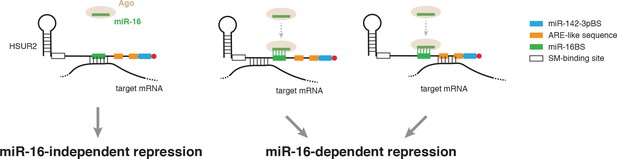
Model for alternative use of miR-16 in HSUR2-mediated mRNA repression.
HSUR2 base-pairs with target mRNAs through different regions and/or arrangements of base pairs in each case. Engagement of the miR-16 binding site in HSUR2 in the interaction with the target mRNA prevents binding of miR-16 to HSUR2, resulting in miR-16-independent target mRNA repression. When the miR-16 binding site in HSUR2 is not engaged in the interaction with the target mRNA, miR-16 binds to HSUR2 which now represses the mRNA in a miR-16-dependent manner.
Additional files
-
Supplementary file 1
This file contains a list of HSUR2 targets identified by iRICC.
Gene name, genomic coordinates, and ratios of HSUR2 sample pulldown versus Control sample pulldown.
- https://doi.org/10.7554/eLife.50530.024
-
Supplementary file 2
This file contains the sequences in HSUR2 and in target mRNAs involved in base-pairing, binding energies, patterns of interactions identified by RNAcofold in dot-bracket notation, lengths and location of the binding sites in mRNAs, and overlap with regulatory elements in HSUR2.
- https://doi.org/10.7554/eLife.50530.025
-
Supplementary file 3
This table contains the sequences of all oligonucleotides used in this study.
- https://doi.org/10.7554/eLife.50530.026
-
Supplementary file 4
This table contains all results from our Gene Ontology (GO)-term enrichment analysis.
- https://doi.org/10.7554/eLife.50530.027
-
Supplementary file 5
Key Resources Table.
- https://doi.org/10.7554/eLife.50530.028
-
Transparent reporting form
- https://doi.org/10.7554/eLife.50530.029





